TO THE EDITOR:
Systemic mastocytosis (SM) is a subset of rare neoplasms characterized by clonal proliferation of aberrant mast cells (MCs) and their accumulation in ≥1 extracutaneous organs, which typically associates with activating KIT mutations.1 SM with an associated hematological neoplasm (AHN) is the second most common type of SM in Mayo series (∼40%),2 with challenging diagnosis because the mast cell infiltration may hide behind the AHN component and vice versa.3-5 This is particularly applicable to acute myeloid leukemia (AML) because the histological examination of bone marrow (BM) has not been established as a standard diagnostic tool. Moreover, the concomitant hematological neoplasm often shares a KIT mutation and/or other clonal genetic abnormalities with neoplastic MCs. AML with t(8;21) (q22;q22)/RUNX1::RUNX1T1 [t(8;21) AML] represents 4% to 8% of all AMLs, which is categorized to the genetically favorable risk group.6 Meanwhile, KIT mutations, most frequently at position D816, are detectable in up to 20% to 47% of patients with t(8;21) AML and associate with decreased remission duration and inferior overall survival (OS).7-10 Previous case reports, case-series, and/or literature reviews have described the potential association of KIT mutant t(8;21) AML [KITpos t(8;21) AML] with underlying SM.11,12 Owing to a lack of literature on investigating similarities and differences between SM-t(8;21) AML and KITpos t(8;21) AML, we carried out this retrospective cohort study from multiple Chinese centers to compare clinical and molecular features between these 2 groups.
Between January 2009 and December 2022, 24 patients with SM-t(8;21) AML and 212 with KITpos t(8;21) AML diagnosed and treated in 16 Chinese centers were eligible. All patients were screened for SM by BM smear, flow cytometry, and/or BM histological examination. Details of the study design, patient population, treatment regimen, and statistical analysis have been described in supplemental Data. This study was approved by the responsible ethics committees and performed in accordance with the Declaration of Helsinki.
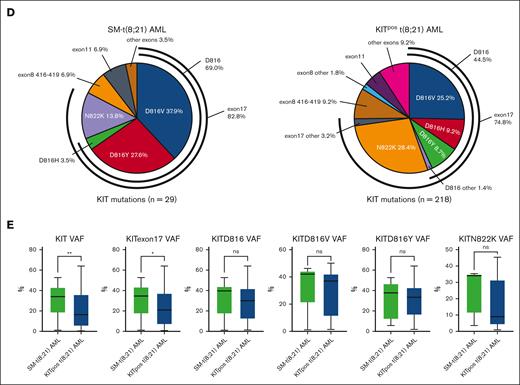
As summarized in supplemental Table 1, the clinical and laboratory characteristics of patients with SM-t(8;21) AML (n = 24) were similar to those of KITpos t(8;21) AML (n = 212), except for a lower platelet levels (P = .001) and a higher burden of MCs in BM. Meanwhile, comparison between the SM-t(8;21) AML and KITpos t(8;21) AML cohort revealed several interesting findings on molecular features (supplemental Table 1 and Figure 1) as follows: (1) the incidence of KIT D816V (45.83% vs 27.83%; P = .046) and KITD816 mutations (69.57% vs 45.75%; P = .030) were much higher in SM-t(8;21) AML; (2) the frequency of the most common somatic mutations (FLT3, ASXL1/2, TET2, ZBTB7A, NRAS, and KDM6A) in de novo AML was comparable between the 2 groups, except that DNMT3A mutations were more frequent in SM-t(8;21) AML (8.70% vs 0.96%; P = .050); (3) in contrast to data from large SM cohorts, the frequency of SRSF2/ASXL1/RUNX1 (S/A/R) aberrations in the 2 groups was relatively low [KITpos t(8;21) AML: 10.07% ; SM-t(8;21) AML: 4.35%; P = .622]; (4) for all known KIT mutation subtypes detected in 2 groups, the proportion of D816Y mutation (27.6% vs 8.7%, P = .002) and D816 mutations (69.0% vs 44.5%; P = .013) were significantly higher in patients with SM-t(8;21) AML. Additionally, N822K and D816V were the most common subtype in KITpos t(8;21) AML and SM-t(8;21) AML, respectively (Figure 1D); and (5) patients with SM-t(8;21) AML had higher median variant allele frequency (VAF) of all KIT mutations (34.00% vs 16.40%; P = .008) and KIT exon17 mutations (34.70% vs 21.05%; P = .025) (Figure 1E). Moreover, no significant association between several pairs of mutations was identified in patients with SM-t(8;21) AML (supplemental Figure 2).
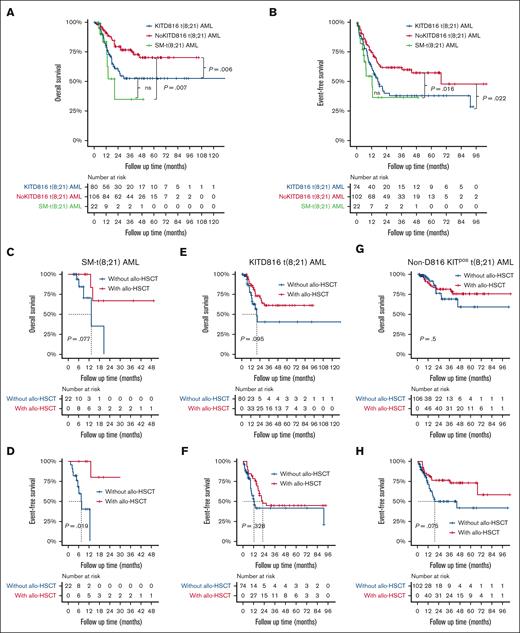
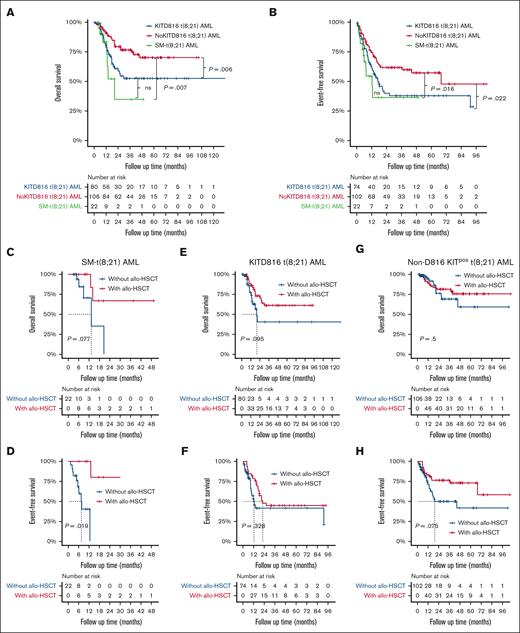
Considering that KITD816 mutations have been reported to be associated with inferior prognosis for t(8;21) AML, we further divided KITpos t(8;21) AML cohort into 2 subgroups: KITD816 mutant t(8;21) AML (KIT816 t[8;21] AML) and non-D816 KIT mutant t(8;21) AML (non-D816 KITpos t[8;21] AML), to better make comparison with SM-t(8;21) AML. As to 208 patients (SM-t[8;21] AML, n = 22; KITD816 t[8;21] AML, n = 80; non-D816 KITpos t[8;21] AML, n = 106) evaluable for therapeutic effect (supplemental Table 2), the overall complete remission (CR) or CR with incomplete count recovery (CRi) rate in the AML component after the first induction therapy of SM-t(8;21) AML was significantly lower than that of KITD816 t(8;21) AML (50.0% vs 80.0%; P = .005) and non-D816 KITpos t(8;21) AML (50.0% vs 81.1%; P = .002), regardless of treatment intensity. Besides, both patients with SM-t(8;21) AML and those with KITD816 t(8;21) AML were more refractory to chemotherapy than those with non-D816 KITpos t(8;21) AML (50.0% vs 43.8% vs 20.8%; P = .001). Additionally, the incidence of leukemia relapse of KITD816 t(8;21) AML was comparable to that of SM-t(8;21) AML (46.3% vs 27.3%; P = .110) but significantly higher than non-D816 t(8;21) AML (46.3% vs 24.5%; P = .002). The median follow-up time of our cohort was 20.6 months (range, 1.0-132.4). Both the OS (P = .330) and event-free survival (EFS) (P = .383) were close between patients with KITD816 t(8;21) AML and those with SM-t(8;21) AML, which were inferior to those of non-D816 KITpos t(8;21) AML (supplemental Table 2; Figure 2A-B). Allogeneic hematopoietic stem cell transplantation (allo-HSCT) was performed in 11 of 22 patients with SM-t(8;21) AML, 49 of 80 with KITD816 t(8;21) AML, and 58 of 106 with non-D816 KITpos t(8;21) AML. When considering allo-HSCT as a time-dependent variable, it significantly improved EFS (median, not reached [95% confidence interval (CI), 13.0-not reached] vs 7.5 [95% CI, 5.6-not reached]; P = .019) and appeared to bring OS benefit (median, not reached [95% CI, 13.0-not reached] vs 13.3 [95% CI, 6.2-not reached]; P = .077) for patients with SM-t(8;21) AML (Figure 2C-D). However, allo-HSCT did not show the favorable impact on survival of patients with neither KITD816 t(8;21) AML nor those with non-D816 KITpos t(8;21) AML by Mantel-Byar test (Figure 2E-H).
Survival curves of patients with KITD816 t(8;21) AML, non-D816 KITpos t(8;21) AML, and SM-t(8;21) AML. (A-B) Kaplan-Meier curves of OS and EFS of patients with KITD816 t(8;21) AML, non-D816 KITpos t(8;21) AML, and SM-t(8;21) AML, respectively. The P value was calculated using log-rank test. (C-D) Simon-Makuch curves showing the impact of allo-HSCT on OS and EFS of patients with SM-t(8;21) AML, respectively. (E-F) Simon-Makuch curves showing the impact of allo-HSCT on OS and EFS of patients with KITD816 t(8;21) AML, respectively. (G-H) Simon-Makuch curves showing the impact of allo-HSCT on OS and EFS of patients with non-D816 KITpos t(8;21) AML, respectively. The P value was calculated using Mantel-Byar test. ns, not significant.
Survival curves of patients with KITD816 t(8;21) AML, non-D816 KITpos t(8;21) AML, and SM-t(8;21) AML. (A-B) Kaplan-Meier curves of OS and EFS of patients with KITD816 t(8;21) AML, non-D816 KITpos t(8;21) AML, and SM-t(8;21) AML, respectively. The P value was calculated using log-rank test. (C-D) Simon-Makuch curves showing the impact of allo-HSCT on OS and EFS of patients with SM-t(8;21) AML, respectively. (E-F) Simon-Makuch curves showing the impact of allo-HSCT on OS and EFS of patients with KITD816 t(8;21) AML, respectively. (G-H) Simon-Makuch curves showing the impact of allo-HSCT on OS and EFS of patients with non-D816 KITpos t(8;21) AML, respectively. The P value was calculated using Mantel-Byar test. ns, not significant.
For all patients in the cohort, risk factors at diagnosis were included in the univariate and multivariate COX analysis (supplemental Table 3). Age >60 years (OS: hazard ratio [HR], 3.71 [95% CI, 1.20-11.54]; P = .023; EFS: HR, 7.45 [95% CI, 2.44-22.79]; P<.001), KITD816 mutation (OS: HR, 3.02 [95% CI, 1.46-6.26]; P = .003; EFS: HR, 3.20 [95% CI, 1.45-7.06], P = .004) and BCOR mutation (OS: HR, 8.18 [95% CI, 1.02-65.73]; P = .048; EFS: HR, 29.92 [95% CI, 2.15-415.70]; P = .011) were independent risk factors. Although univariate analysis identified concomitant SM as significant risk factor for OS (HR, 2.27 [95% CI, 1.02-5.05]; P = .044), it was not an independent predictor (HR, 1.39 [95% CI, 0.52-3.72]; P = .518) in multivariate analysis.
Our results were consistent with a small cohort study from South Korea,13 which found that patients with SM-t(8;21) AML (n = 4) shared similar clinical characteristics with patients with t(8;21) AML (n = 19). Besides, KIT D816V frequency, response rate after induction and OS of patients with SM-t(8;21) AML reported by another case-series study from South Korea were all close to our data.12 Furthermore, our results demonstrated that, compared to those with non-D816 KITpos t(8;21) AML, patients with SM-t(8;21) AML or KITD816 t(8;21) AML had more dismal prognosis. Notably, concomitant SM failed to be identified as independent risk factor for OS by multivariate analysis. This finding might be explained by a high correlation between SM and KITD816 mutation or result from a small number of SM cases. In addition, our study showed that patients with SM-t(8;21) AML, rather than those with KITpos t(8;21) AML, could get survival benefit from allo-HSCT, which favored the previous opinion that allo-HSCT was not the first treatment choice for patients with t(8;21) AML, whether they had KITD816 mutations or not.14-16 Although the role of allo-HSCT is still undetermined for SM, our findings favored a consensus opinion that patients with SM-AHN should receive allo-HSCT when the AHN component warrants HSCT for treatment.17
In our cohort, SM-t(8;21) AML showed a mutation profile similar to that of KITpos t(8;21) AML, which was also close to previous studies.18,19 Meanwhile, we had some interesting findings. First, patients with SM-t(8;21) AML had higher VAF in KIT mutations or KIT exon17 mutations than those with KITpos t(8;21) AML, which might result from additional KIT mutations brought by neoplastic MCs. Second, DNMT3A mutations were more common in SM-t(8;21) AML than in KITpos t(8;21) AML, which favored the opinion that epigenetic changes might play an important role in pathogenesis of mast cell neoplasms.20,21 Last, the incidence of S/A/R mutations in SM-t(8;21) AML was significantly lower than previously reported in KITD816mut CBF-negative SM-AML and other advanced SM subtypes but close to the frequency in t(8;21) AML, which indicated that the molecular features of patients with SM-AHN may be mainly determined by the AHN component.22-24
In summary, our results revealed that SM-t(8;21) AML shared many similarities both in clinical characteristics and molecular features, which created obstacles to distinguish 2 diseases. Compared with non-D816 KITpos t(8;21) AML, SM-t(8;21) AML and KITD816 t(8;21) AML were high-risk subtypes with refractory or relapsed trend and poor prognosis. Allo-HSCT was shown to be an effective method to improve survival outcomes of SM-t(8;21) AML but invalid for the survival of KITpos t(8;21) AML. Our research is based on a retrospective cohort from multiple centers from China, which is limited by shortcomings of the study method; more characteristics of SM-t(8;21) need to be further investigated in a large prospective study.
Acknowledgments: The re-evaluation of BM morphology and histopathology was performed by Feng Zhu and Qian Wang, respectively.
This study was financially supported by grants from the National Key R&D Program of China (2019YFA0111004, 2022YFC2502701), the National Natural Science Foundation of China (82170158, 81970142, 82100175), the Translational Research Grant of NCRCH (2021WSB01, 2020WSB03, 2020WSB11, 2020WSB13), and the Open Project of Jiangsu Biobank of Clinical Resources (SBK202003001, SBK202003003).
Contribution: Z.Z., J.Y., G.L., and X.B. were involved in analysis and interpretation of the data; G.L., M.H., Z.L., Y.Y., R.M., Y.Z., P.S., W.Z., Q.J., H.C., P.X., W.Y., Y.H., Y.W., Y.Z., and D.W. contributed to the collection and interpretation of the data; X.Y. and S.C. conceived the study and participated in its design and coordination; Z.Z. and J.Y. drafted the manuscript; X.Y. and S.C. revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Suning Chen, National Clinical Research Center for Hematologic Diseases, Jiangsu Institute of Hematology, First Affiliated Hospital of Soochow University, Institute of Blood and Marrow Transplantation, Collaborative Innovation Center of Hematology, Soochow University, Suzhou 215006, China; email: chensuning@suda.edu.cn; and Xiaofei Yang, National Clinical Research Center for Hematologic Diseases, Jiangsu Institute of Hematology, First Affiliated Hospital of Soochow University, Institute of Blood and Marrow Transplantation, Collaborative Innovation Center of Hematology, Soochow University, Suzhou 215006, China; email: yangxiaofei1977@163.com.
References
Author notes
Z.Z., J.Y., G.L., and X.B. contributed equally to this work.
The datasets used and/or analyzed during this study are available on reasonable request from the corresponding authors, Suning Chen (chensuning@suda.edu.cn) and Xiaofei Yang (yangxiaofei1977@163.com).
The full-text version of this article contains a data supplement.