Key Points
Thromboinflammatory marker H3Cit-DNA is enhanced in STEMI.
It is associated with clinical outcome and identifies patients at high risk of MACE.
Visual Abstract
Platelets are crucial in thrombus formation during ST-elevation myocardial infarction (STEMI). In addition, they also play an important role in postischemic thromboinflammation, which is determined by the interplay between activated platelets and neutrophils. The latter form neutrophil extracellular traps, which are detectable in plasma as citrullinated histone H3–deoxyribonucleic acid-DNA complexes. Prediction of the risk of recurrent events is important in precision medicine. Therefore, we investigated whether circulating thromboinflammatory markers predict clinical outcome after STEMI. We performed a prospective, multicentric, observational, all-comer study of patients with STEMI (n = 361). Thromboinflammation, measured as H3Cit-DNA complexes, was assessed on day 1 after presentation with STEMI as well as 5 days and 6 months after STEMI by enzyme-linked immunosorbent assay. Twelve months of clinical follow-up was conducted. Multivariate analysis was performed investigating which variables were independently associated with major adverse cardiac events (MACEs). Patients were aged 64 ± 12 years; 80% were male; and 40% had diabetes mellitus. Thromboinflammation was enhanced during index hospitalization compared with 6-months follow-up (137.4 ± 100.0 μg/L vs 53.7 ± 54.7 μg/L; P < .001). Additionally, patients within the highest tertile of thromboinflammation at day 1 after STEMI showed worse outcome during follow-up (hazard ratio, 2.57; 95% confidence interval, 1.72-3.85; P < .001). Receiver operating characteristic analysis revealed a cutoff value of 219.3 μg/L. In multivariate logistic regression analysis, thromboinflammation was independently associated with outcome after STEMI. To sum it up, thromboinflammation is enhanced in STEMI. It identifies patients at high risk of MACE. Therefore, thromboinflammation might be a promising target and marker in precision medicine. The trial was registered at www.clinicaltrials.gov as #NCT03539133.
Introduction
Platelet driven thrombus formation in the coronary artery is the origin of a ST-elevation myocardial infarction (STEMI).1,2 Therefore, antiplatelet therapy is crucial in acute treatment and secondary prevention of patients with STEMI.3 Ideal duration and antiplatelet regime are still a matter of discussion.4 Clinical benefit by reduction of ischemic events by more potent regimens or extended therapy is jeopardized by an enhanced risk of bleeding.5,6 Therefore, the identification of patients at high risk of recurrent ischemic events is of great importance in precision medicine.
In recent years, it has been shown that platelets orchestrate a deleterious liaison between coagulation and immune cells called thromboinflammation in acute and postischemic setting during myocardial infartion.7-10 This dysregulated interplay cumulates in the formation of neutrophil extracellular traps (NETs).11 NETs are web-like prothrombogenic structures that are rapidly released from neutrophils after activation12 and can be initiated by platelets through direct cell-cell contacts or secreted soluble mediators.13 Additionally, there have been some studies showing that antiplatelet therapy can attenuate the formation of NETs (NETosis) improving outcome in other noncardiovascular diseases.14,15 In case of a STEMI, NETosis leads to excess thrombin generation, tissue factor release, and enhanced platelet activation,11 and all of these factors are associated with impaired outcome.16,17 Enhanced thrombin generation after STEMI initiates apoptosis in cardiomyocytes and promotes cardiac fibroblast proliferation.18 Increased release of tissue factor reflects enhanced intravascular procoagulant activity,19 and enhanced platelet activation showed various negative effects on outcome after STEMI, including increased fibrosis20 and an elevated extent of microvascular obstruction.21
Hence, it seems reasonable that the extent of thromboinflammation associates with clinical outcome of patients with STEMI.
Methods
Study population
This is an explorative, prospective, observational, multicentric, cohort study of all-comer patients with STEMI. Inclusion criteria was informed consent. Patients aged <18 years were excluded. We included patients between January 2020 and December 2022 with a follow-up of 12 months. Major adverse cardiac events (MACEs) are defined as death, reinfarction, and rehospitalization due to heart failure. Ethics approval was given by the local ethics committees (Heinrich-Heine-University, Düsseldorf, Germany; and Landesärztekammer Nordrhein), complying with the Declaration of Helsinki under reference numbers 5961R and 2021.055 (trial registration number: NCT03539133).
Blood sampling
Blood was drawn 24 hours after presentation with STEMI into 2.7-mL, 0.1-M sodium citrate tubes (BD Vacutainer; Becton Dickinson). The samples were immediately centrifuged at 500g for 10 minutes to obtain platelet-rich plasma. Platelet-rich plasma was centrifuged at 1700g for 20 minutes to gain platelet-poor plasma. Additional blood samples were taken during routine examinations 5 days after STEMI and 6 months later.
Determination H3Cit-DNA complexes
Determination of citrullinated histone H3–deoxyribonucleic acid DNA complexes as a marker for NET formation in thromboinflammation was performed as previously described and validated.22 Briefly, anti-Histone H3 (citrulline R8) antibody (abR8Cit-1c; Abcam) was coated at a concentration of 5 μg/mL overnight and in the dark at 4°C on clear high bind 96-well microplates (Thermo Fisher Scientific). After that, microplates were washed 3 times with phosphate-buffered saline with 0.05 % (volume/volume) Tween-20 and blocked with phosphate-buffered saline supplemented with 1% (mass/volume) bovine serum albumin for 2 hours at room temperature. Next, microplates were washed 3 times, and 20-μL plasma was added with 80 μL of detection antibody anti-DNA-peroxidase (Cell Death Detection enzyme-linked immunosorbent assay [ELISA] PLUS kit; Roche) and incubated for 2 hours at room temperature on a 200-rounds per minute shaker. After 3 final washing steps, 100-μL horseradish peroxidase substrate (Thermo Fisher Scientific) was added to each well and incubated in the dark for 10 to 15 minutes. Optical density was measured at 650 nm using a multimode plate reader (FLUOstar omega; BMG Labtech). Different concentrations of recombinant mononucleosomes (H3Cit2,8,17; EpiCypher) diluted in standard diluent (50 mmol/L Tris-HCl pH 7.5; 300 mmol/L NaCl; 0.01 % [w/v] bovine serum albumin; and 0.01 % [v/v] Tween-20) served as calibration controls. H3Cit-DNA complexes concentrations were assessed according to the established standard curves.
Statistics
Statistical analyses were conducted with IBM SPSS Software (New York, NY) and version 3.8 of the programming language Python (Centrum voor Wiskunde en Informatica, Amsterdam, The Netherlands). The normality of distribution was tested with Shapiro-Wilk test. Normally distributed variables were analyzed with a t test, and nonnormally distributed variables were analyzed using Mann-Whitney U test. Fisher exact test was used for dichotomous variables, and χ2 test was used for categorical variables. For the multivariate analysis, the odds ratios, P values, and confidence intervals were derived from a logistic regression model from Python’s library statsmodels. The P values were considered significant if P values < .05.
The survival curves and MACE curves were created fitting a Cox’s Proportional Hazard model from the Python package lifelines. To find the optimal threshold, a receiver operating characteristic (ROC) analysis including calculation of the area under the curve and the Youden index were done using the module Metrics from Python’s library scikit-learn.
Results
Patient characteristics
We enrolled patients presenting with an acute STEMI, and sample recruitment was done within 24 hours, after 5 days, and after 6 months. In our study, 80% of patients were men, and the mean age was 64 ± 12 years (Table 1). Average body mass index was 27.0 ± 4.2 kg/m2, and the prevalence of comorbidities were typical for such cohorts. Approximately 64 % had arterial hypertension, and ∼40% had diabetes mellitus. Blood parameter corresponded to the usual risk factors for STEMI. For example, total cholesterol was 143.86 ± 36.99 mg/dL, whereas high-density lipoprotein was 38.24 ± 11.59 mg/dL, and low-density lipoprotein was 84.72 ± 32.46 mg/dL.
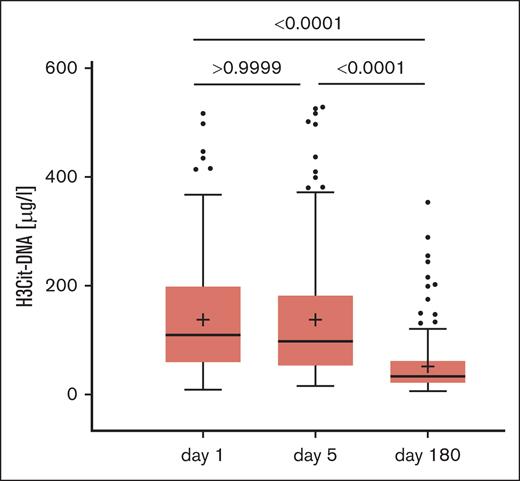
Kinetic of H3Cit-DNA
Besides blood sampling 24 hours after STEMI (day 1), additional blood samples were taken during routine examinations 5 days and 6 months (day 180) after STEMI. Paired analysis of H3Cit-DNA complexes between day 1 and day 5 (137.4 ± 100.0 μg/L vs 137.9 ± 113.5 μg/L; P > .9999) revealed no difference, suggesting a stable thromboinflammatory state within the first days after STEMI (Figure 1). In contrast, the H3Cit-DNA level after 6 months was only just above the detection limit of the assay and, with 53.7 ± 54.7 μg/L, significantly lower than that on day 1 and day 5 after STEMI (Figure 1).
Kinetics of thromboinflammatory marker H3Cit-DNA after STEMI (Kruskal-Wallis test with post hoc Dunn multiple comparison test, Tukey whiskers).
Kinetics of thromboinflammatory marker H3Cit-DNA after STEMI (Kruskal-Wallis test with post hoc Dunn multiple comparison test, Tukey whiskers).
H3Cit-DNA level predicts survival and occurrence of MACE after STEMI
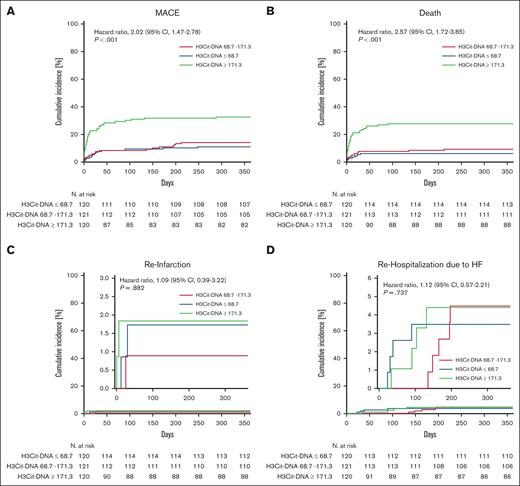
Given that H3Cit-DNA values were stable within 5 days after STEMI, we tested whether H3Cit-DNA level at day 1 (ie, hospital admission) associate with cardiovascular (CV) end points. For this, we divided our cohort of 361 patients into 3 tertiles according to their H3Cit-DNA levels. First, we questioned whether H3Cit-DNA levels are associated with outcome over 1 year. The risk of MACE was significantly increased in the tertile with the highest values of H3Cit-DNA >171.3 μg/L (tertile 3 = T3) compared with the lowest (T1; H3Cit-DNA <68.7 μg/L) and medium (T2) values (Figure 2A; T1 = 10.8 %; T2 = 13.2 %; T3 = 31.7 %; hazard ratio, 2.02; 95% confidence interval, 1.47-2.78). This was primarily driven by an increased mortality rate, which was twice as high in patients with H3Cit-DNA levels >171.3 μg/L compared with patients with low (T1) and medium (T2) levels (Figure 2B; T1 = 5.8%; T2 = 8.3%; T3 = 26.7%; hazard ratio, 2.57; 95% confidence interval, 1.72-3.85). This effect is primarily determined by the significantly increased mortality in the first 30 days. In contrast, only a minor reduction in mortality was found between the low and middle tertiles. However, other end points of occurrence of reinfarction (Figure 2C) and rehospitalization due to heart failure (Figure 2D) did not differ between the groups. Because ∼30% of our cohort had preexisting antiplatelet therapy and these antiplatelet agents might have an impact on thromboinflammation during STEMI, we compared patients with and without preexisting antiplatelet therapy. However, we did not find any difference in H3Cit-DNA level between the 2 groups, suggesting no relevant impact of antiplatelet agents on our results (supplemental Figure 1).
Time-to-event curves for each tertile of H3Cit-DNA. (A) Patients in the tertile with highest H3Cit-DNA plasma values (green line, T3) showed increased occurrence of MACE compared with patients in medium (red line, T2) and low tertile (blue line, T1). (B) Patients in the tertile with highest H3Cit-DNA plasma values did also show an increased mortality within 1 year after STEMI. (C-D) No significant difference was observed between the groups regarding reinfarction (C) and rehospitalization (D) due to heart failure. Numbers under the y-axis indicate patients at risk against the days listed on x-axis.
Time-to-event curves for each tertile of H3Cit-DNA. (A) Patients in the tertile with highest H3Cit-DNA plasma values (green line, T3) showed increased occurrence of MACE compared with patients in medium (red line, T2) and low tertile (blue line, T1). (B) Patients in the tertile with highest H3Cit-DNA plasma values did also show an increased mortality within 1 year after STEMI. (C-D) No significant difference was observed between the groups regarding reinfarction (C) and rehospitalization (D) due to heart failure. Numbers under the y-axis indicate patients at risk against the days listed on x-axis.
H3Cit-DNA level is a specific prognostic marker
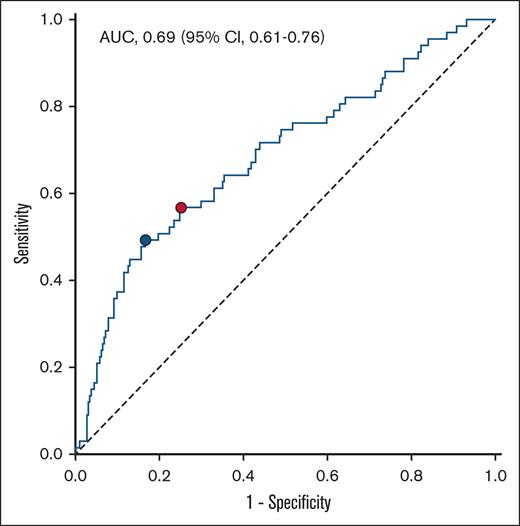
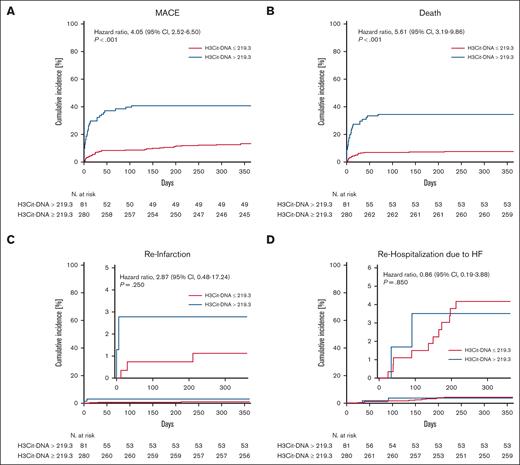
ROC analysis was conducted to evaluate the predictive value for 1-year occurrence of MACE (Figure 3). The area under the curve of H3Cit-DNA was 0.69, which showed a moderate diagnostic quality.23 According to Youden index,24 the ideal threshold was set at 219.3 μg/L, and the cohort was divided into 2 groups (Table 2). After dividing the cohort according to the cutoff value of 219.3 μg/L, there was a clear difference in the occurrence of MACE between the groups. The risk of MACE was increased by a factor of >3 in the group with H3Cit-DNA values >219.3 μg/l (Figure 4A; 39.5% vs 12.5%). Additionally, patients above an H3Cit-DNA level of 219.3 μg/L showed a 4.6-fold increase in mortality within 1 year after STEMI (Figure 4B; 34.6% vs 7.5%). This effect was also mainly due to the increased mortality within the first 30 days. The increase in reinfarctions (Figure 4C) and rehospitalization due to heart failure was just numerical and not statistically significant.
ROC curve for the prediction of MACE within 1 year after STEMI. Area under the curve (AUC) of H3Cit-DNA plasma level was 0.69. Youden index revealed H3Cit-DNA level of 219.3 μg/L as the ideal threshold (blue dot). Corresponding values for sensitivity and specificity are 0.493 and 0.833, respectively. Threshold at the highest third of H3Cit-DNA plasma level showed sensitivity and specificity values of 0.567 and 0.748, respectively (red dot).
ROC curve for the prediction of MACE within 1 year after STEMI. Area under the curve (AUC) of H3Cit-DNA plasma level was 0.69. Youden index revealed H3Cit-DNA level of 219.3 μg/L as the ideal threshold (blue dot). Corresponding values for sensitivity and specificity are 0.493 and 0.833, respectively. Threshold at the highest third of H3Cit-DNA plasma level showed sensitivity and specificity values of 0.567 and 0.748, respectively (red dot).
Time-to-event curves after dividing the cohort corresponding to threshold determined in ROC analysis. (A) Patients with H3Cit-DNA plasma >219.3 μg/L (blue line) showed increased mortality compared with patients with plasma H3Cit-DNA <219.3 μg/L (red line). (B) Patients with H3Cit-DNA plasma >219.3 μg/L did also show an increased risk of MACE within 1 year after STEMI. (C-D) No difference was observed between the groups regarding reinfarction (C) and rehospitalization (D) due to heart failure. Numbers under y-axis indicate patients at risk against the days listed on x-axis.
Time-to-event curves after dividing the cohort corresponding to threshold determined in ROC analysis. (A) Patients with H3Cit-DNA plasma >219.3 μg/L (blue line) showed increased mortality compared with patients with plasma H3Cit-DNA <219.3 μg/L (red line). (B) Patients with H3Cit-DNA plasma >219.3 μg/L did also show an increased risk of MACE within 1 year after STEMI. (C-D) No difference was observed between the groups regarding reinfarction (C) and rehospitalization (D) due to heart failure. Numbers under y-axis indicate patients at risk against the days listed on x-axis.
Thromboinflammatory marker H3Cit-DNA is an independent predictor for outcome after STEMI
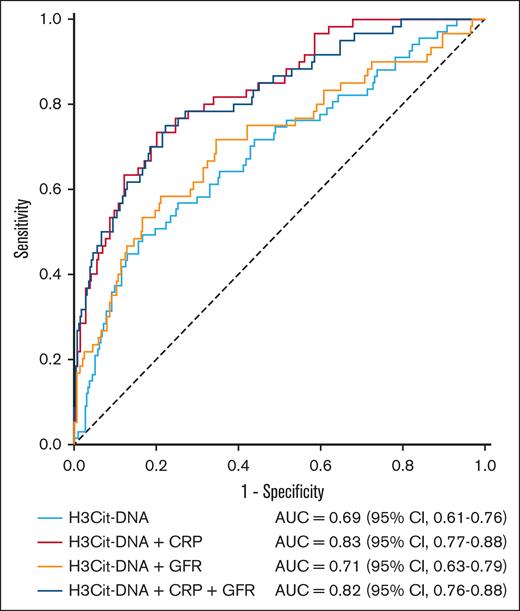
To further elucidate the predictive power of thromboinflammatory marker H3Cit-DNA, we conducted logistic multivariate regression analysis. We first compared the groups from our ROC analysis (Table 2) to find parameters that might have had an impact on outcome after STEMI in our cohort. We then performed univariate analyses of demographic parameters (sex, age, and body mass index) and (almost) significant prognostic parameters (diabetes mellitus, smoker, total cholesterol, high-density lipoprotein, low-density lipoprotein, glomerular filtration rate (GFR), C-reactive protein (CRP), and troponin T). These univariate analyses revealed significant results for several parameters in their power to predict MACE (Table 3). As a next step, we performed a logistic multivariate analysis with all parameters that reached a P value < .1 in univariate analyses. In this process, thromboinflammatory marker H3Cit-DNA was confirmed to have independent predictive power, whereas the other parameters, with the exception of CRP, could not reach statistical significance (Table 3). For this reason, we performed further ROC analyses to investigate the predictive performance by combining the H3Cit-DNA level with other biomarkers, such as CRP and GFR (Figure 5). This showed that a combination of H3Cit-DNA and CRP further improved the predictive power of thromboinflammation. The addition of GFR as a third marker did not lead to any further improvement.
ROC curve for the prediction of MACE within 1 year after STEMI, combination of thromboinflammation with CRP and GFR. AUC of H3Cit-DNA plasma level alone was 0.69. The combination of H3Cit-DNA level to CRP and GFR improved AUC and, therefore, the predictive power for MACE.
ROC curve for the prediction of MACE within 1 year after STEMI, combination of thromboinflammation with CRP and GFR. AUC of H3Cit-DNA plasma level alone was 0.69. The combination of H3Cit-DNA level to CRP and GFR improved AUC and, therefore, the predictive power for MACE.
Discussion
The main findings of this study are as follows: (1) thromboinflammation measured by H3Cit-DNA is enhanced in STEMI; and (2) it independently predicts outcome after STEMI. Of note, H3Cit-DNA associated with early mortality (within 30 days), reflecting the extent of thromboinflammation in the acute setting.
It is well-known that inflammatory processes orchestrate infarct size and outcome after STEMI.25 Inflammatory cells such as neutrophils can already be found in myocardial tissue a few hours after infarction.26 They do not only contribute to wound healing but are also able to form NETs. NETs are web-like structures that consist of decondensed chromatin coated with histones, proteases, and granular proteins, which are rapidly released from neutrophils after activation.27 They were first found to trap and disarm pathogens as a part of innate immune system.12,28 However, the impact of NETs goes far beyond defense against pathogens, because they contribute to noninfectious inflammation as well. Recently, the pivotal role of NETosis in sterile inflammation during ischemia was revealed.29 Additionally, it was demonstrated that they accumulate in coronary thrombi, causing the ischemic event.30
The systemic thromboinflammatory milieu can be detected by several plasma markers including NETosis.31 Double-stranded DNA (dsDNA) is a key component of NETs and was supposed to be a sensitive biomarker for CV risk. Therefore, it was used in several studies analyzing the association between thromboinflammation and outcome after CV events. Indeed, dsDNA was increased after STEMI,32 and first outcome studies revealed a correlation between dsDNA in plasma and outcome after STEMI.33,34 However, the authors of these studies themselves point out several disadvantages that hamper interpretation of these data. Therefore, circulating dsDNA as a marker for thromboinflammation might not be specific enough. Another marker regularly used to determine thromboinflammation in human plasma are myeloperoxidase-DNA (MPO-DNA) complexes, which are released in NETosis. Although several studies used circulating MPO-DNA complexes to find an association with CV outcome,35,36 the quantification of MPO-DNA complexes is not yet very standardized, which makes interpretation difficult. At least the studies that used a commercial ELISA kit for the detection of MPO-DNA complexes do not seem to be ideal, because a recent study revealed commercial ELISA kits as “highly questionable regarding specificity of NET detection.”37 Taken together, the optimal method of detection has not been identified yet.
For this reason, we investigated H3Cit-DNA as a well-characterized marker of NET formation during thromboinflammation using a validated and robust protocol.22 This assay carries several advantages. First, the specificity to detected NET is increased because the assays detect only CitH3 bound DNA, excluding other sources of DNA and CitH3 in blood. Second, H3Cit-DNA complexes are stable to allow for a reliable accurate quantification of thromboinflammatory response.38 Third, ELISA standardization (ie, standard curves), required to reliable determine absolute values, is highly reproducible by using artificially designed and commercially available nucleosomes. Fourth, we analyzed plasma samples, which are significantly more suitable than serum samples.39
Of note, a recent study revealed the feasibility of using a multiomic factor analysis to predict CV outcomes in patients with acute coronary syndrome.40 However, such analysis involving single-cell sequencing technology are elaborative and resource demanding and remain far away from a routinely and widely diagnostics.
The scientific view on thromboinflammation and NETosis was often very focused on neutrophils and the effects of NETs. However, activated platelets were shown to contribute to NETosis.41 Thereby, it does not matter which of the classical stimuli activates the platelets, because all stimuli (thrombin, adenosine diphosphate, collagen, and arachidonic acid) were shown to trigger platelet-driven NETosis.41 We found that H3Cit-DNA complexes as a marker of thromboinflammation after STEMI predicts outcome. This was independent of comorbidities or demographic characteristics.
However, thromboinflammation might be directly targetable as well. Currently used antiplatelet drugs14,42-44 and novel strategies such as glycoprotein VI inhibition or interleukin-6 receptor inhibition were shown to reduce thromboinflammation.45,46 In our cohort, antiplatelet therapy had no impact on H3Cit-DNA levels. However, our study was not designed and powered to answer this question. The optimal antithrombotic regimen after STEMI is still under discussion. The risk of recurrent ischemic events is weighted against the risk of bleeding using different scores.47
In summary, we introduced a novel, robust, and easily accessible marker that associated with enhanced risk of MACE and mortality in a high-risk STEMI cohort. The combination of thromboinflammation with CRP could further improve predictive power. Therefore, in light of precision medicine, thromboinflammation might be very promising in guiding the optimal antiplatelet regimen after STEMI.
Limitations
This study has several limitations. It is exploratory; therefore, sample size is limited, and patients are not randomized to a treatment. Hence, the results might be biased. However, multivariate analyses were conducted to reduce this risk.
Conclusion
This study demonstrates that H3Cit-DNA is a reliable marker to determine the thromboinflammatory state in STEMI. It allows for the identification of patients at high risk of mortality. It might be a promising marker and target in precision medicine.
Acknowledgments
The authors acknowledge the support of the Susanne-Bunnenberg-Stiftung at the Düsseldorf Heart Center.
This work was supported by Deutsche Forschungsgemeinschaft (German Research Foundation) grants 530690968 (M.B.), 493400536, 413659045, and 510844896 (A.P.), and PE2704/3-1, PE2704/4-1, and PE2704/5-1 (T.P.). This study was also supported by German Heart Foundation grant F/32/22 (A.P.) and Medical Faculty, Heinrich-Heine-University grant 18-2019 (A.P.).
Authorship
Contribution: M.B., K.A., T.P., and A.P. designed the study, analyzed the data, and drafted the manuscript; V.E. and J.D. conducted the measurements of thromboinflammation with the help of J.M.H. and D.A.D.; D.M. aided in statistical analysis; together with M.B. and A.P., C.J., M.K., and T.P. revised the manuscript and discussed all data; and all authors critically reviewed and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Amin Polzin, Klinik für Kardiologie, Pneumologie und Angiologie, Universitätsklinikum Düsseldorf, Moorenstraße 5, 40225 Düsseldorf, Germany; email: amin.polzin@med.uni-duesseldorf.de.
References
Author notes
M.B., K.A., V.E., T.P., and A.P. contributed equally to this study.
Data are available upon request from the corresponding author, Amin Polzin (amin.polzin@med.uni-duesseldorf.de).
The full-text version of this article contains a data supplement.