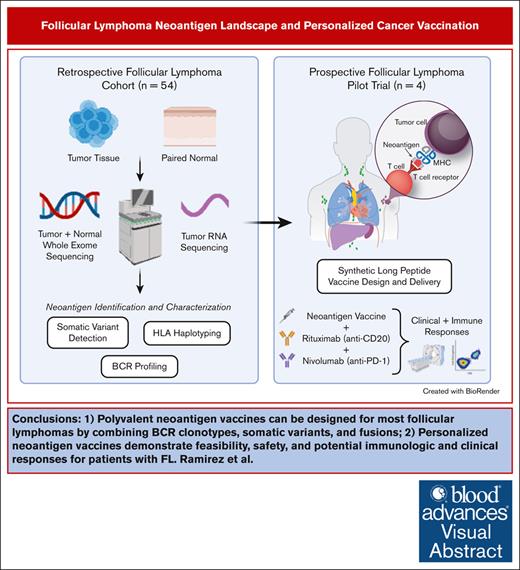
Key Points
Polyvalent neoantigen vaccines can be designed for most FLs by combining BCR clonotypes, somatic variants, and fusions.
Personalized neoantigen vaccines demonstrate feasibility, safety, and potential immunologic and clinical responses for patients with FL.
Visual Abstract
Personalized cancer vaccines designed to target neoantigens represent a promising new treatment paradigm in oncology. In contrast to classical idiotype vaccines, we hypothesized that “polyvalent” vaccines could be engineered for the personalized treatment of follicular lymphoma (FL) using neoantigen discovery by combined whole-exome sequencing (WES) and RNA sequencing (RNA-seq). Fifty-eight tumor samples from 57 patients with FL underwent WES and RNA-seq. Somatic and B-cell clonotype neoantigens were predicted and filtered to identify high-quality neoantigens. B-cell clonality was determined by the alignment of B-cell receptor (BCR) CDR3 regions from RNA-seq data, grouping at the protein level, and comparison with the BCR repertoire from healthy individuals using RNA-seq data. An average of 52 somatic mutations per patient (range, 2-172) were identified, and ≥2 (median, 15) high-quality neoantigens were predicted for 56 of 58 FL samples. The predicted neoantigen peptides were composed of missense mutations (77%), indels (9%), gene fusions (3%), and BCR sequences (11%). Building off of these preclinical analyses, we initiated a pilot clinical trial using personalized neoantigen vaccination combined with PD-1 blockade in patients with relapsed or refractory FL (#NCT03121677). Synthetic long peptide vaccines targeting predicted high-quality neoantigens were successfully synthesized for and administered to all 4 patients enrolled. Initial results demonstrate feasibility, safety, and potential immunologic and clinical responses. Our study suggests that a genomics-driven personalized cancer vaccine strategy is feasible for patients with FL, and this may overcome prior challenges in the field. This trial was registered at www.ClinicalTrials.gov as #NCT03121677.
Introduction
Follicular lymphoma (FL) is the most common indolent non-Hodgkin lymphoma1,2 and remains predominantly incurable with conventional therapies.3 The clinical course of patients with FL is heterogenous; some patients experience minimal annual progression over decades,4 whereas others rapidly progress or transform into aggressive diffuse large B-cell lymphoma. In an attempt to identify less toxic alternatives to traditional chemotherapy-based approaches, the anti-CD20 monoclonal antibody (mAb) rituximab was approved as the first immunotherapy for the treatment of FL and demonstrated improvements in progression-free and overall survival in clinical trials.5-7 Even with the combination of rituximab plus chemotherapy, ∼35% to 45% of patients with advanced disease relapse within 5 years of initial treatment.8 Although more recent advances include the use of the immunomodulatory agent lenalidomide, T-cell–engaging bispecific antibodies, various targeted small molecule inhibitors, and chimeric antigen receptor T cells, there remains a need to develop novel therapies to optimally balance response rates, duration of treatment and responses, as well as short- and long-term toxicities.9,10
Personalized cancer vaccines stimulate a patient’s own immune system to specifically attack cancer cells. Such vaccines typically target neoantigens, which are short, mutated peptide sequences uniquely expressed by tumor cells. Major histocompatibility complex (MHC) class I and/or II molecules have demonstrated the ability to present neoantigens to cytotoxic and/or helper T cells for recognition and antitumor immune responses.11-14 Reports of neoantigen vaccination showed promise in various solid cancers,15-31 and CD8+ T-cell responses were identified against 10 driver mutations in a retrospective cohort of patients with FL.32 Prior efforts to develop cancer vaccines for FL focused on targeting only the single dominant B-cell receptor (BCR) idiotype, inducing an anti-idiotype antibody response and variable T-cell responses. Furthermore, they were not administered in conjunction with potentially synergistic immune checkpoint inhibitors, which may have contributed to the negative results from previous phase 3 clinical trials.33-35 We hypothesized that a polyvalent approach that collectively identifies multiple cancer vaccine targets (ie, BCR clonotypes, small somatic mutations, and gene fusions) and uses long peptides to overcome potential immune tolerances could aid in personalized immunotherapy for patients with FL.
Defining the feasibility of neoantigen vaccines, optimizing vaccine design informed by next-generation sequencing, and identifying factors that contribute to the success of personalized cancer vaccines are important areas of inquiry.36,37 FL typically has a low-to-medium mutation burden,38,39 and in a report that evaluated 117 FL tumors,40 the median number of nonsilent somatic variants per individual was 55 (range, 2-169). However, the proportion of somatic FL-specific mutations that create high-quality predicted neoantigens is not well described. Therefore, given the modest correlation between mutation burden and immunotherapy response, and that even low numbers of highly immunogenic neoantigens can precipitate a detectable antitumor immune response, testing for neoantigens within FL is supported.32 Here, we report on neoantigen vaccine design in FL using a comprehensive genomic approach (Figure 1). We applied whole-exome sequencing (WES) and RNA sequencing (RNA-seq) to a retrospective cohort of 54 samples from 53 patients with FL and then performed neoantigen prediction and in silico vaccine design. This strategy was then applied prospectively to 4 patients with relapsed or refractory (R/R) FL in a pilot clinical trial to assess the safety and efficacy of treatment with personalized neoantigen vaccination combined with programmed cell death protein 1 (PD-1) blockade (www.ClinicalTrials.gov identifier #NCT03121677).
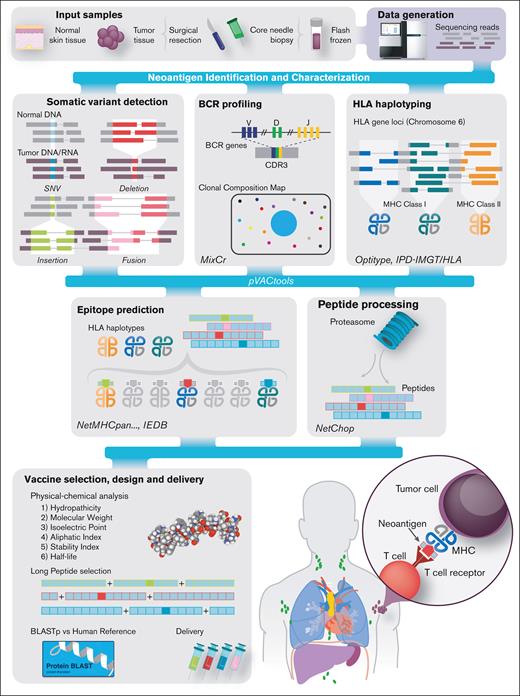
Overview of the FL personalized cancer vaccine pipeline. Patient samples are acquired and then sequenced (top left). Somatic variants of various types, including SNVs (blue), deletions (red), insertions (green), and fusions (pink), are predicted. Sequence data are analyzed to determine HLA types and B-cell clonotypes for each patient. Variant and clonal B-cell peptide sequences are inferred from variants and analyzed with respect to their predicted expression, proteasome processing, and ability to bind the patient’s MHC class I complexes. Candidates are then selected for vaccine design, and additional analyses are performed to assess manufacturability. Bioinformatic tools used for each step are indicated in italics. CDR3, complementarity-determining region 3; IEDB, Immune Epitope Database. Adapted, per CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/), from Richters et al.37
Overview of the FL personalized cancer vaccine pipeline. Patient samples are acquired and then sequenced (top left). Somatic variants of various types, including SNVs (blue), deletions (red), insertions (green), and fusions (pink), are predicted. Sequence data are analyzed to determine HLA types and B-cell clonotypes for each patient. Variant and clonal B-cell peptide sequences are inferred from variants and analyzed with respect to their predicted expression, proteasome processing, and ability to bind the patient’s MHC class I complexes. Candidates are then selected for vaccine design, and additional analyses are performed to assess manufacturability. Bioinformatic tools used for each step are indicated in italics. CDR3, complementarity-determining region 3; IEDB, Immune Epitope Database. Adapted, per CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/), from Richters et al.37
Methods
Fresh-frozen tumor samples with paired nonmalignant tissue (skin or blood) were retrospectively collected. All patients provided written informed consent for the use of their samples in sequencing, and all clinical characteristics are summarized in supplemental Table 1. Samples with known FL status underwent WES and RNA-seq. Somatic and B-cell clonotype neoantigens were predicted and filtered to identify high-quality neoantigens. B-cell clonality was determined by alignment of BCR CDR3 regions from RNA-seq data, grouping at the protein level, and comparison with the BCR repertoire of RNA-seq data from healthy individuals. The analysis pipeline considered all small somatic mutations (single nucleotide variants [SNVs] and indels), gene fusions, and BCR dominant clonotypes that passed the filtering criteria and manual inspection as neoantigen candidates. Neoantigen candidates were considered for both clonal and subclonal populations. Subsequently, a pilot clinical trial was initiated using personalized neoantigen vaccination combined with PD-1 blockade in patients with relapsed FL (#NCT03121677). Pretreatment biopsies were used to identify multiple unique high-quality tumor-specific neoantigens (12-19 neoantigens per patient) for all enrolled patients. Synthetic long peptide (SLP) vaccines were successfully synthesized for all 4 patients, and for each patient, ∼20 peptides (including alternate registers for the same neoantigen, as needed) were pooled into 4 groups of 5 to minimize competition for the same MHC molecule. Each of the 4 pools were administered to patients via subcutaneous injection into different limbs with concurrent IV nivolumab administration. See supplemental Methods for additional details on patient characteristics/sample acquisition, library preparation, sequencing, retrospective neoantigen analysis, and pilot trial neoantigen analysis including vaccine candidate design, pooling strategy, and manufacturing.
All samples were collected within protocols approved by the Washington University School of Medicine (WUSM) institutional review board.
Results
WES/transcriptome sequencing reveals the diverse mutational landscape of FL
To predict high-quality neoantigen candidates for FL personalized cancer vaccines, we performed WES on 54 retrospectively and 4 prospectively collected fresh-frozen tumor samples with paired nonmalignant tissue from 57 unique patients. RNA-seq was performed on 57 of the 58 total tumor samples. In total, our cohort included 28 patients with treatment-naive FL, 21 with relapsed FL, and 8 with transformed FL (supplemental Table 1). WES for all samples (tumor and normal) achieved >20× coverage for >75% of the targeted region, with a mean coverage of 76×. RNA-seq for all samples averaged 145 million total reads (range, 48 million to 545 million), with an average of 67% of reads mapped (range, 36%-98%). For the RNA-seq data, the average breakdown of aligned bases was 6.3% ribosomal (range, ∼0%-45%), 11% untranslated region (UTR; range, ∼9%-27%), 6% intronic (range, ∼1%-42%), 1% intergenic (range, ∼0%-7%), and 76% coding (range, ∼14%-87%). After filtering, the number of nonsynonymous coding variants per sample ranged from 2 to 172 (mean, 52; median, 36). Of the 1787 affected genes, 264 were mutated in >1 patient. Thirty-two of 39 genes that were previously identified as significantly mutated by Krysiak et al40 were also found to be mutated within our cohort. For our cohort, many genes with established relevance to FL were recurrently mutated (KMT2D [67%], CREBBP [41%], TNFRSF14 [41%], BCL2 [36%], ATP6V1B2 [16%], STAT6 [12%], EZH2 [10%], IRF8 [10%], CD79B [10%], BCL7A [9%], EP300 [9%], MEF2B [9%], CARD11 [7%], TP53 [7%], and GNA13 [7%]; Figure 2; supplemental Table 2). Many individuals harbored >1 KMT2D mutation (57 mutations observed in 39 individuals) and/or >1 BCL2 mutation (31 mutations observed in 21 individuals). No novel hot spot mutations were identified. Additionally, of 57 patients, 23 (40%) harbored novel fusion markers (supplemental Table 3; supplemental Figure 1).
Recurrently mutated genes and mutation burden observed for all patients. The bar graph on the top corresponds to the number of total mutations per patient and is colored by mutation type. The bar graph on the left corresponds to the percentage of mutations for a given gene for the entire cohort. Columns represent each sample in the cohort (1 patient, LYM120, with both tumor [t] and relapse [r] samples is shown) and are ordered by the presence of mutations in the most to least frequently mutated gene. The third plot indicates the presence or absence of a mutation for each patient and gene combination, colored by mutation type. If a patient has multiple mutations for an individual gene, it is colored according to the priority order as indicated in the mutation type legend, from top to bottom. A white star indicates which mutations are predicted to result in high-quality neoantigen vaccine candidates.
Recurrently mutated genes and mutation burden observed for all patients. The bar graph on the top corresponds to the number of total mutations per patient and is colored by mutation type. The bar graph on the left corresponds to the percentage of mutations for a given gene for the entire cohort. Columns represent each sample in the cohort (1 patient, LYM120, with both tumor [t] and relapse [r] samples is shown) and are ordered by the presence of mutations in the most to least frequently mutated gene. The third plot indicates the presence or absence of a mutation for each patient and gene combination, colored by mutation type. If a patient has multiple mutations for an individual gene, it is colored according to the priority order as indicated in the mutation type legend, from top to bottom. A white star indicates which mutations are predicted to result in high-quality neoantigen vaccine candidates.
Dominant BCR clonotypes could be inferred from most patients with FL using MiXCR
Given that distinct cases of FL express unique cell surface BCRs and selectively retain expression of the BCR for survival,41 FL neoantigen candidates can be derived from the immunoglobulin heavy (IgH) and/or light (lambda/kappa [IgL/K]) chains in addition to somatic variants (SNVs/indels/fusions). To investigate B-cell clonal composition in normal lymph nodes and FL, we first compared our tumor RNA-seq data with publicly available RNA-seq data of B-cell–enriched samples.42-50 In total, 53 normal and 57 tumor samples had sufficient BCR sequencing coverage for analysis. An average of 5083 reads (range, 136-76 742) were used in clonotyping each patient’s BCR repertoire (supplemental Figure 3C). Each unique amino acid sequence was defined as a distinct clonotype (supplemental Methods). The mean number of total clonotypes (IgH, IgL, and IgK) detected within each patient’s FL BCR repertoire was 1400 (median, 550; range, 20-22 052). A mean of 361 unique IgH clonotypes (range, 6-7762) and a mean of 770 unique IgL/K clonotypes (range, 6-10 029) were identified.
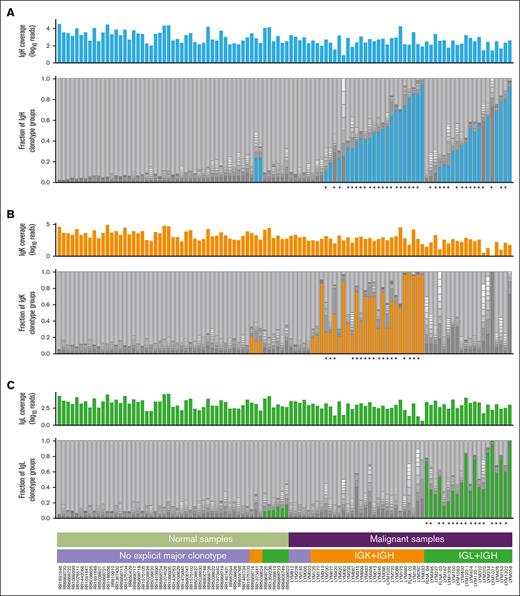
Comparison of the reconstructed BCR repertoire predicted by MiXCR51 revealed that normal samples showed diverse polyclonality (median IgH clonality, 0.04), whereas FL samples predictably showed evidence of a clonal malignant process (median IgH clonality, 0.25; Figure 3; supplemental Figure 2). Based on ad hoc analyses to maximize sensitivity and specificity for correctly identifying malignant from normal samples by BCR diversity, we defined dominant sequences for IgH and IgL/K using a 9% threshold (a more stringent cutoff than some existing clinical BCR clonality assays). Among the 53 normal samples, only 2 (3.8%) had a dominant IgH clonotype exceeding the 9% cutoff, whereas most FL samples (78%) had at least 1 likely malignant, dominant clonotype exceeding the 9% cutoff. Analyses of clonotype composition for IgL and IgK revealed similar patterns. A total of 116 dominant (>9%) IgH or IgL/K clonotypes were identified (Figure 3; supplemental Figure 3A-B,F). Of the 57 patients with RNA-seq data, 46 had at least 1 dominant IgH and 1 dominant IgL/K clonotype, 2 patients had dominant IgL/K clonotypes only, 1 patient had dominant IgH clonotypes only, and 8 patients had no dominant IgH or IgL/K clonotype.
Clonality analysis of BCR populations within healthy normal samples and FL samples. The plots show the composition of BCR repertoire of both normal (left half) and malignant (right half) samples for each of Ig chains separately: panels A-C correspond to heavy (blue), kappa (orange), and lambda (green) chains, respectively. In each panel, the upper histogram shows the coverage of the given Ig chain in the sample (log10 of read counts), whereas the lower stacked bar plot shows BCR repertoire structure. Each bar from bottom to top is composed of 10 dark sections representing the fraction of repertoire for the 10 largest clones in the sample and a single top light gray section representing all other (minor) clones. The colored bottom sections depict the single most dominant clone that exceeded the cutoff value of fraction in the sample (9%) and had sufficient overall coverage (>40 reads for malignant samples whereas normal samples were preselected having >100 reads for each chain, see “Methods”). When both light chains passed the cutoff, only the one with the larger fraction was selected and colored. Stars below each panel indicate major clonotypes predicted to result in one or more high-quality neoantigen vaccine candidates.
Clonality analysis of BCR populations within healthy normal samples and FL samples. The plots show the composition of BCR repertoire of both normal (left half) and malignant (right half) samples for each of Ig chains separately: panels A-C correspond to heavy (blue), kappa (orange), and lambda (green) chains, respectively. In each panel, the upper histogram shows the coverage of the given Ig chain in the sample (log10 of read counts), whereas the lower stacked bar plot shows BCR repertoire structure. Each bar from bottom to top is composed of 10 dark sections representing the fraction of repertoire for the 10 largest clones in the sample and a single top light gray section representing all other (minor) clones. The colored bottom sections depict the single most dominant clone that exceeded the cutoff value of fraction in the sample (9%) and had sufficient overall coverage (>40 reads for malignant samples whereas normal samples were preselected having >100 reads for each chain, see “Methods”). When both light chains passed the cutoff, only the one with the larger fraction was selected and colored. Stars below each panel indicate major clonotypes predicted to result in one or more high-quality neoantigen vaccine candidates.
Interestingly, 17 patients had ≥2 dominant (>9%) IgH and/or IgL/K clonotypes. To determine the relationship between these clonotypes, pairwise nucleotide and protein sequence alignments between all dominant clonotypes within a patient’s BCR repertoire were assessed (supplemental Figure 3G-H; supplemental Tables 4 and 5). In 1 notable case with multiple dominant IgH sequences (LYM720), 4 dominant IgH clonotypes at 25% (clone 1), 24% (clone 2), 17% (clone 3), and 13% (clone 4) were identified. Three of these (clones 1-3) share the exact same variable–diversity–joining (VDJ) alleles and nearly identical CDR3 sequences (only 1-3 amino acid differences, with best protein alignment score percentages of 93%-98%; supplemental Table 4). However, when these 3 dominant clonotypes were compared with the fourth clonotype with a differing VDJ allele, the best alignment score percentage was only 49%. This finding, in conjunction with somatic variant allele frequencies (VAFs) from WES data, indicates that clonotypes 1 to 3 from LYM720 are highly related and likely represent subclonotypes with the same rearrangement and same malignant cell population. Clonotype 4 shares little sequence similarity to the other clonotypes and may represent B-cell expansion from an unrelated immune response or, less likely, an independent malignant clone.
A similar pattern was observed for patients with multiple IgL/K dominant clonotypes (supplemental Table 5). For a patient with multiple dominant IgL sequences (LYM120; supplemental Figure 3G-H), 2 dominant IgL clonotypes were identified at 42% and 19%. Both clonotypes share the exact same VJ alleles and have nearly identical CDR3 amino acid sequences; furthermore, at least 3 additional minor clonotypes (1%-3%) also appear highly related to these dominant sequences. We hypothesize that Ig somatic hypermutation likely accounts for the most of the changes in BCR clonality observed within our cohort (supplemental Figure 3H). Notably, the BCR repertoire in 5 of 8 patients with ≥2 dominant IgH clonotypes and 3 of 9 patients with ≥2 dominant IgL/K clonotypes could be clearly reconciled as subclonal evolution in this context. To further evaluate our BCR clonality analysis, we compared MiXCR results with an orthogonal algorithm, TRUST4,52 and found highly concordant results (supplemental Figure 12; supplemental Table 14). In summary, most patients with FL have a clearly dominant BCR clonotype, in contrast to normal samples. In cases in which multiple dominant clonotypes are observed, these are usually highly related and likely represent subclones with the same underlying rearrangement and malignant cell population. Only those clonotypes defined as dominant in the tumor sample were considered in vaccine design.
High-quality personalized neoantigen cancer vaccine candidates are predicted for most patients
Using the WashU analysis pipeline (pVACtools),53 3065 high-quality somatic variants were identified (Figure 4A). Of these, 783 were predicted to be small, high-quality somatic variants with neoantigen potential (Figure 4A; supplemental Table 6). Peptides suitable for cancer vaccine generation were identified for all but 2 patients from our cohort, and 54 of 57 patients (95%) had at least 2 vaccine candidates (Figure 4B-C). Many but not all of these predicted high-quality neoantigens were associated with putatively oncogenic, recurrently mutated genes in FL (including BCL2, CARD11, CREBBP, EP300, EZH2, FOXO1, HIST1H1B, HIST1H1C, HIST1H1D, IGLL5, IRF8, KMT2D, MEF2B, PIM1, RRAGC, STAT6, and TNFRSF14), whereas others were presumed passenger mutations. In total, 68 variants in recurrently mutated genes were observed in 36 of 56 unique samples (64%; supplemental Figure 4). For the 42 fusion variants identified across all patients, 25 gene fusions (median, 2; range, 0-4) were predicted to generate high-quality neoantigen vaccine candidates (supplemental Figure 1; supplemental Table 3). These fusions were identified in 14 of 57 patients (25%) in our FL cohort (Figure 4C; supplemental Figure 5; supplemental Table 7). In total, 26 of 116 BCR clonotypes (22%) identified as dominant clones included short peptide sequences predicted to sufficiently bind to their respective MHCs (500 nm < half-maximal inhibitory concentration [IC50] binding affinity ≤1000 nm), whereas 71 of 116 (61%) included short peptide sequences that are particularly strong predicted binders to their respective MHCs (IC50 binding affinity ≤500 nm). A total of 97 dominant BCRs (mode, 2; range, 0-6) were predicted to be high-quality idiotype-derived epitopes, representing 79% (45/57) of our FL cohort (Figure 4C; supplemental Figure 3F; supplemental Table 8). Both somatic variants and BCR-derived epitopes are collectively referred to as neoantigens for vaccine design.
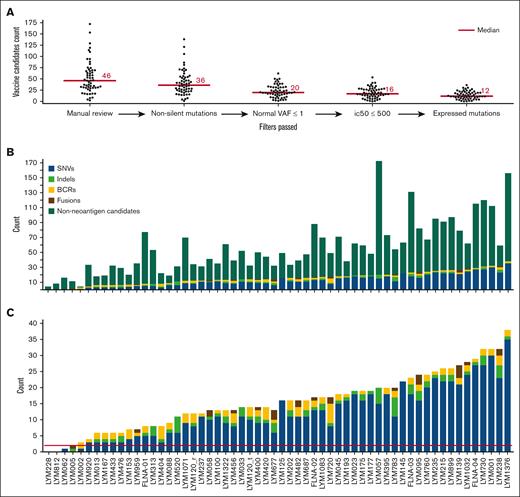
Personalized neoantigen cancer vaccine identification and prioritization. (A) Swarm plots display the number of vaccine candidates on the y-axis for the entire cohort at each stage of filtering (x-axis), moving from left to right. The bar graphs depict the numbers of final neoantigen vaccine candidates for each patient, colored according to source (SNV, indel, BCR, and fusion) and are sorted from least to most total candidates. Bar graph (B) includes nonneoantigen mutations that did not pass neoantigen filtering, whereas bar graph (C) only contains final candidates. The red line in panel C depicts the minimum cutoff of 2 candidates required for potential vaccine design.
Personalized neoantigen cancer vaccine identification and prioritization. (A) Swarm plots display the number of vaccine candidates on the y-axis for the entire cohort at each stage of filtering (x-axis), moving from left to right. The bar graphs depict the numbers of final neoantigen vaccine candidates for each patient, colored according to source (SNV, indel, BCR, and fusion) and are sorted from least to most total candidates. Bar graph (B) includes nonneoantigen mutations that did not pass neoantigen filtering, whereas bar graph (C) only contains final candidates. The red line in panel C depicts the minimum cutoff of 2 candidates required for potential vaccine design.
Two or more neoantigen candidates were identified for 55 of 58 samples (95%), with a mean of 16 predicted neoantigens per patient (range, 0-38). Overall, 77% (702/906) of the total predicted patient peptides arose from missense mutations, 9% (81/906) from indels, 3% (25/906) from gene fusions, and 11% (98/906) from dominant BCR sequences (supplemental Tables 6-8). Furthermore, 56 of 58 samples (97%) had at least 1 missense or indel somatic vaccine candidate, 14 of 57 patients (25%) had at least 1 fusion vaccine candidate, 45 of 57 patients (79%) had at least 1 BCR vaccine candidate, and only 2 patients (LYM228 and LYM812) had no vaccine candidates. These 2 cases had very low mutation burden (4 and 0 SNVs/indels respectively, 0 fusions, and 0 dominant BCR sequences). When compared with an orthogonal approach for neoantigen prediction (BostonGene Vaccine Module), the WashU approach yielded more predicted neoantigen candidates, which is likely attributable to the use of a less conservative requirement surrounding RNA-seq support (supplemental Tables 9 and 10). Twenty-one percent of predicted vaccine candidates were shared of the total 1179 collectively predicted vaccine candidates from the 2 approaches. For 44 of 58 samples (76%), a set of common neoantigens was generated by both approaches (supplemental Figure 6).
APM assessment
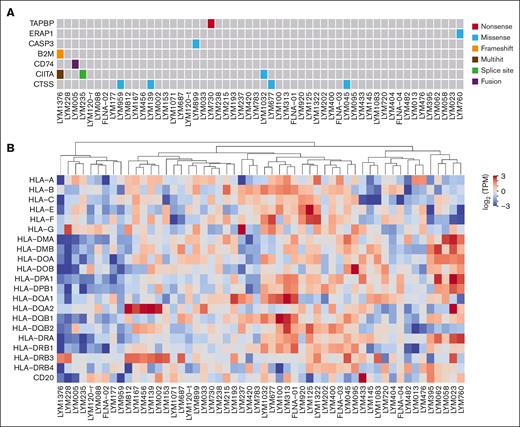
Neoantigens are recognized by antigen-specific T cells via presentation by MHC on the tumor cell surface. When investigating mutations in antigen-presenting machinery (APM) genes, we identified 2 samples bearing likely loss-of-function mutations (Figure 5A). A novel frameshift insertion in the β2 microglobulin (B2M) gene was observed in the beginning of the transcript (ENST00000558401:p.Ala6ArgfsTer52) for sample LYM1376. Another likely loss-of-function (nonsense) variant was found in the tapasin (TAPBP) gene (ENST00000426633:p.Gln477Ter) in LYM730. When investigating mutations in genes associated with MHC, no nonsynonymous mutations were identified; however, some samples had low expression of class I genes (eg, LYM395, LYM760, and LYM1071), whereas others had low expression of class II genes (eg, LYM228, LYM1376, and LYM235) (Figure 5B). Similar patterns have been previously described.54
Molecular functional profiling of APM and MHC. (A) Somatic mutations in APM genes. Only 7 mutated genes of the total 32 APM genes considered. (B) Depicts median transformed log2(TPM + 1) RNA expression levels of MHC class I and II genes. ssGSEA, single sample gene set enrichment analysis; TPM, transcripts per million.
Molecular functional profiling of APM and MHC. (A) Somatic mutations in APM genes. Only 7 mutated genes of the total 32 APM genes considered. (B) Depicts median transformed log2(TPM + 1) RNA expression levels of MHC class I and II genes. ssGSEA, single sample gene set enrichment analysis; TPM, transcripts per million.
FL clonal architecture informs vaccine design
We also investigated the impact of clonal architecture on vaccine design for the 54 retrospectively collected FL tumor samples. The average variant read depth per patient was 173 (range, 60-554; supplemental Figure 7). The average VAF per patient was 23% (range, 7.6%-40.9%; supplemental Figure 8). The average number of tumor (sub)clonal populations identified per patient was 2 (range, 1-4; supplemental Figure 9). For 43 patients (80%), their VAF distributions supported the presence of at least 1 clear subclonal population. The average tumor DNA VAF for predicted high-quality neoantigens was 26.3% (range, 5.0%-97.7%). Most patients (94%) had at least 1 predicted high-quality neoantigen in the dominant/founding clone (median, 11; range, 0-30). Most patients (72%) with evidence of subclonality also had evidence of subclonal neoantigens. For example, patient LYM1376, with the most predicted high-quality neoantigens (36 small somatic mutations), had 5 and 25 high-quality neoantigens predicted within all clonal and subclonal populations, respectively (supplemental Figure 9; supplemental Table 6). Of note, the clonality analysis used only SNVs/indels as input, which underestimates the number of high-quality neoantigens observed.
A pilot clinical trial shows preliminary safety and efficacy of neoantigen vaccine plus anti–PD-1 mAb therapy in R/R FL
A feasibility pilot clinical trial (#NCT03121677) was initiated using personalized neoantigen vaccine therapy combined with PD-1 blockade in patients with R/R FL (supplemental Figures 10 and 11; supplemental Table 13). Four patients were enrolled, and analyses of their pretreatment biopsies identified 12 to 19 neoantigens per patient. All 4 patients had at least 1 mutation in a putatively oncogenic, recurrently mutated gene in FL (STAT6, CARD11, EZH2, BCL2, and RRAGC), targeted in the vaccine design (supplemental Table 11). Peptide vaccines were synthesized using the predicted neoantigens (supplemental Table 12) and administered concurrently with nivolumab, an anti–PD-1 mAb. In total, each of the 4 patients received up to 30 long peptides targeting neoantigens arising from somatic mutations or BCR clonotypes, including register shifting around the key MHC-binding short peptides. Overall, of 120 long peptides, 72 (60%) gave rise to class I epitopes with strong predicted binding affinities (IC50 binding affinity ≤500 nm). Given the emerging evidence of the importance of CD4 T-cell responses to neoantigen vaccines,55 we also examined the potential for our SLPs to elicit class II responses, despite our vaccine design being focused on class I epitope prediction. Intriguingly, 82 of 120 (68%) gave rise to class II epitopes with strong predicted binding affinities (IC50 binding affinity ≤500 nm).
In our pilot clinical trial, time from biopsy to treatment ranged from 5 to 7 months. Vaccination and nivolumab treatment were well tolerated with no grade 3 to 5 adverse events being observed. Responses assessed by positron emission tomography and/or computerized tomography scanning after cycle 2 (C2) included 1 complete response (CR), 1 stable disease, and 2 instances of progressive disease. The patient who achieved a CR after C2, whose disease had previously responded to and relapsed after rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) as well as R-bendamustine, remained disease free at 32 months after their last dose of study treatment. The 2 patients with progressive disease after C2 received 4 weekly doses of rituximab, per protocol, in combination with additional neoantigen vaccine and nivolumab doses. Of these 2 patients, 1 achieved a partial response, and 1 achieved a CR. Both continued on study with vaccine, nivolumab, and 4 additional doses of rituximab.
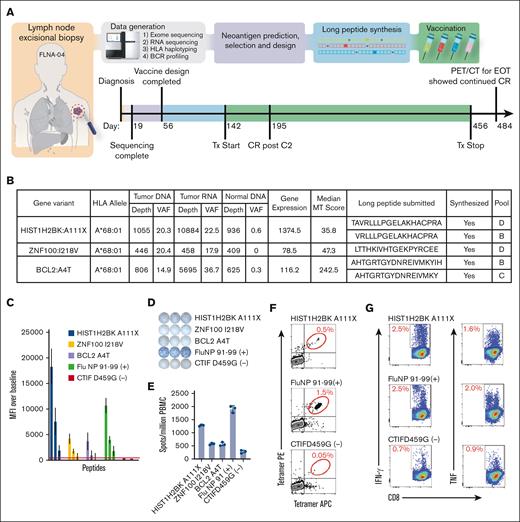
Correlative studies from the patient with CR after C2 of vaccine plus nivolumab revealed a neoantigen-specific immune response (Figure 6A-F). For this patient, we identified 19 predicted high-quality neoantigen vaccine candidates using pVACtools (Figure 6B). A total of 16 peptides were formulated into the SLP vaccine, of which 13 were predicted to bind to HLA-A∗68:01, and 5 were predicted to bind to HLA-A∗23:01. After completing therapy, 9 unique HLA-A∗68:01 candidate peptides were screened with a newly created transporter associated with antigen processing (TAP) negative cell line to assess whether neoantigens stabilized cognate MHC class I molecules. Four neoantigen candidates stabilized HLA-A∗68:01 in a dose-dependent manner (3/4 positives shown: HIST1H2BK ENSP00000349430.1:p.A111X, ZNF100 ENSP00000445201.3:p.I218V, and BCL2 ENSP00000329623.3:p.A4T; Figure 6C). One of the candidates (CTIF ENSP00000256413.3:p.D495G) that did not stabilize HLA-A∗68:01 on the TAP– cell line was selected as a nonbinding control for subsequent assays. The long vaccine peptides were used to stimulate and expand peripheral blood mononuclear cells (PBMCs) taken from the patient 4 months after initiation of the vaccine study therapy (Figure 6D-E). The greatest interferon gamma (IFN-γ) response by enzyme-linked immunosorbent spot (ELISPOT) was observed for HIST1H2BK A111X (1282 ± 30 spot forming units [SPUs] per million PBMCs), whereas the other 2 candidates ZNF100 I218V and BCL2 A4T showed 573 ± 35 and 566 ± 70 SPUs per million PBMCs, respectively. Both of the latter candidate SPUs were above the nonbinding control peptide CTIF D459G (278 ± 50 SPU per million), indicating a positive IFN-γ response. To determine whether circulating antigen-specific CD8+ T cells could be detected in postvaccination PMBCs, MHC class I tetramers were prepared with the 3 candidate peptides shown to be enriched by ELISPOT assay and used to probe 10-day, SLP-stimulated PBMCs. We found 0.5% of the CD8+ T cells were specific for HIST1H2BK A111X phycoerythrin (PE)/allophycocyanin (APC) tetramers, which was greater than the CTIF D459G nonbinding control (Figure 6F). Lastly, 10-day postvaccination PBMC cultures were stimulated with HIST1H2BK A111X SLP and then restimulated with artificial antigen presenting cells loaded with the candidate SLP. IFN-γ/tumor necrosis factor (TNF) expression from CD8+ T cells was comparable with the positive control (Figure 6G). The same cultures, pulsed with the CTIF D459G nonbinding control peptide, demonstrated a low cytokine response. This patient’s prevaccine PBMCs were not available for evaluation in these assays. Together, these results indicate that lymphoma-specific antigens can be used to stimulate antigen-specific T cells for a patient with FL.
Implementation of neoantigen vaccines in pilot clinical trial. (A) Trial overview and timeline for patient FLNA-04 (see supplemental Figures 11 and 12 for more detailed versions). (B) The table lists all variants that were predicted to result in high-quality neoantigen vaccine candidates along with corresponding HLA allele, selection criteria, long peptide sequence submitted, synthesis success status, and vaccination pool (supplemental Tables 11 and 12). Short epitope sequences that binding predictions were based on are bolded within the long peptide sequences submitted column. (C) Peptide stabilization of selected candidate neoantigens. Various concentrations of peptides were incubated overnight with ICP-47 expressing (TAP deficient) B-cell line expressing HLA A∗68:01 heavy chain, washed and then stained with W6/32 APC. Mean fluorescence intensity (MFI) of cells pulsed with decreasing peptide concentrations were compared with no peptide pulsed control cells to validate predicted peptide stabilization of MHC class I molecule. (D) PBMCs from C6 apheresis were pulsed with vaccinating peptides and cultured for 12 days in vitro and then challenged with predicted short peptides overnight on IFN-γ–coated ELISPOT plates. (E) Bar graph shows triplicate value SPUs per million PBMCs from D12 culture. (F) An example positive antigen-specific CTL enrichment detected by peptide loaded tetramers stained with tetramer PE and tetramer APC gated on live, CD3/CD8DP CTL from D12 CTL. (G) Antigen-specific CTL from D12 cultures were challenged with artificial APCs pulsed with specific peptides, incubated for 6 hours, and then IFN-γ or TNF–expressing CD3/CD8 DP cells were detected by FACS. APC, antigen-presenting cells; CTL, cytotoxic T lymphocyte; D12, day 12; EOT, end of treatment; FACS, fluorescence-activated cell sorting; MT, mutant; PE, phycoerythrin; PET/CT, positron emission tomography-computed tomography; TAP, transporter associated with antigen processing; TNF, tumor necrosis factor; Tx, treatment.
Implementation of neoantigen vaccines in pilot clinical trial. (A) Trial overview and timeline for patient FLNA-04 (see supplemental Figures 11 and 12 for more detailed versions). (B) The table lists all variants that were predicted to result in high-quality neoantigen vaccine candidates along with corresponding HLA allele, selection criteria, long peptide sequence submitted, synthesis success status, and vaccination pool (supplemental Tables 11 and 12). Short epitope sequences that binding predictions were based on are bolded within the long peptide sequences submitted column. (C) Peptide stabilization of selected candidate neoantigens. Various concentrations of peptides were incubated overnight with ICP-47 expressing (TAP deficient) B-cell line expressing HLA A∗68:01 heavy chain, washed and then stained with W6/32 APC. Mean fluorescence intensity (MFI) of cells pulsed with decreasing peptide concentrations were compared with no peptide pulsed control cells to validate predicted peptide stabilization of MHC class I molecule. (D) PBMCs from C6 apheresis were pulsed with vaccinating peptides and cultured for 12 days in vitro and then challenged with predicted short peptides overnight on IFN-γ–coated ELISPOT plates. (E) Bar graph shows triplicate value SPUs per million PBMCs from D12 culture. (F) An example positive antigen-specific CTL enrichment detected by peptide loaded tetramers stained with tetramer PE and tetramer APC gated on live, CD3/CD8DP CTL from D12 CTL. (G) Antigen-specific CTL from D12 cultures were challenged with artificial APCs pulsed with specific peptides, incubated for 6 hours, and then IFN-γ or TNF–expressing CD3/CD8 DP cells were detected by FACS. APC, antigen-presenting cells; CTL, cytotoxic T lymphocyte; D12, day 12; EOT, end of treatment; FACS, fluorescence-activated cell sorting; MT, mutant; PE, phycoerythrin; PET/CT, positron emission tomography-computed tomography; TAP, transporter associated with antigen processing; TNF, tumor necrosis factor; Tx, treatment.
Discussion
In this study, we performed comprehensive exome and transcriptome profiling to define the feasibility of a neoantigen vaccine approach for the treatment of patients with FL. This included HLA typing, mutation calling, BCR clonotyping, fusion calling, neoantigen prediction, assessment of mutations in APM/MHC complexes, and sequencing-based in silico vaccine design for 58 FL samples from 57 patients. Our analyses indicated that the modest mutational burden of FL does not preclude patients from harboring immunogenic neoantigens that could be used for a personalized vaccine strategy. We further demonstrated that “polyvalent” vaccine design is possible using our broad genomics-driven strategy. In the retrospective component of our study, 97% of patients had at least 1 peptide suitable for cancer vaccine development, and 95% of patients had multiple vaccine candidates. A subset of these high-quality neoantigens were associated with genes implicated in FL pathogenesis, suggesting that known driver variants (as well as passenger variants) may play important roles in vaccine design. We translated these preclinical proof-of-principle efforts into a pilot clinical trial to examine the safety and preliminary efficacy of neoantigen vaccines for patients with relapsed FL. This clinical trial was further corroborated by correlative studies of patient T-cell responses.
Previous attempts to develop personalized tumor vaccines for patients with FL used a strategy that targeted lymphoma-specific immunoglobulins (ie, BCRs). These approaches used a single cancer target, and patients often received cytotoxic chemotherapy in the interim, which may have contributed to key clinical trials failing to meet their primary efficacy end points.33-35 Emerging evidence indicates that simultaneously targeting multiple neoantigens improves immune elimination of a tumor,12 and recent studies have increased the number of neoantigens incorporated into cancer vaccines.16,17,56,57 These “polyvalent” personalized neoantigen cancer vaccines may enhance the antitumor immune response and improve clinical efficacy of cancer vaccines.58
In this study, cancer vaccine design was informed by FL’s unique mutational landscape. In total, 67% of our cohort possessed at least 1 predicted high-quality neoantigen vaccine candidate specifically within a previously identified set of recurrently mutated genes in FL.40,59-63 For example, 25% of our cohort contained at least 1 KMT2D predicted high-quality neoantigen vaccine candidate. KMT2D mutations are present in ∼50% of FLs40 and play a clear role in FL pathogenesis, synergizing with BCL2 overexpression in mouse models.64 They have also been identified as some of the earliest mutations acquired after the canonical IgH::BCL2 initiating event,64 which makes KMT2D neoantigens particularly attractive for FL cancer vaccines, because they are typically associated with the founding clone.64,65 Additionally, 40% (23/57) of the cohort harbored unique fusion genes (supplemental Figure 1; supplemental Table 3). The most recurrently identified fusion gene pair was a frameshift ARL17A::KANSL1, which has been implicated in other disease types but not in FL.66-68 However, this lesion has also been described in normal cohorts,69 and its significance remains uncertain. BCL6 was also involved in fusions with different gene partners occurring within 4 patients. A total of 25 gene fusions were predicted to be high-quality neoantigen vaccine candidates within 25% of the entire cohort (Figure 4C). Overall, somatic point mutations and fusions in genes with known pathogenic roles in FL contributed significantly to vaccine design.
Although mutations in putatively oncogenic, recurrently mutated genes were regularly featured in our vaccine designs, hot spot alterations were not predicted to be high-quality neoantigens, which implies that FL vaccine designs will require patient-specific customization. Of the 783 predicted high-quality neoantigens resulting from small somatic mutations, only 6 were shared between 2 patients, and only 1 was shared between 3 patients. Furthermore, due to differences in HLA alleles between individuals, even a shared variant does not imply a shared neoantigen. For example, of the 6 variants shared between 2 patients, only 3 were predicted to be high-quality neoantigens when accounting for patient HLA alleles. Similarly, the BCL2 variant R129H shared by 3 different patients (LYM783, LYM235, and LYM730) all resulted in different neoantigen candidates (TPFTARG-H-F, FTARG-H-FATV, and H-FATVVEEL) due to each patient’s specific HLA alleles. These observations suggest that patient-specific vaccines will be needed to improve patient outcomes compared with a universal approach.
Furthermore, personalized cancer vaccines must consider both clonal and subclonal tumor populations. Tumors are heterogeneous and frequently evolve, creating subclones that are spatially and temporally separated.70-74 Vaccines targeting clonal neoantigens should induce a comprehensive antitumor response to eliminate the entire tumor population, which requires targeting both tumor-specific neoantigens, including driver mutations when possible, as well as idiotype-derived epitopes from BCR-based sequences. However, targeting subclonal-specific neoantigens may protect against escape mechanisms (immune editing) in which a subclone has selectively lost immunogenic neoantigens present in the founding clone.75,76 We show preliminary data suggesting that high-quality neoantigens can be identified in both clonal and subclonal tumor cell populations for most patients with FL.
Several limitations and future work remain to be addressed to improve personalized cancer vaccines for FL. It is currently unclear what factors result in the ultimate success or failure of personalized cancer vaccines,36,37 and clinical trial results from other cancers published to date show a low accuracy for neoantigen prediction pipelines.77 Factors that could influence vaccine response include the following: (1) neoantigen identification pipelines; (2) HLA typing predictions, expression, and mutations; (3) potential selection of BCR clonotypes derived from nonmalignant B cells; (4) peptide processing prediction; (5) MHC-binding predictions; (6) vaccine design/delivery approach including neoantigen vaccine form; (7) T-cell recognition; and (8) tumor microenvironment. Existing tools either require further optimization or have yet to be incorporated into our pipeline (eg, considering TRUST4 instead of MiXCR for BCR analysis). Further limitations include evaluation of only class I epitopes for our formal vaccine design (although many were predicted to have strong binding affinities for class II), inability to evaluate T-cell responses before vaccine delivery, and evaluation of only 4 patients for efficacy data. Addressing these limitations, along with optimizing checkpoint inhibitor timing,78 will be the focus of future preclinical and clinical studies.
Overall, our retrospective analysis suggests that nearly all patients with FL may be candidates for personalized cancer vaccine clinical trials. These preclinical results, along with recent studies16,57 supporting polyvalent neoantigen vaccination combined with immune checkpoint blockade, led, to our knowledge, to the first-in-human pilot trial of personalized neoantigen vaccines combined with nivolumab in relapsed FL (#NCT03121677) reported in this study. Preliminary feasibility data are encouraging, no serious adverse events were observed, and 1 of 4 evaluable patients achieved a CR after initial vaccination. This experience supports further clinical study of neoantigen vaccines in patients with lymphoma. These pilot clinical trials are the ultimate test of bioinformatic predictions and will permit improvement of future neoantigen prediction pipelines. This study supports ongoing early phase clinical trial assessment of neoantigen vaccines in lymphoma, which match the goal of a chemotherapy-free immunotherapy without serious adverse events.
Acknowledgments
The authors thank the patients and their families for the donation of their samples and participation in clinical trials. The authors also thank the Siteman Cancer Center at Washington University School of Medicine for the use of the Siteman Flow Cytometry Core, which provided cell sorting service and the Bursky Center for Human Immunology and Immunotherapy Immunomonitoring Laboratory.
This work was supported by the Michael and Ena Feinberg Lymphoma Research Fund (T.A.F.), the Jamie Erin Follicular Lymphoma Research Consortium (T.A.F), the Lymphoma Research Foundation (T.A.F.), the Larry and Winnie Chiang Lymphoma Fellowship (T.A.F. and F.G.), and the Steinback fund of the Barnes-Jewish Hospital Foundation (N.L.B.). Team science support was from the Alvin J. Siteman Cancer Center Siteman Investment Program (supported by The Foundation for Barnes-Jewish Hospital, Cancer Frontier Fund, and Barnard Trust) and the Leukemia and Lymphoma Society Specialized Center of Research. The Siteman Cancer Center is supported in part by an National Cancer Institute (NCI) Cancer Center Support Grant (P30CA091842). O.L.G was supported by the NCI of the National Institutes of Health (NIH; awards U01CA209936, U01CA231844, U24CA237719, and U01CA248235). M.G. was supported by the National Human Genome Research Institute of the NIH (award R00HG007940), the NCI (awards U01CA209936, U01CA231844, U24CA237719, and U01CA248235), and the V Foundation for Cancer Research (award V2018-007). T.A.F. was also supported by the NCI of the National Institutes of Health (NIH; awards P50CA171963 and P30CA91842). D.A.R.-G. is supported by funding from the Lymphoma Research Foundation, Institute for Follicular Lymphoma Innovation, and American Society of Clinical Oncology Young Investigator Award.
Authorship
Contribution: R.D.S., N.M.-S., A.F.C., N.L.B., B.S.K., R.A., M.G., O.L.G., and T.A.F. conceptualized the study; C.A.R., F.F., M.B.-H., O.K., R.D.S., N.L.B., M.G., O.L.G., and T.A.F. developed the experimental design; C.A.R., M.B.-H., E.D.M., S.D., T.S., and M.P.W. acquired samples and clinical data; T.B.M, J.R.W., C.C.F., and R.S.F. led sequence library construction and data acquisition; C.A.R., F.F., M.B.-H., E.D.M., S.D., O.K., V.B., A.B., E.O., K.S., D.A.R.-G., and Z.L.S. performed data analysis; S.K., J.H., V.B., E.O., F.F., and J.R.W. developed software for neoantigen analysis; M.B.-H., E.D.M., S.D., T.S., and O.C.O. performed immunologic response studies; C.A.R., F.F., O.K., M.B.-H., and O.L.G. prepared figures and tables; C.A.R., F.F., M.B.-H., O.K., E.K.B., K.S., D.A.R.-G., and O.L.G. wrote the manuscript with input from K.K., F.G., M.G., and T.A.F.; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: N.M.-S. has served as a consultant for Kyowa Hakka Kirin, Daiichi Sankyo, Karyopharm Therapeutics, and C4 Therapeutics; and has institutional research funding from Celgene, Bristol Myers Squibb, Verastem Oncology, Innate Pharmaceuticals, Corvus Pharmaceuticals, and Genentech/Roche. F.F., O.K., V.B., A.B., E.O., and R.A. are full time employees of BostonGene Corporation. D.A.R.-G. has institutional research funding from Genentech; and has served on an advisory board for AstraZeneca. N.L.B. has research funding from ADC Therapeutics, Affimed, Autolus, Bristol Myers Squibb, Celgene, Forty Seven Immune Design, Janssen, Kite Pharma, Merck, Millennium, Pfizer, Pharmacyclics, Roche/Genentech, and Seagen; and has been on the advisory board for ADC Therapeutics, Roche/Genentech, Seagen, BTG, and Acerta. T.A.F. has research funding from ImmunityBio, Affimed, Wugen, and HCW Biologics; consults for Wugen, Gamida Cell, Takeda, Nkarta, Indapta, and Orca Bio; and has equity and potential royalty interest in Wugen. M.G. and O.L.G. have received consulting fees from Rare Cancer Research Foundation and H37 Foundation. M.G., O.L.G., and K.S. have received consulting fees from Jaime Leandro Foundation for their work in neoantigen vaccine design. The remaining authors declare no competing financial interests.
Correspondence: Malachi Griffith, McDonnell Genome Institute, Campus Box 8501, 4444 Forest Park Ave, St. Louis, MO 63108; email: mgriffit@wustl.edu; Obi L. Griffith, McDonnell Genome Institute, Campus Box 8501, 4444 Forest Park Ave, St. Louis, MO 63108; email: obigriffith@wustl.edu; and Todd A. Fehniger, Washington University School of Medicine, Campus Box 8007, 660 S. Euclid Ave, St. Louis, MO 63110; email: tfehnige@wustl.edu.
References
Author notes
Sequencing data have been deposited in the database of Genotypes and Phenotypes (accession number phs001229).
The full-text version of this article contains a data supplement.

![Recurrently mutated genes and mutation burden observed for all patients. The bar graph on the top corresponds to the number of total mutations per patient and is colored by mutation type. The bar graph on the left corresponds to the percentage of mutations for a given gene for the entire cohort. Columns represent each sample in the cohort (1 patient, LYM120, with both tumor [t] and relapse [r] samples is shown) and are ordered by the presence of mutations in the most to least frequently mutated gene. The third plot indicates the presence or absence of a mutation for each patient and gene combination, colored by mutation type. If a patient has multiple mutations for an individual gene, it is colored according to the priority order as indicated in the mutation type legend, from top to bottom. A white star indicates which mutations are predicted to result in high-quality neoantigen vaccine candidates.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/15/10.1182_bloodadvances.2022007792/3/m_blooda_adv-2022-007792-gr2.jpeg?Expires=1769402456&Signature=iixBXT-IVzbPA0M6shJw0OBjrJeyvNfugcmBmpPMfUf0QQhuzcJax1InFBueginQuT0N15abl4s0Jgszjr6EZzfSGcSjVtR-PBuDDazdLSNGS-YxAVDXaTCWNXD6Z5cbp0XQZ-1Ls9HgKS0vEPgVCfXNr95Ft80mBb9NCKhUQWLEOSL0eE6kMNbdvM-cYiKCKKOZJKO52OiWKwnTeeXO40-MR~Fdb2wu1l~R5wp1blX5Rui3L32cXuLZUMwZoRcz2UgDV~u6HpGfOSnW0d~nRQ6zvu6OdbHmXdOnY7CFRsKVkKW3i1ShrSo7anZZwnjJJR~A8peUiwHYyAeYbOFy2g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)