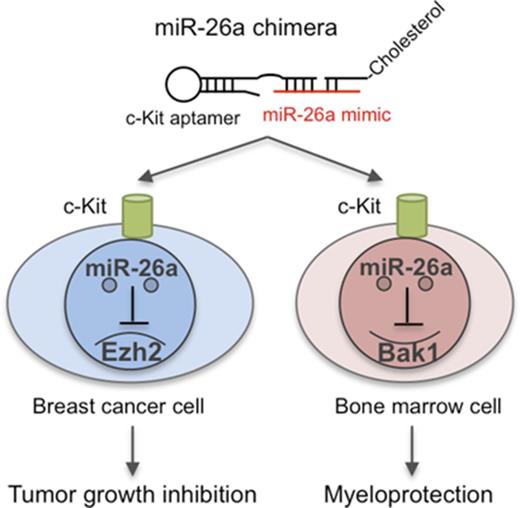
Key Points
miR-26a is a key regulator for apoptosis of cancer cells and hematopoietic toxicity of chemotherapy.
A novel chimera with an anti-KIT aptamer and miR-26a can selectively deliver microRNA to cancer cells and HSPCs.
Abstract
The efficacy of traditional chemotherapy is limited by its toxicity, especially with regard to hematopoiesis. Here we show that miR-26a plays a critical role in protecting mice against chemotherapy-induced myeloid suppression by targeting a proapoptotic protein (Bak1) in hematopoietic stem/progenitor cells (HSPCs). Because c-Kit is expressed at high levels in HSPCs, we designed a microRNA-aptamer chimera that contains miR-26a mimic and c-Kit–targeting aptamer and successfully delivered miR-26a into HSPCs to attenuate toxicity of 5′ fluorouracil (5-FU) and carboplatin. Meanwhile, our in silico analysis revealed widespread and prognosis-associated downregulation of miR-26a in advanced breast cancer and also showed that KIT is overexpressed among basal-like breast cancer cells and that such expression is associated with poor prognosis. Importantly, the miR-26a aptamer effectively repressed tumor growth in vivo and synergized with 5-FU or carboplatin in cancer therapy in the mouse breast cancer models. Thus, targeted delivery of miR-26a suppresses tumor growth while protecting the host against myelosuppression by chemotherapy.
Introduction
Because of the highly regenerative nature of the hematopoietic system,1 commonly used cytotoxic chemotherapy often causes myelosuppression, as manifested by neutropenia and severe anemia.2 Myelosuppression is a cause of dose-limiting toxicity; therefore, it limits the intensity of chemotherapy for multiple cancer types and thus likely contributes to the reduced therapeutic effect.3
On the basis of analysis of hematopoietic stem-cell (HSC) niches4 and the kinetics of HSC proliferation,5,6 it has been suggested that distinct HSCs are responsible for homeostatic and injury-induced hematopoiesis.4,5,7,8 Actively cycling HSCs mediate homeostatic hematopoiesis, whereas dormant HSCs are responsible for injury-induced hematopoiesis.4,5,7,8 Although HSCs responsible for injury-induced but not homeostatic hematopoiesis are vulnerable to destruction by macrophages unless protected by CD47,9 it is unclear whether the HSCs that mediate homeostatic versus injury-induced homeostatic proliferation are maintained by distinct molecular programs.
Because chemotherapeutic drugs10-12 cause massive cell loss and trigger injury-induced hematopoiesis, understanding regulators that selectively regulate injury-induced hematopoiesis may lead to new approaches in minimizing myelosuppression associated with chemotherapy. In this context, it is of great interest to consider the role of microRNA (miRNA). With exception of natural killer T cells, most lineages of hematopoietic cells developed normally when the Dicer1 gene, a critical regulator for most miRNA, was deleted in both vascular endothelial stem cells and HSCs.13 However, under conditions that could have elicited injury-induced hematopoiesis, including treatment that induced inflammation and bone marrow transplantation, the Dicer1 gene as well as specific miRNA, such as miR125, played a major role in hematopoiesis.14 It is therefore of interest to explore whether injury-induced hematopoiesis under the condition of cancer chemotherapy can be protected by manipulation of specific miRNA.
In this study, we show that miR-26a mediates a converging pathway in chemotherapy-related myelosuppression and tumor suppression and describe an miRNA aptamer as a platform to deliver miRNA to enhance the therapeutic effect of chemotherapy while abrogating myelosuppression.
Methods
Bioinformatic analyses
Level 3 data of miRNA sequencing and RNA sequencing (version 2) measured by Illumina HiSeq and clinical annotation tables of breast tumors from The Cancer Genome Atlas (TCGA) were downloaded from the University of California Santa Cruz Cancer Genomics Browser. miRNA expression value was measured as counts normalized to reads per million mapped reads, and Wilcoxon rank sum tests were used for comparisons between basal-like breast tumors and normal breast tissue, with a false-discovery rate < 0.05 and log2 ratio > 0.5 (<0.5) considered significantly upregulated (or downregulated). The target prediction of miR-26a was performed using TargetScan 3.0 software (http://www.targetscan.org/) and miRBase (http://microrna.sanger.ac.uk/).
Aptamer and miRNA chimera preparation
DNA sequences for anti-human KIT aptamer15 and mouse c-Kit aptamers16 conjugated with biotin at 3′ were functionalized by short denaturation-renaturation step (95°C for 10 minutes; 5 minutes of snap cooling on ice). These DNA sequences were 5′-GAGGCATACCAGCTTATTCAAGGGGCCGGGGCAAGGGGGGGGTACCGTGGTAGGACATAGTAAGTGCAATCTGCGAA-3′ for human KIT and 5′-GCTCAACGCGGGACGGCTCTCCCATTGAC-3′ for mouse c-Kit. For chimera preparation, a human KIT–aptamer or mouse c-Kit–aptamer miR-26a chimera was assembled by 3 compartments of DNA/RNA hybrid sequences. The sequences were 5′-GAGGCATACCAGCTTATTCAAGGGGCCGGGGCAAGGGGGGGGTACCGTGGTAGGACATAGTAAGTGCAATCTGCGAA/C3 spacer/CCUAUUCUGG-3′ for the human KIT aptamer plus part of the passenger sequence for miR-26a-5p, 5′-GCTCAACGCGGGACGGCTCTCCCATTGAC/C3 spacer/CCUAUUCUGG-3′ for the mouse c-Kit aptamer plus part of the passenger sequence for miR-26a-5p, 5′-GUUACUUGCACG/TEG (triethylene glycol)-Cholesterol-3′ for part of the sequence for miR-26a-5p plus cholesterol, and 5′-UUCAAGUAAUCCAGGAUAGGCU-3′ for the guide sequence for miR-26a-5p (italics indicate RNA sequences). The guide sequence for scramble control was GGUUCGUACGUACACUGUUCA. The RNA sequences were modified with 2’-fluoro-uridines. The conjugation with cholesterol at 3′ oligonucleotide improved in vivo pharmacokinetic properties, enhanced the permeation of cellular membranes, and protected the RNA from in vivo degradation.17-19 All oligonucleotides were synthesized and purified by Integrated DNA Technologies (Coralville, IA).
Chimera treatment in vivo
For tumor models, the NSG mice and BALB/c mice were subcutaneously injected with 2 × 106 viable MDA-MB-231 cells and TUBO cells in their right hind limbs, respectively. After the tumors grew to 1 cm in diameter, mice were randomly divided into the following groups: the control group, with saline 100 μL containing 2% serum, or the treatment group, with 670 pmol per 20g of chimeras and/or 5-fluorouracil (5-FU) 50 mg/kg or carboplatin 40 mg/kg IV injected into the tail vein. Tumor sizes were measured in 2 dimensions every 3 days. Tumor volume was calculated as: V = (1/2)S2 × L (where S was the shortest dimension and L the longest dimension). For chimera treatment in normal mice, C57BL/6 mice were treated with miR-26a chimera (670 pmol per 20 g) daily for 3 days by IV injection. At day 2, 5-FU 150 mg/kg and/or carboplatin 120 mg/kg was injected with the chimera. Peripheral blood was collected at days 5 and 10 after 5-FU treatment. Complete blood counts were measured by Hemavet 950FS (Drew Scientific, Miami Lakes, FL). The animal protocol was approved by the Institutional Animal Care and Use Committee at Children’s National Medical Center.
Statistics
For statistical test selection, distribution fitting and variance testing were determined to justify test selection. Data were analyzed using a Student t test to compare between 2 groups, and 2-way repeated-measures analysis of variance, followed by the Bonferroni post hoc procedure, was used for follow-up pairwise comparison. Survival data were analyzed by a Kaplan-Meier survival analysis with log-rank test. Statistical calculations were performed using GraphPad Prism software (GraphPad Software, San Diego, CA) and R software (https://www.r-project.org/).
Data availability
All data generated or analyzed during this study were included in this article and its supplemental information.
Results
Identification and validation of KIT-targeting miR-26a chimera that inhibits human breast cancer growth in vitro and in vivo
To identify an miRNA for treatment of advanced breast cancer, we analyzed the breast cancer miRNA data set with information from 826 patients in TCGA program. Our in silico analysis revealed that miR-26a-2 (mature miR-26a-1 and miR-26a-2 have the same sequence despite being encoded by distinct genes, and the mature sequence is referred to as miR-26a) was significantly downregulated in basal-like breast cancer in comparison with paired normal tissue; it also showed that the downregulation was significantly associated with shorter overall survival of patients with basal-like breast cancer (Figure 1A; supplemental Figure 1A).
Identification and validation of KIT-targeting miR-26a chimera that inhibits human breast cancer growth in vitro and in vivo. (A) Overall survival of patients with basal-like breast cancer with higher or lower expression of miR-26a-2 based on expression scores compared with the mean value in TCGA cohort. (B) The binding of anti-KIT antibody and KIT DNA aptamer to MDA-MB-231 cell line. Other cell-targeting aptamer is specific for Ramos cells. (C) The secondary structure of KIT aptamer–miR-26a chimera. KIT DNA aptamer was linked with C3 linker to complementary RNA passenger sequence that was bound to miR-26a mimic sequence. Another RNA passenger sequence binding to the 5′ of miR-26 mimic was conjugated with triethylene glycol (TEG)–cholesterol. (D) Specific miR-26a delivery by the miR-26a chimera. Two days after incubation with the miR-26a chimera, significant increase of miR-26a expression in the MDA-MB-231 cells compared with control (ctrl) chimera treatment was detected by quantitative polymerase chain reaction. Data (mean ± standard deviation [SD]) were pooled from 3 experiments. (E) miR-26a chimera suppressed the growth and induced apoptosis of MDA-MB-231 cells in a dose-dependent manner. After 3 days of culture with miR-26a chimera or ctrl chimera, MDA-MB-231 cell numbers (left) were counted using hemocytometer, and percentage of annexin V+ cells (right) was determined by flow cytometry. Data (mean ± SD) were pooled from 2 experiments. (F) Significant suppression of EZH2 protein in miR-26a chimera–treated MDA-MB-231 cells detected by immunoblot. (G) Relative expression of miR-26a in tumor harvested from NSG mice bearing human breast tumor with MDA-MB-231 cells. Significant increase of miR-26a was observed in KIT+ tumor cells 3 days after IV injection with 670 pmol per 20 g of miR-26a chimera. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (H) Therapeutic effect of miR-26a chimera. The tumor-bearing mice were treated daily with miR-26a chimera (670 pmol per 20 g) for 5 or 10 days (first injection defined as day 0). Data (mean ± SD) were pooled from 2 experiments involving a total of 5 mice per group. Tumor volume over time (left). Significant difference between miR-26 chimera (×5) versus miR26 chimera (×10; 2-way repeated-measures analysis of variance followed by Bonferroni post-test for day 0 to day 18 detected the significant difference (interaction P = .0012). Kaplan-Myer survival curve (right). Log-rank test detected significant differences between ctrl chimera group and miR-26a chimera groups (ctrl chimera vs miR-26 chimera [×5], P = .002; ctrl chimera vs miR-26 chimera [×10], P = .002). The difference in survival between miR-26 chimera (×5) and miR26 chimera (×10) did not reach statistical significance (P = .077). *P < .05, **P < .01. Error bars indicate SD.
Identification and validation of KIT-targeting miR-26a chimera that inhibits human breast cancer growth in vitro and in vivo. (A) Overall survival of patients with basal-like breast cancer with higher or lower expression of miR-26a-2 based on expression scores compared with the mean value in TCGA cohort. (B) The binding of anti-KIT antibody and KIT DNA aptamer to MDA-MB-231 cell line. Other cell-targeting aptamer is specific for Ramos cells. (C) The secondary structure of KIT aptamer–miR-26a chimera. KIT DNA aptamer was linked with C3 linker to complementary RNA passenger sequence that was bound to miR-26a mimic sequence. Another RNA passenger sequence binding to the 5′ of miR-26 mimic was conjugated with triethylene glycol (TEG)–cholesterol. (D) Specific miR-26a delivery by the miR-26a chimera. Two days after incubation with the miR-26a chimera, significant increase of miR-26a expression in the MDA-MB-231 cells compared with control (ctrl) chimera treatment was detected by quantitative polymerase chain reaction. Data (mean ± standard deviation [SD]) were pooled from 3 experiments. (E) miR-26a chimera suppressed the growth and induced apoptosis of MDA-MB-231 cells in a dose-dependent manner. After 3 days of culture with miR-26a chimera or ctrl chimera, MDA-MB-231 cell numbers (left) were counted using hemocytometer, and percentage of annexin V+ cells (right) was determined by flow cytometry. Data (mean ± SD) were pooled from 2 experiments. (F) Significant suppression of EZH2 protein in miR-26a chimera–treated MDA-MB-231 cells detected by immunoblot. (G) Relative expression of miR-26a in tumor harvested from NSG mice bearing human breast tumor with MDA-MB-231 cells. Significant increase of miR-26a was observed in KIT+ tumor cells 3 days after IV injection with 670 pmol per 20 g of miR-26a chimera. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (H) Therapeutic effect of miR-26a chimera. The tumor-bearing mice were treated daily with miR-26a chimera (670 pmol per 20 g) for 5 or 10 days (first injection defined as day 0). Data (mean ± SD) were pooled from 2 experiments involving a total of 5 mice per group. Tumor volume over time (left). Significant difference between miR-26 chimera (×5) versus miR26 chimera (×10; 2-way repeated-measures analysis of variance followed by Bonferroni post-test for day 0 to day 18 detected the significant difference (interaction P = .0012). Kaplan-Myer survival curve (right). Log-rank test detected significant differences between ctrl chimera group and miR-26a chimera groups (ctrl chimera vs miR-26 chimera [×5], P = .002; ctrl chimera vs miR-26 chimera [×10], P = .002). The difference in survival between miR-26 chimera (×5) and miR26 chimera (×10) did not reach statistical significance (P = .077). *P < .05, **P < .01. Error bars indicate SD.
Because the ectopic expression of miR-26a inhibited the proliferation and metastasis of basal-like breast cancer cells,20,21 we sought to develop a method to deliver miR-26a into cancer cells. To deliver the miR-26a mimic selectively to the basal-like breast cancer, we sought for the cell-surface proteins enriched on the basal-like breast cancer cells. Bioinformatic analysis using TCGA database identified the KIT gene as the most highly expressed cell-surface protein on the basal-like breast cancer cells compared with other subtypes of breast cancer (supplemental Figure 1B). The higher expression of the KIT gene was significantly associated with poor clinical outcomes in patients with basal-like breast cancer, although not in those with non–basal-cell breast cancer (supplemental Figure 2A-C). A majority of breast cancer samples, including both basal and nonbasal types, expressed high KIT and low miR-26a (supplemental Figure 2D). Therefore, we decided to target KIT proteins for the delivery of the miR-26a mimic breast cancer cells to correct the abnormally suppressed miR-26a expression. Because basal types expressed higher levels of the KIT gene (supplemental Figure 2D), they were the primary candidates for the approach.
To evaluate the binding ability of the human KIT aptamer to basal-like breast cancer cells, we performed flow cytometry analysis and confirmed the binding to human basal-like breast cancer cell line (MDA-MB-231). As shown in Figure 1B, the anti-KIT antibody and aptamer revealed nearly identical distribution of KIT expression among cancer cells, with higher levels on a small subset of cancer cells. Using this aptamer, we designed a KIT-targeting miR-26a chimera comprising (1) miR-26a-5p RNA mimic sequence, (2) anti-KIT DNA aptamer linked with a part of the miR-26a complementary sequence, and (3) triethylene glycol–cholesterol linked with the rest of the complementary sequence (Figure 1C). The KIT receptor was chosen because this receptor-targeting aptamer enhanced the uptake of chimera via receptor-mediated internalization22 ; the conjugation of triethylene glycol–cholesterol improved in vivo pharmacokinetic properties, enhanced the permeation of cellular membranes, and protected the RNA from in vivo degradation17-19 while an internal nicking in the complementary sequence of miR-26a prevented nonspecific miRNA targeting by RNA-induced silencing complex complex.23 Using this system, we confirmed that this chimera successfully delivered miR-26a into the MDA-MB-231 cells (Figure 1D). Importantly, the chimera consisting of miR-26a, but not scrambled sequence, inhibited cell growth by the induction of apoptosis in vitro (Figure 1E). Correspondingly, the chimera treatment in MDA-MD-231 cells significantly suppressed EZH2 protein expression (Figure 1F), which is a major oncogene for basal-like breast cancer.21
To evaluate the therapeutic potential of the miR-26a chimera, we injected the chimera IV into immune-compromised NSG mice bearing large MDA-MB-231 xenograft tumors daily either 5 or 10 times. To confirm specific targeting, we isolated single-cell suspension from the tumors at 3 days after single injection, sorted into KIT+ and KIT− populations and measured miR-26a levels by quantitative polymerase chain reaction. As shown in Figure 1G, significant elevation of miR-26a was detected in KIT+ tumor cells isolated from mice that received the miR-26a chimera. No increase of miR-26a was observed in control chimera–treated KIT+ tumor cells or in KIT− tumor cells from mice that received either miR-26a or control chimera. Therefore, the aptamer-based delivery allowed specific targeting of KIT+ tumor cells. Remarkably, 5 or 10 injections with the miR-26a chimera, but not the control chimera, significantly suppressed tumor growth and extended survival (Figure 1H). These results suggest the therapeutic potential of our miR-26a chimera for basal-like breast cancer treatment.
miR-26a protects hematopoiesis from chemotherapeutic agent–induced myelosuppression
Because c-Kit is expressed at high levels in HSCs/progenitor cells (HSPCs),24 we tested the effect of miR-26a on myelosuppression by 5-FU. As expected, 5-FU induced significant defects in hematopoiesis, as revealed by reduction in leukocytes (white blood cells [WBCs]; (Figure 2A; supplemental Figure 3A) and total BM cellularity (Figure 2B). Remarkably, the miR-26a chimera significantly ameliorated myelosuppression, as revealed by increased leukocyte counts and BM cellularity. Consistent with this notion, we found that it was transiently and significantly increased in HSPCs after 5-FU treatment in vivo (supplemental Figure 3B), which suggests that this acute expression of miR-26a might be involved in the recovery of BM cells from 5-FU–induced myelosuppression.
miR-26a protects hematopoiesis from chemotherapeutic agent–induced myelosuppression. The mice received IV injection of 670 pmol per 20g of miR-26a or control (ctrl) chimera daily for 3 days. On day 2 of chimera treatment, 5-FU 150 mg/kg was injected. (A) The numbers of WBCs, red blood cells (RBCs), and platelets (PLTs) 10 days after 5-FU treatment. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (B) The number of bone marrow (BM) cells 10 days after 5-FU treatment. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (C) Representative picture of LSK population 5 days after 5-FU treatment with or without miR-26a chimera treatment (left). Percentages of LSK population (right). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (D) Apoptosis. Representative picture of annexin V+ in LSK population 5 days after 5-FU treatment (left). Percentages of annexin V+ in the LSK population (right). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. *P < .05, **P < .01. Error bars indicate SD.
miR-26a protects hematopoiesis from chemotherapeutic agent–induced myelosuppression. The mice received IV injection of 670 pmol per 20g of miR-26a or control (ctrl) chimera daily for 3 days. On day 2 of chimera treatment, 5-FU 150 mg/kg was injected. (A) The numbers of WBCs, red blood cells (RBCs), and platelets (PLTs) 10 days after 5-FU treatment. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (B) The number of bone marrow (BM) cells 10 days after 5-FU treatment. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (C) Representative picture of LSK population 5 days after 5-FU treatment with or without miR-26a chimera treatment (left). Percentages of LSK population (right). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (D) Apoptosis. Representative picture of annexin V+ in LSK population 5 days after 5-FU treatment (left). Percentages of annexin V+ in the LSK population (right). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. *P < .05, **P < .01. Error bars indicate SD.
Give the preferential expression of c-Kit in the HSPCs, we evaluated the protective effect of miR-26a on the HSPCs (LSK population) against 5-FU. As shown in Figure 2C, 5-FU caused nearly 10-fold reduction in the LSK population (CD3e−/B220−/CD11b−/Gr-1−/Ter119−/Sca-1+/c-Kit+) at 5 days after treatment. This was largely prevented by the miR-26a chimera (P = .0063) but not the control chimera. Correspondingly, the percent of apoptotic LSK was massively increased by 5-FU and specifically protected by the miR-26a but not the control chimera (Figure 2D).
Bak1 is a target of miR-26a for its myeloprotection from chemotherapeutic agent
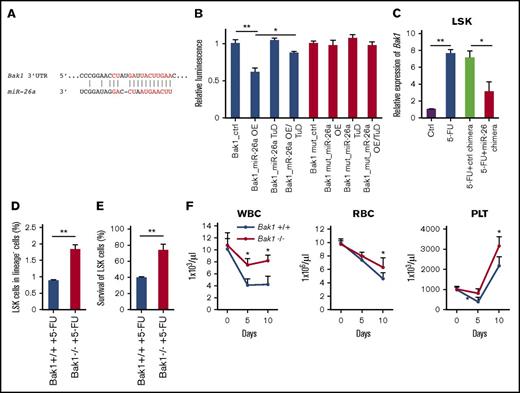
Using an in silico approach, we searched for miR-26a target genes that potentially regulate apoptosis. Among them, Bak1 (Bcl-2 antagonist/killer1) is a putative target (Figure 3A). To directly test whether miR-26a targets Bak1, we cloned the 3′UTR of the Bak1 downstream sequence into a luciferase reporter vector and cotransfected it with miR-26a precursors miR-26a OE or miR-26a TuD, which inhibits miRNA function by acting like a sponge for miRNAs.25 The luciferase activity was significantly downregulated by the overexpression of miR-26a. Further cotransfection of miR-26a TuD inhibitor significantly rescued the downregulation of luciferase activity caused by miR-26a overexpression. The depletion of miR-26a binding site on the Bak1 3′UTR abolished the inhibitory effects of miR-26a OE on the luciferase expressions, indicating that Bak1 is a direct target of miR-26a (Figure 3B).
Bak1 is a target of miR-26a for its myeloprotection from chemotherapeutic agent. (A) Putative miR-26a target site in the 3′UTR of mouse Bak1. Red characters denote the sites with highly probable preferential conservation between mammals. (B) Relative luciferase activity of reporter constructs containing the wild-type or mutant (mut) 3′UTR of mouse Bak1 in HEK293 cells cotransfected with either miR-26a precursor transducing lentivirus (miR-26a OE), miR-26a tough-decoy inhibitor transducing virus (miR-26a TuD) inhibitor, or negative control (ctrl). Data (mean ± SD) were pooled from 2 experiments. (C) Relative expression level of Bak1 messenger RNA in LSK population 5 days after 5-FU treatment. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (D) Targeted mutation of the Bak1 gene increased survival of LSK in BM at day 5 after 5-FU 150 mg/kg treatments. Data (mean ± SD) shown are percentages of LSK in BM (n = 3 for Bak1+/+ mice and n = 4 for Bak1−/− mice). (E) Percent survival of Bak1+/+ and Bak1−/− LSK in BM. Data (mean ± SD) shown are percentages of LSK, after normalization using means of LSK percentages from BM of 2 untreated Bak1+/+ and Bak1−/− mice as 100%. (F) The numbers of WBCs, red blood cells (RBCs), and platelets (PLTs) 10 days after 5-FU 150 mg/kg treatments between Bak1−/− mice and wild-type mice. Data (mean ± SD) were pooled from 2 experiments involving a total of 7 mice per group. *P < .05, **P < .01. Error bars indicate SD.
Bak1 is a target of miR-26a for its myeloprotection from chemotherapeutic agent. (A) Putative miR-26a target site in the 3′UTR of mouse Bak1. Red characters denote the sites with highly probable preferential conservation between mammals. (B) Relative luciferase activity of reporter constructs containing the wild-type or mutant (mut) 3′UTR of mouse Bak1 in HEK293 cells cotransfected with either miR-26a precursor transducing lentivirus (miR-26a OE), miR-26a tough-decoy inhibitor transducing virus (miR-26a TuD) inhibitor, or negative control (ctrl). Data (mean ± SD) were pooled from 2 experiments. (C) Relative expression level of Bak1 messenger RNA in LSK population 5 days after 5-FU treatment. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (D) Targeted mutation of the Bak1 gene increased survival of LSK in BM at day 5 after 5-FU 150 mg/kg treatments. Data (mean ± SD) shown are percentages of LSK in BM (n = 3 for Bak1+/+ mice and n = 4 for Bak1−/− mice). (E) Percent survival of Bak1+/+ and Bak1−/− LSK in BM. Data (mean ± SD) shown are percentages of LSK, after normalization using means of LSK percentages from BM of 2 untreated Bak1+/+ and Bak1−/− mice as 100%. (F) The numbers of WBCs, red blood cells (RBCs), and platelets (PLTs) 10 days after 5-FU 150 mg/kg treatments between Bak1−/− mice and wild-type mice. Data (mean ± SD) were pooled from 2 experiments involving a total of 7 mice per group. *P < .05, **P < .01. Error bars indicate SD.
To determine whether Bak1 is a target of miR-26a in vivo, we compared Bak1 levels among LSK cells in mice that received 5-FU with or without miR-26a or control chimeras. As shown in Figure 3C, Bak1 expression was massively induced by 5-FU treatment in LSK cells. miR-26a but not control chimera treatment significantly diminished the 5-FU–induced Bak1 elevation in vivo (Figure 3C). To examine the role of Bak1 in chemotherapy-induced hematopoietic injury, we treated Bak1 knockout mice and wild-type mice with 5-FU and analyzed the frequency of LSK in bone marrow at 5 days after treatment. As shown in Figure 3D, 5-FU–treated wild-type mouse BM had a significantly lower percentage of LSK when compared with BM of Bak1 knockout mice. When normalized against the percentage of LSK in BM of the untreated mice, it was clear that 5-FU caused more dramatic reduction of Bak1+/+ LSK than Bak1−/− LSK (Figure 3E). The myeloprotective effect against 5-FU by Bak1 deletion was further revealed by increased leukocyte counts and platelet counts in peripheral blood of the 5-FU–treated Bak1−/− mice (Figure 3F). These results demonstrate that exogenously delivered miR-26a mediates myeloprotection from chemotherapy at least in part by inhibiting Bak1-induced proapoptotic signaling.
miR-26a plays an essential role in hematopoietic reconstitution by targeting Bak1 after BMT
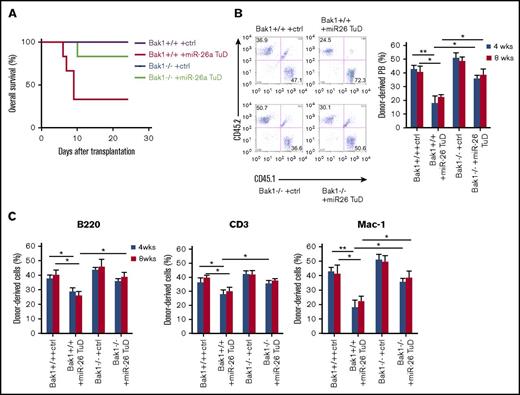
To investigate the role of endogenous miR-26a on stress hematopoiesis, we tested the effect of miR-26a TuD inhibitor on hematopoietic recovery from BM transplantation (BMT). Although mice receiving control inhibitor–treated BM survived the observation period, >70% of mice receiving transplants of miR-26a TuD BM cells died between 7 and 10 days after BMT (Figure 4A), suggesting that miR-26a plays a major role in stress hematopoiesis during the BMT. Furthermore, we compared the miR-26a TuD–transduced and control inhibitor–transduced BM cells for their ability to compete with recipient-type BM. Briefly, donor-type BM cells (CD45.2) were transduced with control inhibitor or miR-26a TuD, mixed with equal number of recipient-type BM cells (CD45.1), and then transplanted into lethally irradiated CD45.1 recipients. At 8 weeks, control inhibitor–transduced BM cells contributed roughly 40% of leukocytes in the peripheral blood, whereas miR-26a TuD–transduced BM cells contributed only ∼20% (Figure 4B left panel). The significant reduction was observed throughout 20 weeks of observation period (Figure 4B right panel). Similar defects were observed in B cells, T cells, and myeloid cells (Figure 4C). Likewise, the miR-26a TuD BM-derived leukocytes were significantly reduced in BM, spleen, and thymus (Figure 4D). The severe defects were reflected among LSK cells (CD3e−/B220−/CD11b−/Gr-1−/Ter119−/Sca-1+/c-Kit+) and HSCs (CD3e−/B220−/CD11b−/Gr-1−/Ter119−/Sca-1+/c-Kit+/CD48−/CD150+; Figure 4E). As shown in Figure 4F, annexin V+ cells were significantly increased both in miR-26a TuD–treated LSK and HSC populations. As expected, Bak1 expression levels were also significantly increased in these populations (Figure 4G). These results suggest that miR-26a plays an essential role against hematopoietic stresses by inhibiting an apoptotic pathway.
miR-26a plays an essential role in hematopoietic reconstitution after BMT. BM cells (5 × 105) transduced with miR-26a TuD inhibitor (miR-26 TuD) or control (ctrl) were transplanted into lethally irradiated congenic recipients. (A) Data shown are percent survival of recipients after BMT. Data were pooled from 2 experiments involving a total of 7 mice per group. (B) BM cells (CD45.2) transduced with ctrl or miR-26a TuD inhibitor (miR-26 TuD) were harvested and mixed with equal number of recipient-type BM cells (CD45.1) before transplantation into lethally irradiated congenic recipients (CD45.1). Representative plots of recipient peripheral blood (PB) leukocytes for ctrl or miR-26a TuD cells at 8 weeks after BMT (left). Reconstitution ratio of ctrl or miR-26a TuD donor cells in the recipients’ PB at 4, 8, and 20 to 30 weeks after transplantation (right). Data (mean ± SD) were pooled from 2 experiments involving a total of 10 mice per group. (C) Reconstitution ratio of ctrl or miR-26a TuD donor cells in the recipients’ PB B220+, CD3+, and Mac-1+ populations at 4, 8, and 20 to 30 weeks after BMT. Data (mean ± SD) were pooled from 2 experiments involving a total of 10 mice per group. (D) Reconstitution ratio of ctrl or miR-26 TuD donor cells in the BM, spleen, and thymus of recipients at 20 to 30 weeks after transplantation. Data (mean ± SD) were pooled from 2 experiments involving a total of 10 mice per group. (E) Reconstitution ratio of ctrl or miR-26a TuD donor cells in the LSK and HSC populations of the recipients’ BM at 20 to 30 weeks after BMT. Data (mean ± SD) were pooled from 2 experiments involving a total of 10 mice per group. (F) The percentages of annexin V+ in donor-derived LSK and HSC populations 5 days after BMT. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (G) Relative expression level of Bak1 messenger RNA in donor-derived LSK and HSC populations. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. *P < .05, **P < .01. Error bars indicate SD.
miR-26a plays an essential role in hematopoietic reconstitution after BMT. BM cells (5 × 105) transduced with miR-26a TuD inhibitor (miR-26 TuD) or control (ctrl) were transplanted into lethally irradiated congenic recipients. (A) Data shown are percent survival of recipients after BMT. Data were pooled from 2 experiments involving a total of 7 mice per group. (B) BM cells (CD45.2) transduced with ctrl or miR-26a TuD inhibitor (miR-26 TuD) were harvested and mixed with equal number of recipient-type BM cells (CD45.1) before transplantation into lethally irradiated congenic recipients (CD45.1). Representative plots of recipient peripheral blood (PB) leukocytes for ctrl or miR-26a TuD cells at 8 weeks after BMT (left). Reconstitution ratio of ctrl or miR-26a TuD donor cells in the recipients’ PB at 4, 8, and 20 to 30 weeks after transplantation (right). Data (mean ± SD) were pooled from 2 experiments involving a total of 10 mice per group. (C) Reconstitution ratio of ctrl or miR-26a TuD donor cells in the recipients’ PB B220+, CD3+, and Mac-1+ populations at 4, 8, and 20 to 30 weeks after BMT. Data (mean ± SD) were pooled from 2 experiments involving a total of 10 mice per group. (D) Reconstitution ratio of ctrl or miR-26 TuD donor cells in the BM, spleen, and thymus of recipients at 20 to 30 weeks after transplantation. Data (mean ± SD) were pooled from 2 experiments involving a total of 10 mice per group. (E) Reconstitution ratio of ctrl or miR-26a TuD donor cells in the LSK and HSC populations of the recipients’ BM at 20 to 30 weeks after BMT. Data (mean ± SD) were pooled from 2 experiments involving a total of 10 mice per group. (F) The percentages of annexin V+ in donor-derived LSK and HSC populations 5 days after BMT. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (G) Relative expression level of Bak1 messenger RNA in donor-derived LSK and HSC populations. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. *P < .05, **P < .01. Error bars indicate SD.
To further determine whether Bak1 is a target of miR-26a on stress hematopoiesis, we inhibited miR-26a function in Bak−/− mouse BM cells and tested the effect on recovery from BMT. Although >60% of mice receiving transplants of miR-26a TuD–treated Bak1+/+ BM cells died after BMT, all but 1 recipient of miR-26a TuD–treated Bak1−/− BM cells recovered (Figure 5A). The synthetic lethality suggests that Bak1 is the critical target gene of miR-26a in stress hematopoiesis. To critically evaluate the impact of the miR-26a-Bak1 axis on stress hematopoiesis, donor-type BM cells derived from Bak+/+ or Bak1−/− mice (CD45.2) were transduced with either control TuD or miR-26 TuD, mixed with equal number of recipient-type BM cells (CD45.1), and then transplanted into lethally irradiated CD45.1 recipients; we then analyzed the donor contribution at 8 weeks after BMT. As shown in Figure 5B, miR-26a TuD–transduced Bak1−/− BM cells contributed ∼40% of leukocytes in the peripheral blood, whereas miR-26a TuD–transduced Bak1+/+ BM cells contributed only ∼20% (Figure 5B left panel). The defects were significantly rescued by using Bak1−/− BM cells throughout 8 weeks of observation period (Figure 5B right panel). Similar effects were observed in B cells, T cells, and myeloid cells (Figure 5C).
Bak1 is a critical target gene of miR-26a in hematopoietic reconstitution after BMT. BM cells (5 × 105) transduced with miR-26a TuD inhibitor (miR-26 TuD) or control (ctrl) were transplanted into lethally irradiated congenic recipients. (A) Data shown are percent survival of recipients after BMT. Data were pooled from 2 experiments involving a total of 6 mice per group. (B) Bak1+/+ or Bak1−/− BM cells (CD45.2) transduced with ctrl or miR-26a TuD inhibitor (miR-26 TuD) were harvested and mixed with equal number of recipient-type BM cells (CD45.1) before transplantation into lethally irradiated congenic recipients (CD45.1). Representative plots of recipient peripheral blood (PB) leukocytes at 8 weeks after BMT (left). Reconstitution ratio of donor cells in the recipients’ PB at 4 and 8 weeks (wks) after transplantation (right). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (C) Reconstitution ratio of donor cells in the recipients’ PB B220+, CD3+, and Mac-1+ populations at 4 and 8 wks after BMT. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. *P < .05, **P < .01. Error bars indicate SD.
Bak1 is a critical target gene of miR-26a in hematopoietic reconstitution after BMT. BM cells (5 × 105) transduced with miR-26a TuD inhibitor (miR-26 TuD) or control (ctrl) were transplanted into lethally irradiated congenic recipients. (A) Data shown are percent survival of recipients after BMT. Data were pooled from 2 experiments involving a total of 6 mice per group. (B) Bak1+/+ or Bak1−/− BM cells (CD45.2) transduced with ctrl or miR-26a TuD inhibitor (miR-26 TuD) were harvested and mixed with equal number of recipient-type BM cells (CD45.1) before transplantation into lethally irradiated congenic recipients (CD45.1). Representative plots of recipient peripheral blood (PB) leukocytes at 8 weeks after BMT (left). Reconstitution ratio of donor cells in the recipients’ PB at 4 and 8 weeks (wks) after transplantation (right). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (C) Reconstitution ratio of donor cells in the recipients’ PB B220+, CD3+, and Mac-1+ populations at 4 and 8 wks after BMT. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. *P < .05, **P < .01. Error bars indicate SD.
miR-26a chimera inhibits mouse breast cancer growth and protects from 5-FU–induced myelosuppression
We used the mouse TUBO breast cancer model to evaluate the therapeutic potential of the miR-26a chimera for the myeloprotection and antitumor growth in breast cancer chemotherapy. We first tested the binding ability of an aptamer targeting mouse c-Kit to the TUBO cells by flow cytometry. As shown in Figure 6A, the biotinylated c-Kit aptamer bound to TUBO cells at nearly identical levels to an anti–c-Kit antibody. We also confirmed that this c-Kit aptamer bound to exogenously expressed mouse c-Kit protein on mouse MEF cells, comparable to a c-Kit antibody (supplemental Figure 4A). To determine whether the mouse c-Kit–miR-26a chimera could inhibit the growth of mouse breast cancer cells in vitro, we treated the TUBO cells with increasing doses of the miR-26a or control chimera and counted the viable cells under microscope at 2 days after the treatment. As shown in Figure 6B (left panel), the miR-26a chimera alone caused a dose-dependent reduction in tumor-cell number, with 50% inhibitory concentration of approximately 83 nM. This is considerably more effective than 7 μM of 5-FU alone, which cause <20% reduction in tumor-cell number. In combination, 5-FU and the miR-26a chimera synergistically reduced tumor cells. The reduction resulted from induction of apoptosis as the percentage of apoptotic TUBO cells were increased by both 5-FU and the miR-26a chimera and their combination (Figure 6B right panel).
miR-26a chimera inhibits mouse breast cancer growth and protects from chemotherapy-induced myelosuppression. (A) The binding of c-Kit aptamer to TUBO cells. (B) miR-26a chimera suppressed the growth and induced apoptosis of TUBO cells. Additional 5-FU treatment (1 μg/ml) enhanced these effects of miR-26a chimera. Cell count (left) and percentage of annexin V+ cells (%) (right) of the cells cultured with miR-26a chimera or the control (ctrl) were determined by flow cytometry. Asterisks denote the significant difference between ctrl chimera vs miR-26a chimera and between ctrl chimera plus 5-FU vs miR-26a chimera plus 5FU. Data (mean ± SD) were pooled from 2 experiments. (C-E) miR-26a, Ezh2, and Bak1 expressions in c-Kit+ or c-Kit− cells harvested from TUBO-derived tumors in BALB/c mice treated with miR-26a chimera (670 pmol per 20 g) for 3 days. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (F,G) miR-26a and Bak1 expressions in BM detected by quantitative polymerase chain reaction at 3 days after IV injection with the miR-26a chimera (670 pmol per 20 g). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (H) Bak1 expression in c-Kit+ BM cells harvested from tumor (TUBO)-bearing BALB/c mice treated with miR-26a chimera (670 pmol per 20 g; 3 times) and 5-FU 50 mg/kg. Representative plots of Bak1 levels in cKit+ BM cells at day 3 after 5-FU treatment (left). Mean fluorescence intensity (MFI) of Bak1 staining in cKit+ BM cells (right). Data (mean ± SD) were pooled from 2 experiments involving a total of 3 mice per group. (I) MFI of Bak1 staining in cKit+ tumor cells harvested from the tumor (TUBO)-bearing BALB/c mice treated with miR-26a chimera (670 pmol per 20 g; 3 times) and 5-FU 50 mg/kg. Data (mean ± SD) were pooled from 2 experiments involving a total of 3 mice per group. (J) Tumor volume over time. BALB/c mice bearing TUBO cells were treated with miR-26a chimera (670 pmol per 20 g; 5 times, gray arrows) and 5-FU 50 mg/kg (3 times, blue arrows). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. There was significant difference between the 5-FU–only group vs miR-26a chimera–only group (2-way repeated-measures analysis of variance [ANOVA] followed by Bonferroni post-test for day 0 to day 18; interaction P < .001). (K) Tumor volume over time after combinational treatment with 5-FU and chimeras. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. There was a significant difference between 5-FU plus ctrl chimera group vs 5-FU plus miR-26a chimera group (2-way repeated-measures ANOVA followed by Bonferroni post-test for day 0 to day 21; interaction P < .0001). (L) The numbers of WBCs and platelets (PLTs) in the tumor bearing mice 5 days after 5-FU treatment. Data (mean ± s.d.) pooled from 2 experiments involving a total of 6 mice per group. *P < .05, **P < .01. Error bars indicate SD.
miR-26a chimera inhibits mouse breast cancer growth and protects from chemotherapy-induced myelosuppression. (A) The binding of c-Kit aptamer to TUBO cells. (B) miR-26a chimera suppressed the growth and induced apoptosis of TUBO cells. Additional 5-FU treatment (1 μg/ml) enhanced these effects of miR-26a chimera. Cell count (left) and percentage of annexin V+ cells (%) (right) of the cells cultured with miR-26a chimera or the control (ctrl) were determined by flow cytometry. Asterisks denote the significant difference between ctrl chimera vs miR-26a chimera and between ctrl chimera plus 5-FU vs miR-26a chimera plus 5FU. Data (mean ± SD) were pooled from 2 experiments. (C-E) miR-26a, Ezh2, and Bak1 expressions in c-Kit+ or c-Kit− cells harvested from TUBO-derived tumors in BALB/c mice treated with miR-26a chimera (670 pmol per 20 g) for 3 days. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (F,G) miR-26a and Bak1 expressions in BM detected by quantitative polymerase chain reaction at 3 days after IV injection with the miR-26a chimera (670 pmol per 20 g). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (H) Bak1 expression in c-Kit+ BM cells harvested from tumor (TUBO)-bearing BALB/c mice treated with miR-26a chimera (670 pmol per 20 g; 3 times) and 5-FU 50 mg/kg. Representative plots of Bak1 levels in cKit+ BM cells at day 3 after 5-FU treatment (left). Mean fluorescence intensity (MFI) of Bak1 staining in cKit+ BM cells (right). Data (mean ± SD) were pooled from 2 experiments involving a total of 3 mice per group. (I) MFI of Bak1 staining in cKit+ tumor cells harvested from the tumor (TUBO)-bearing BALB/c mice treated with miR-26a chimera (670 pmol per 20 g; 3 times) and 5-FU 50 mg/kg. Data (mean ± SD) were pooled from 2 experiments involving a total of 3 mice per group. (J) Tumor volume over time. BALB/c mice bearing TUBO cells were treated with miR-26a chimera (670 pmol per 20 g; 5 times, gray arrows) and 5-FU 50 mg/kg (3 times, blue arrows). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. There was significant difference between the 5-FU–only group vs miR-26a chimera–only group (2-way repeated-measures analysis of variance [ANOVA] followed by Bonferroni post-test for day 0 to day 18; interaction P < .001). (K) Tumor volume over time after combinational treatment with 5-FU and chimeras. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. There was a significant difference between 5-FU plus ctrl chimera group vs 5-FU plus miR-26a chimera group (2-way repeated-measures ANOVA followed by Bonferroni post-test for day 0 to day 21; interaction P < .0001). (L) The numbers of WBCs and platelets (PLTs) in the tumor bearing mice 5 days after 5-FU treatment. Data (mean ± s.d.) pooled from 2 experiments involving a total of 6 mice per group. *P < .05, **P < .01. Error bars indicate SD.
To test the therapeutic effect of the miR-26a chimera, we injected the miR-26a or control chimera intravenously into TUBO tumor–bearing mice. As shown in supplemental Figure 5A, significant levels of the miR-26a chimera could be detected in the blood of tumor-bearing mice that received 670 pmol of chimera up to 8 hours after injection. Moreover, the accumulation of the miR-26a chimera in the tumor was visualized by in vivo imaging using Alexa Fluor 647 dye-conjugated miR-26a chimera (supplemental Figure 5B). To further confirm the targeting delivery of miR-26a in vivo, we measured the levels of miR-26a among sorted c-Kit+ and c-Kit− TUBO cells at 3 days after injection by quantitative polymerase chain reaction. As shown in Figure 6C, a significant increase of miR-26a was detected among c-Kit+ but not c-Kit− tumor cells, thus confirming specific delivery of miR-26a to c-Kit+ tumor cells. Functional delivery was further confirmed by the specific decrease in the miR-26a target gene Ezh2 and the protein among c-Kit+ tumor cells (Figure 6D; supplemental Figure 6A). In contrast, Bak1 was barely expressed among c-Kit+ tumor cells (supplemental Figure 6B), and such expression was not significantly regulated by the miR-26a chimera (P = .49; Figure 6E).
The c-Kit aptamer has been reported to bind to a cell population in mouse BM cells like a c-Kit antibody.16 As shown in supplemental Figure 4B, we observed that this c-Kit aptamer bound to the lineage− population (CD3e−/B220−/CD11b−/Gr-1−/Ter119−) in BM cells. Consistent with these results, a selective increase in miR-26a levels was observed in c-Kit+ BM cells from mice treated with the miR-26a chimera on day 3 after single treatment in non–tumor-bearing C57BL/6 mice (Figure 6F). Interestingly, in mice that received no 5-FU treatment, c-Kit+ HSPCs also expressed low levels of Bak1, and its expression was not significantly downregulated by the miR-26a chimera (Figure 6G). Therefore, it seems that miR-26a regulates only abnormally induced Bak1 expression during stress-induced hematopoiesis, such as within hosts receiving irradiation or chemotherapy.4 To determine whether Bak1 is a target gene of miR-26a in BM and tumor cells during chemotherapy in vivo, we treated mice bearing mammary tumors (TUBO) with the miR-26a chimera and 5-FU (Figure 6H). We observed a significant elevation of Bak1 by 5-FU treatment in c-Kit+ BM cells, but not in c-Kit+ tumor cells (Figure 6H-I). Further miR-26a chimera treatment suppressed the elevation of Bak1 in c-Kit+ BM cells, suggesting a different sensitivity against 5-FU treatment in this dose between tumor and BM cells for initiating the Bak1-related apoptotic signaling pathway, which may explain the different effects of the miR-26a chimera between tumor and BM cells in vivo.
Regarding antitumor effect of the miR-26a chimera, we observed a significant decrease in tumor size after 5 daily injections of the miR-26a chimera (Figure 6J). Although either 5-FU or miR-26a chimera monotherapy was sufficient to cause growth retardation, the miR-26a chimera seemed more effective than 5-FU treatment (Figure 6J). Remarkably, combining 5-FU with the miR-26a chimera achieved the most effective growth retardation (Figure 6K).
Notably, the number of leukocytes in TUBO tumor–bearing mice were nearly 3 times more than the normal range of 8.05+/− 1.04 × 103/μL,26 which is consistent with tumor-induced leukocytosis. As expected, 5-FU treatment not only eliminated leukocytosis but also caused significant leukopenia and thrombocytopenia (Figure 6L; supplemental Figure 7). Importantly, combination treatment with the miR-26a chimera nearly doubled the number of leukocytes (WBCs) and thrombocytes (platelets) and largely prevented leukopenia (Figure 6L; supplemental Figure 7).
miR-26a chimera protects mice from carboplatin-induced myelosuppression and inhibits human breast cancer growth
The addition of neoadjuvant carboplatin to the regimen of taxane and anthracycline significantly increases the proportion of patients achieving a pathological complete response.27 However, adverse effects such as anemia, neutropenia, and thrombocytopenia (grade 3 or 4 hematological events) occur more frequently in the patient group administered carboplatin. We therefore tested the protective effect of miR-26a on myelosuppression by carboplatin treatment. As observed in the clinical trial,27 high-dose carboplatin treatment induced significant defects in hematopoiesis. Compared with vehicle, a single dose of carboplatin significantly reduced WBC and platelet counts as early as day 5 (Figure 7A). The defect persisted and was exacerbated for at least 10 days, when a reduction of red blood cells was also observed (Figure 7B). Remarkably, 3 daily injections of the miR-26a chimera, but not the control chimera, prevented reductions in WBCs and red blood cells. Although reduction in platelets on day 5 was not prevented, the miR-26a chimera prevented a further drop in platelets from day 5 to day 10. Therefore, the myeloprotective effect of the miR-26a chimera is not limited to 5-FU.
miR-26a protects mice against carboplatin-induced myelosuppression. The mice received IV injection of 670 pmol per 20 g of miR-26a or control (ctrl) chimera daily for 3 days. On day 2 of the chimera treatment, carboplatin 120 mg/kg was injected. The numbers of WBCs, red blood cells (RBCs), and platelet (PLTs) 5 days (A) and 10 days (B) after carboplatin treatment. Data (mean ± SD) were pooled from 2 experiments involving a total of 3 mice per group. (C) Therapeutic effect of miR-26a chimera with 5-FU. NSG mice bearing MDA-MB-231 cells were treated with miR-26a chimera (670 pmol per 20 g; 5 times) and/or 5-FU 50 mg/kg (3 times). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. There was a significant difference between the 5-FU plus ctrl chimera group vs 5-FU plus miR-26a chimera group (2-way repeated-measures analysis of variance [ANOVA] followed by Bonferroni post-test for day 0 to day 21; interaction P < .0001). (D) Therapeutic effect of miR-26a chimera with carboplatin. NSG mice bearing MDA-MB-231 cells were treated with miR-26a chimera (670 pmol per 20 g; 5 times) and/or carboplatin 40 mg/kg (3 times). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. There was a significant difference between the carboplatin plus ctrl chimera group vs carboplatin plus miR-26a chimera group (2-way repeated-measures ANOVA followed by Bonferroni post-test for day 0 to day 24; interaction P < .0001). *P < .05, **P < .01. Error bars indicate SD.
miR-26a protects mice against carboplatin-induced myelosuppression. The mice received IV injection of 670 pmol per 20 g of miR-26a or control (ctrl) chimera daily for 3 days. On day 2 of the chimera treatment, carboplatin 120 mg/kg was injected. The numbers of WBCs, red blood cells (RBCs), and platelet (PLTs) 5 days (A) and 10 days (B) after carboplatin treatment. Data (mean ± SD) were pooled from 2 experiments involving a total of 3 mice per group. (C) Therapeutic effect of miR-26a chimera with 5-FU. NSG mice bearing MDA-MB-231 cells were treated with miR-26a chimera (670 pmol per 20 g; 5 times) and/or 5-FU 50 mg/kg (3 times). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. There was a significant difference between the 5-FU plus ctrl chimera group vs 5-FU plus miR-26a chimera group (2-way repeated-measures analysis of variance [ANOVA] followed by Bonferroni post-test for day 0 to day 21; interaction P < .0001). (D) Therapeutic effect of miR-26a chimera with carboplatin. NSG mice bearing MDA-MB-231 cells were treated with miR-26a chimera (670 pmol per 20 g; 5 times) and/or carboplatin 40 mg/kg (3 times). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. There was a significant difference between the carboplatin plus ctrl chimera group vs carboplatin plus miR-26a chimera group (2-way repeated-measures ANOVA followed by Bonferroni post-test for day 0 to day 24; interaction P < .0001). *P < .05, **P < .01. Error bars indicate SD.
To determine the combinational effect of chemoreagents and miR-26a chimera against human breast cancer growth in vivo, we treated the mice bearing mammary tumors (MDA-MB-231) with the miR-26a chimera in combination with either 5-FU or carboplatin (Figure 7C-D). Although either 5-FU, carboplatin, or miR-26a chimera monotherapy was sufficient to cause growth retardation, the combination of these chemoreagents with the miR-26a chimera achieved substantial tumor regression (Figure 7C-D).
Discussion
Taken together, on the basis of our finding that miR-26a mediated a converging pathway in cancer progression and chemotherapy-induced myelosuppression, we developed a new combination therapy that improved efficacy of 5-FU and carboplatin while ameliorating their main adverse effect. The dual benefit of combination therapy will likely extend to other chemotherapies, because miR-26a has been shown to act synergistically with paclitaxel in killing breast cancer cells in vitro.28 Apart from 5-FU, our data show remarkable efficacy of the miR-26a chimera in protecting mice against hematopoietic toxicity from carboplatin. Carboplatin is a platinum-based and interstrand cross-linking antineoplastic agent. Platinum-based compounds remain in use for chemotherapy drugs despite toxicity.27 By preventing its hematopoietic toxicity, our new approach may allow even broader use of this class of chemotherapeutic drugs.
Although general and specific inactivations of miRNA play a major role in cancer pathogenesis,29-31 difficulties in miRNA delivery to cancer cells have limited the potential utility of miRNA in cancer therapy. Although an adenovirus-associated virus-based miRNA delivery system was reported for miR-26a administration in a liver cancer model,32 it is unclear whether this is generally applicable to other cancer types. Here we have demonstrated that an aptamer can be used for specific delivery of miRNA to targeted cells, providing the advantages of effectiveness at a lower dose, low immunogenicity, and high scalability for miRNA delivery with no risk of genomic integration.33 Given the existence of the large bank of aptamers for cell-surface markers, the new approach described herein will likely have a broad impact in the study of the biological function of miRNA in vivo and in cancer therapy. Although our study needs further examination to validate the effect of the miR-26a chimera in breast cancer therapy using primary cells and other cell lines derived from other subtypes of breast cancer, it has demonstrated the therapeutic potential of our miR-26a chimera for basal-like breast cancer treatment.
Furthermore, although our study focused on breast cancer models, miR-26a as a tumor suppressor has been observed in other cancer types, including prostate cancer, pancreatic cancer, and lung cancer.34-36 Likewise, KIT is also widely expressed among human cancers, including gastrointestinal stromal tumors, myeloid leukemia, small-cell lung cancer, prostate cancer, pancreatic cancer, ovarian cancer, and glioblastoma.37-43 Therefore, the clinical significance of our miR-26a chimera targeting KIT+ cancers could extend well beyond breast cancer.
The full-text version of this article contains a data supplement.
Acknowledgments
The results presented here were based in part on data generated by The Cancer Genome Atlas Research Network.
This work was supported by grants from the National Institutes of Health, National Institute on Aging (AG036690) (P. Zheng) and National Institute of Allergy and Infectious Diseases (AI064350) (Y.L.) and the US Department of Defense (W81XWH-11-1-0436) (Y.L.).
Authorship
Contribution: T.T., P. Zheng, and Y.L. conceived and designed the study; T.T., P. Zhang, and Y.L. developed the methodology; T.T., P. Zhang, and C.A.L. acquired the data; T.T. and P. Zhang analyzed and interpreted the data; T.T., P. Zheng, and Y.L. wrote and reviewed the manuscript; P. Zhang and C.A.L. provided technical support; and P. Zheng and Y.L. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pan Zheng, Children’s National Medical Center, 111 Michigan Ave NW, M5206, Washington, DC 20010; e-mail: pzheng@cnmc.org.

![Figure 1. Identification and validation of KIT-targeting miR-26a chimera that inhibits human breast cancer growth in vitro and in vivo. (A) Overall survival of patients with basal-like breast cancer with higher or lower expression of miR-26a-2 based on expression scores compared with the mean value in TCGA cohort. (B) The binding of anti-KIT antibody and KIT DNA aptamer to MDA-MB-231 cell line. Other cell-targeting aptamer is specific for Ramos cells. (C) The secondary structure of KIT aptamer–miR-26a chimera. KIT DNA aptamer was linked with C3 linker to complementary RNA passenger sequence that was bound to miR-26a mimic sequence. Another RNA passenger sequence binding to the 5′ of miR-26 mimic was conjugated with triethylene glycol (TEG)–cholesterol. (D) Specific miR-26a delivery by the miR-26a chimera. Two days after incubation with the miR-26a chimera, significant increase of miR-26a expression in the MDA-MB-231 cells compared with control (ctrl) chimera treatment was detected by quantitative polymerase chain reaction. Data (mean ± standard deviation [SD]) were pooled from 3 experiments. (E) miR-26a chimera suppressed the growth and induced apoptosis of MDA-MB-231 cells in a dose-dependent manner. After 3 days of culture with miR-26a chimera or ctrl chimera, MDA-MB-231 cell numbers (left) were counted using hemocytometer, and percentage of annexin V+ cells (right) was determined by flow cytometry. Data (mean ± SD) were pooled from 2 experiments. (F) Significant suppression of EZH2 protein in miR-26a chimera–treated MDA-MB-231 cells detected by immunoblot. (G) Relative expression of miR-26a in tumor harvested from NSG mice bearing human breast tumor with MDA-MB-231 cells. Significant increase of miR-26a was observed in KIT+ tumor cells 3 days after IV injection with 670 pmol per 20 g of miR-26a chimera. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (H) Therapeutic effect of miR-26a chimera. The tumor-bearing mice were treated daily with miR-26a chimera (670 pmol per 20 g) for 5 or 10 days (first injection defined as day 0). Data (mean ± SD) were pooled from 2 experiments involving a total of 5 mice per group. Tumor volume over time (left). Significant difference between miR-26 chimera (×5) versus miR26 chimera (×10; 2-way repeated-measures analysis of variance followed by Bonferroni post-test for day 0 to day 18 detected the significant difference (interaction P = .0012). Kaplan-Myer survival curve (right). Log-rank test detected significant differences between ctrl chimera group and miR-26a chimera groups (ctrl chimera vs miR-26 chimera [×5], P = .002; ctrl chimera vs miR-26 chimera [×10], P = .002). The difference in survival between miR-26 chimera (×5) and miR26 chimera (×10) did not reach statistical significance (P = .077). *P < .05, **P < .01. Error bars indicate SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/15/10.1182_bloodadvances.2017004705/3/m_advances004705f1.jpeg?Expires=1767731783&Signature=lXhTaKGbsabwCV2mCot1OiTQEjQPCAmZ0s-iI3cADqUav1KcEsz0j0nZNG8cDSPo93BLqiuq95AxWVdikbcEgAm8MXFvpCS34OvZRURfM4wa87c5YSxhbqyekaJgez6ZH2qiuePiP5al1-WtjTajJ3qIXhhfFMju3lU2oxMTcBJjVmImqfUVA7aW1aM-i-Da1sv26-y6duBGxoAOIW1lEea8iQ4gojxsKcqDUQDKq~z9diVop14uZzcZlEfxBGEFXN2vl5dsv7bFKT8eb7~UrYf-RCQ2Pukfs~YkCqHG~NFyIJLcMSDIl8gruk-6goCORZIGW6s~o240JOoFhKwoaQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 6. miR-26a chimera inhibits mouse breast cancer growth and protects from chemotherapy-induced myelosuppression. (A) The binding of c-Kit aptamer to TUBO cells. (B) miR-26a chimera suppressed the growth and induced apoptosis of TUBO cells. Additional 5-FU treatment (1 μg/ml) enhanced these effects of miR-26a chimera. Cell count (left) and percentage of annexin V+ cells (%) (right) of the cells cultured with miR-26a chimera or the control (ctrl) were determined by flow cytometry. Asterisks denote the significant difference between ctrl chimera vs miR-26a chimera and between ctrl chimera plus 5-FU vs miR-26a chimera plus 5FU. Data (mean ± SD) were pooled from 2 experiments. (C-E) miR-26a, Ezh2, and Bak1 expressions in c-Kit+ or c-Kit− cells harvested from TUBO-derived tumors in BALB/c mice treated with miR-26a chimera (670 pmol per 20 g) for 3 days. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (F,G) miR-26a and Bak1 expressions in BM detected by quantitative polymerase chain reaction at 3 days after IV injection with the miR-26a chimera (670 pmol per 20 g). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. (H) Bak1 expression in c-Kit+ BM cells harvested from tumor (TUBO)-bearing BALB/c mice treated with miR-26a chimera (670 pmol per 20 g; 3 times) and 5-FU 50 mg/kg. Representative plots of Bak1 levels in cKit+ BM cells at day 3 after 5-FU treatment (left). Mean fluorescence intensity (MFI) of Bak1 staining in cKit+ BM cells (right). Data (mean ± SD) were pooled from 2 experiments involving a total of 3 mice per group. (I) MFI of Bak1 staining in cKit+ tumor cells harvested from the tumor (TUBO)-bearing BALB/c mice treated with miR-26a chimera (670 pmol per 20 g; 3 times) and 5-FU 50 mg/kg. Data (mean ± SD) were pooled from 2 experiments involving a total of 3 mice per group. (J) Tumor volume over time. BALB/c mice bearing TUBO cells were treated with miR-26a chimera (670 pmol per 20 g; 5 times, gray arrows) and 5-FU 50 mg/kg (3 times, blue arrows). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. There was significant difference between the 5-FU–only group vs miR-26a chimera–only group (2-way repeated-measures analysis of variance [ANOVA] followed by Bonferroni post-test for day 0 to day 18; interaction P < .001). (K) Tumor volume over time after combinational treatment with 5-FU and chimeras. Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. There was a significant difference between 5-FU plus ctrl chimera group vs 5-FU plus miR-26a chimera group (2-way repeated-measures ANOVA followed by Bonferroni post-test for day 0 to day 21; interaction P < .0001). (L) The numbers of WBCs and platelets (PLTs) in the tumor bearing mice 5 days after 5-FU treatment. Data (mean ± s.d.) pooled from 2 experiments involving a total of 6 mice per group. *P < .05, **P < .01. Error bars indicate SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/15/10.1182_bloodadvances.2017004705/3/m_advances004705f6.jpeg?Expires=1767731783&Signature=VDTsfi8xSE7PJ8XSpQiP7AxxjulEaQzOhHRQG0H~S5ncmxBeboQTvEZzyAa2w~bcDf2jnSywmU4gsjIEOsEhxx1DiC4d9-58lxL9gwaHuPbkC4PG8YXtJUxcxcZtIOYpK69Ss8AEy55ueTeci-ZRw38pgzWvPhngZ~JiWmdSozALGoTXNxTU2NMxl-vizy~C1OCnw-Enp4ZkkJuS4amFyhY3EzAn1Vh6UBRHuui8n2f85kPY6al6tcq3hvOBnXwOLDRGFw85FfoWtI3wew86AMaN2ZmzGiabZ35xo-YPlzMCYUETYnsnMcW664lx6Nkq3dtrRLlvGJoZ7eSt-uzpHQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. miR-26a protects mice against carboplatin-induced myelosuppression. The mice received IV injection of 670 pmol per 20 g of miR-26a or control (ctrl) chimera daily for 3 days. On day 2 of the chimera treatment, carboplatin 120 mg/kg was injected. The numbers of WBCs, red blood cells (RBCs), and platelet (PLTs) 5 days (A) and 10 days (B) after carboplatin treatment. Data (mean ± SD) were pooled from 2 experiments involving a total of 3 mice per group. (C) Therapeutic effect of miR-26a chimera with 5-FU. NSG mice bearing MDA-MB-231 cells were treated with miR-26a chimera (670 pmol per 20 g; 5 times) and/or 5-FU 50 mg/kg (3 times). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. There was a significant difference between the 5-FU plus ctrl chimera group vs 5-FU plus miR-26a chimera group (2-way repeated-measures analysis of variance [ANOVA] followed by Bonferroni post-test for day 0 to day 21; interaction P < .0001). (D) Therapeutic effect of miR-26a chimera with carboplatin. NSG mice bearing MDA-MB-231 cells were treated with miR-26a chimera (670 pmol per 20 g; 5 times) and/or carboplatin 40 mg/kg (3 times). Data (mean ± SD) were pooled from 2 experiments involving a total of 6 mice per group. There was a significant difference between the carboplatin plus ctrl chimera group vs carboplatin plus miR-26a chimera group (2-way repeated-measures ANOVA followed by Bonferroni post-test for day 0 to day 24; interaction P < .0001). *P < .05, **P < .01. Error bars indicate SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/15/10.1182_bloodadvances.2017004705/3/m_advances004705f7.jpeg?Expires=1767731783&Signature=gAR929HbBk9Pf1HlWv~O9hXHuq5A~uywCMForargPOelISgd~pQJCFPLrjochGhMLaH8uoaFcEhBSuMk89UW1p3c7Kwt2sfY~BQmXTO~8P2etszIN-AMhUH3FoNrnD-GvRmvd374H8ri0GdHd7T3XZYK47-gMKQfAPWadjXbAsGQpTKhzjzY-1l7fzutNlJFgBai-meblL9y8dCM3ETCLBc8V833fTpYJw9P6j8nQOnbyG7wv3jPcEIBjKgAtVQf5YTU3qzSJ58kKutsCRO5I6ZHwScLirn-UA5asbB3scNCQ4oSTpyTY9EYTzW7xTAjaoEs2QQZ7NXLDcL6kmKZYw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)