TO THE EDITOR:
Variants in the C-X-C motif chemokine receptor 4 gene (CXCR4) are associated with a rare autosomal-dominant combined immunodeficiency, WHIM (warts, hypogammaglobulinemia, infections, and myelokathexis) syndrome.1,2 The CXCR4/C-X-C motif chemokine ligand 12 (CXCL12) axis is a master regulator of immune cell bone marrow homing and egress3-5 and B-cell development.6 Previously known pathogenic nonsense, frameshift, and missense variants identified in individuals with WHIM syndrome (CXCR4WHIM) have been localized to the intracellular C-terminal region of CXCR4, a G protein–coupled receptor.2 We describe 4 unrelated patients, designated P1 to P4, with variable clinical presentations including leukopenia, hypercellular bone marrow, recurrent infections, hypogammaglobulinemia, and susceptibility to human papillomavirus, harboring a novel heterozygous CXCR4 variant (c.250G>C; p.D84H), designated as CXCR4D84H, in the second transmembrane domain of the receptor. We performed comprehensive ex vivo and in vitro functional characterization of cells harboring CXCR4D84H in parallel with in silico molecular modeling to determine pathogenicity of the newly discovered variant. See supplemental Methods for detailed methods, including supplemental Table 1A-B.
The studies were approved by institutional review board (IRB) of the respective institutions, including institutional review of Duke University (IRB number Pro00066839), Belarusian Research Center for Pediatric Oncology (IRB number 200809-RD), and University Medical Center Freiburg (IRB number 354/19).
Table 1 and supplemental Table 2A summarize the clinical and laboratory characteristics and immunophenotyping data for P1 to P4.
P1 is a female aged 40 years with plantar warts since childhood, recurrent vulvovaginal and anal dysplasia, and carcinoma in situ for 20 years. Cytopenia, including neutropenia, were first documented at age 15 years. She had Epstein-Barr virus in childhood from which she recovered and was hospitalized briefly for influenza-related pneumonia. P1 has normal immunoglobulin levels. Peripheral blood smear (supplemental Figure 1A) showed hypolobated neutrophils, not typical of myelokathexis.
P2 is a male aged 12 years with trilineage leukopenia, transient thrombocytopenia, recurrent infections, and autoimmune thyroiditis but no warts or hypogammaglobulinemia. His bone marrow biopsy showed abnormal neutrophil retention without classic myelokathexis neutrophil morphology (supplemental Figure 1B). P2’s mother shares the genetic variation but is healthy (supplemental Table 2B).
Limited information is known about P3, a male who died at age 3 years from infectious complications leading to multiple organ failures. He had recurrent infections and was suspected to have immune thrombocytopenic purpura. He did not have peripheral blood leukopenia, although peripheral blood sampling may have occurred during active infection. Family history of P3 and P4 are unknown.
P4 is a female aged 67 years with hypogammaglobulinemia. Immunoglobulin G (IgG) level was 403 mg/dL, and IgG1 subclass was 200 mg/dL; other immunoglobulins were normal. She had upper respiratory tract infections that ceased after IgG replacement therapy initiation and rosacea treated with doxycycline. She does not have cytopenia or human papillomavirus.
The probands are nonrelated from nonconsanguineous unions and were identified from 3 separate countries. All are heterozygous for CXCR4 (c.250G>C), resulting in a single amino acid substitution, D84H. The missense variant is in a highly evolutionarily conserved region in the second transmembrane helix of the receptor (supplemental Figure 1C-D). ClinVar (VCV000808804.12) classified D84H as a variant of uncertain significance. In silico algorithms characterize D84H as deleterious (SIFT score, 0; CADD score, 26.8), probably damaging (PolyPhen2 score, 1) and likely disease causing (REVEL score, 0.951).
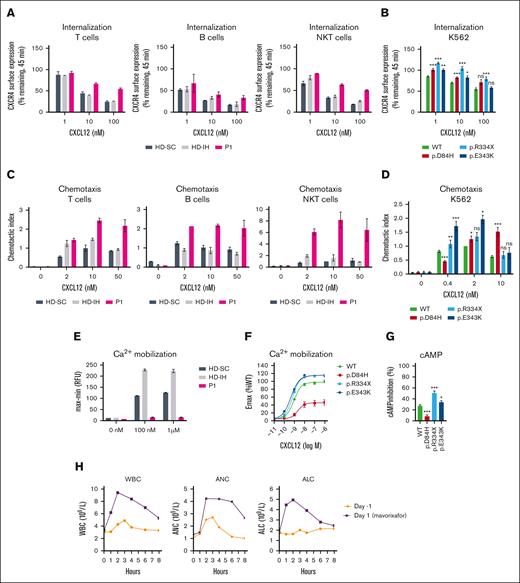
Impaired CXCR4 receptor internalization, amplified signaling responsiveness, and enhanced migration are hallmarks of immune cells expressing CXCR4WHIM receptors.7-10 CXCL12-induced CXCR4 internalization was impaired in peripheral blood mononuclear cells (PBMCs) from P1, P2, and P4 compared with healthy donors (HDs; Figure 1A; supplemental Figure 2A-B). The findings prompted comparative studies of CXCR4D84H with CXCR4R334X, the most common and best-studied CXCR4WHIM variant,2 and CXCR4E343K, the only known pathogenic missense variant.8 The CXCR4-negative K562 cell line9 was transfected to express aforementioned CXCR4 variants for quantitative comparison of variant receptor function (supplemental Figure 2C). CXCR4D84H showed impaired CXCL12-induced receptor internalization (Figure 1B; supplemental Figure 2D) similar to CXCR4E343K. P1 and P2 lymphocyte subpopulations showed enhanced chemotaxis to CXCL12 relative to HDs across tested concentrations (Figure 1C; supplemental Figure 2E), similar to transfected K562 cells expressing CXCR4D84H and other CXCR4WHIM variants compared with K562-CXCR4WT (Figure 1D). CXCR4R334X and CXCR4E343K showed significant increase in pAKT and pERK levels after CXCL12 stimulation compared with CXCR4WT, confirming previous findings9,10; however, no enhancement was observed in pAKT or pERK levels in P1 lymphocytes or K562-CXCRD84H (supplemental Figure 2F-I). Ca2+ mobilization was impaired in P1 PBMCs after CXCL12 stimulation compared with HDs (Figure 1E). This phenotype was confirmed in K562-CXCR4D84H, in which the maximum efficacy of CXCL12 to mobilize calcium decreased to 46%. In contrast, K562-CXCR4R334X and K562-CXCRE343K showed ∼1.2-fold increased Ca2+ mobilization compared with K562-CXCR4WT (Figure 1F). Similar results were obtained when cyclic adenosine monophosphate (cAMP) inhibition was measured in K562-CXCR4D84H compared with K562-CXCR4R334X and K562-CXCR4E343K (Figure 1G). In summary, signaling responses of CXCR4D84H are distinct from other CXCR4WHIM variants.
Impact of CXCR4D84H on receptor function and effects of mavorixafor treatment in P1. (A) PBMCs isolated from HDs (HD-SC and HD-IH) and P1 were stimulated with CXCL12 (vehicle, 1 nM, 10 nM, and 100 nM) for 45 minutes. CXCR4 receptor internalization was assessed by surface expression of CXCR4 by flow cytometry. Values are expressed as percent of remaining CXCR4 expression compared with vehicle-treated cells. PBMCs were subtyped using fluorescent mAbs specific for CD3 (T cells), CD19 (B cell), and CD3 and CD56 (NKT cells). Statistical analysis was not performed owing to low sample numbers. Mean ± standard deviation (SD) of 2 to 4 from 2 biological replicates. (B) K562 cells were transiently transfected with indicated CXCR4 constructs and stimulated with CXCL12 (vehicle, 1 nM, 10 nM, and 100 nM) for 45 minutes. CXCR4 receptor internalization was assessed by surface expression of CXCR4 via flow cytometry. Statistical significance determined by Mann-Whitney tests comparing variants with the WT at each respective concentration of ligand. Values are expressed as percent of remaining CXCR4 compared with vehicle-treated cells. Values represent mean ± standard error of mean (SEM) of 3 to 17. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. (C) Transwell chemotaxis assay: PBMCs isolated from HDs (HD-SC and HD-IH) and P1 migrated toward 2, 10, and 50 nM CXCL12 or medium only for 2.5 hours. The total number of migrated cells was normalized to the HD-SC with 10 nM CXCL12 in each cell type. PBMCs were subtyped using fluorescent mAbs specific for CD3 (T cells), CD19 (B cell), and CD3 and CD56 (NKT cells). Statistical analysis in PBMCs was not performed owing to low sample numbers. Mean ± SD of 2. (D) Transwell chemotaxis assay: K562 cells transiently transfected with indicated CXCR4 constructs migrated toward 0.4, 2, and 10 nM CXCL12 or medium only for 4 hours. The total number of migrated cells was normalized to WT with 2 nM CXCL12 in each assay. Statistical significance was determined by Mann-Whitney tests (0.4 nM and 10 nM) and 1-sample Wilcoxon signed-rank test (2 nM) comparing variants with the WT at each respective concentration of ligand. Mean ± SEM of 5 to 20. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. (E) PBMCs isolated from HDs (HD-SC and HD-IH) and P1 were stimulated with vehicle, 100 nM or 1 µM CXCL12 to measure Ca2+ mobilization. Statistical analysis was not performed owing to low sample numbers; n = 2 technical replicates; the experiment is representative of 2 independent experiments. (F) K562 cells with stable CXCR4 expression were stimulated with serial dilutions of CXCL12 to measure Ca2+ mobilization. Relative fluorescence units measured in WT-expressing cell line at 1 µM CXCL12 represented 100%. Mean ± SEM of 11 to 12. (G) K562 cells with stable CXCR4 expression were stimulated with forskolin ± 100 nM CXCL12 for 30 minutes. cAMP production was measured by enzyme-linked immunosorbent assay. Percent of inhibition of cAMP production by CXCL12 was calculated respective to forskolin-only treated cells. Statistical significance was determined by Mann-Whitney tests comparing variants with the WT. Values represent mean ± SEM of 9 to 12. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. (H) Time course of WBC counts, ANC, and ALC for P1 before receiving oral mavorixafor (day –1) and on the first day of mavorixafor treatment (day 1). ALC, absolute lymphocyte count; AMC, absolute monocyte count; ANC, absolute neutrophil count; CD, cluster of differentiation; HD-IH, healthy donor in-house; HD-SC, healthy donor shipping control; mAb, monoclonal antibody; NK, natural killer; NKT, natural killer T; ns, not significant; WBC, white blood cell; WT, wild type.
Impact of CXCR4D84H on receptor function and effects of mavorixafor treatment in P1. (A) PBMCs isolated from HDs (HD-SC and HD-IH) and P1 were stimulated with CXCL12 (vehicle, 1 nM, 10 nM, and 100 nM) for 45 minutes. CXCR4 receptor internalization was assessed by surface expression of CXCR4 by flow cytometry. Values are expressed as percent of remaining CXCR4 expression compared with vehicle-treated cells. PBMCs were subtyped using fluorescent mAbs specific for CD3 (T cells), CD19 (B cell), and CD3 and CD56 (NKT cells). Statistical analysis was not performed owing to low sample numbers. Mean ± standard deviation (SD) of 2 to 4 from 2 biological replicates. (B) K562 cells were transiently transfected with indicated CXCR4 constructs and stimulated with CXCL12 (vehicle, 1 nM, 10 nM, and 100 nM) for 45 minutes. CXCR4 receptor internalization was assessed by surface expression of CXCR4 via flow cytometry. Statistical significance determined by Mann-Whitney tests comparing variants with the WT at each respective concentration of ligand. Values are expressed as percent of remaining CXCR4 compared with vehicle-treated cells. Values represent mean ± standard error of mean (SEM) of 3 to 17. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. (C) Transwell chemotaxis assay: PBMCs isolated from HDs (HD-SC and HD-IH) and P1 migrated toward 2, 10, and 50 nM CXCL12 or medium only for 2.5 hours. The total number of migrated cells was normalized to the HD-SC with 10 nM CXCL12 in each cell type. PBMCs were subtyped using fluorescent mAbs specific for CD3 (T cells), CD19 (B cell), and CD3 and CD56 (NKT cells). Statistical analysis in PBMCs was not performed owing to low sample numbers. Mean ± SD of 2. (D) Transwell chemotaxis assay: K562 cells transiently transfected with indicated CXCR4 constructs migrated toward 0.4, 2, and 10 nM CXCL12 or medium only for 4 hours. The total number of migrated cells was normalized to WT with 2 nM CXCL12 in each assay. Statistical significance was determined by Mann-Whitney tests (0.4 nM and 10 nM) and 1-sample Wilcoxon signed-rank test (2 nM) comparing variants with the WT at each respective concentration of ligand. Mean ± SEM of 5 to 20. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. (E) PBMCs isolated from HDs (HD-SC and HD-IH) and P1 were stimulated with vehicle, 100 nM or 1 µM CXCL12 to measure Ca2+ mobilization. Statistical analysis was not performed owing to low sample numbers; n = 2 technical replicates; the experiment is representative of 2 independent experiments. (F) K562 cells with stable CXCR4 expression were stimulated with serial dilutions of CXCL12 to measure Ca2+ mobilization. Relative fluorescence units measured in WT-expressing cell line at 1 µM CXCL12 represented 100%. Mean ± SEM of 11 to 12. (G) K562 cells with stable CXCR4 expression were stimulated with forskolin ± 100 nM CXCL12 for 30 minutes. cAMP production was measured by enzyme-linked immunosorbent assay. Percent of inhibition of cAMP production by CXCL12 was calculated respective to forskolin-only treated cells. Statistical significance was determined by Mann-Whitney tests comparing variants with the WT. Values represent mean ± SEM of 9 to 12. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. (H) Time course of WBC counts, ANC, and ALC for P1 before receiving oral mavorixafor (day –1) and on the first day of mavorixafor treatment (day 1). ALC, absolute lymphocyte count; AMC, absolute monocyte count; ANC, absolute neutrophil count; CD, cluster of differentiation; HD-IH, healthy donor in-house; HD-SC, healthy donor shipping control; mAb, monoclonal antibody; NK, natural killer; NKT, natural killer T; ns, not significant; WBC, white blood cell; WT, wild type.
D84 is a critical, conserved residue of the sodium ion binding pocket, essential for signaling of class A G protein–coupled receptors.11,12 Though crystal structures of CXCR4 show no Na+ ion in this location,13,14 site-directed mutagenesis and molecular dynamics studies support its presence.15-17 We performed molecular dynamics simulations analyzing the D84H variant structure, given the change in amino acid charge from negative D to neutral H. Simulations suggested a collapse of the Na+ ion binding pocket, destabilized inactive state, and potential constitutive activity (CA) of a more thermodynamically preferred active-form CXCR4D84H configuration (supplemental Figures 3A-C and 4). To test this hypothesis, CXCR4D84H activity was analyzed under basal conditions. In K562-CXCR4WT, CXCL12 addition led to inhibition of forskolin-stimulated cAMP production, typical for Gαi-coupled receptors (supplemental Figure 3D). In K562-CXCR4D84H, cAMP production was already suppressed in absence of CXCL12, indicative of constitutive Gαi activation. In line with CXCR4D84H effects, other previously known CA CXCR4 variants exhibited enhanced chemotactic CXCL12 response.18 Due to the already high receptor basal activity, CA CXCR4 cannot be further stimulated with CXCL12 and show decreased responses in ligand-induced calcium mobilization.17 These observations explained the decreased signaling observed with CXCR4D84H in calcium mobilization and cAMP assays.
CXCR4D84H represents a novel disease-causing variant with distinct functional properties that could mechanistically explain the phenotypic differences of P1 to P4 compared with previously described C-terminal variants. Patients with WHIM syndrome display variable phenotypes and incomplete penetrance of the classic tetrad,2,19 and the CXCR4D84H patients described have milder infections, cytopenia, and hypogammaglobulinemia. Further, 3 of 4 patients had thrombocytopenia, and 2 of 4 displayed low natural killer cell counts, less typical for WHIM syndrome.
Mavorixafor, a small-molecule CXCR4 antagonist,20,21 inhibited CXCL12 binding with comparable potency (IC50, 3.9-5.6 nM) across K562-CXCR4D84H, K562-CXCR4R334X, K562-CXCR4E343K, and K562-CXCR4WT (supplemental Figure 5A). Mavorixafor abrogated enhanced CXCL12-induced chemotaxis in patient-derived lymphocytes (supplemental Figure 5B-E). P1 was enrolled in a phase 1b/2 open-label trial evaluating safety, tolerability, and early efficacy of mavorixafor (400 mg once daily) across chronic neutropenic disorders (NCT04154488).22 Hematologic parameter monitoring showed increased white blood cell count, absolute neutrophil count, and absolute lymphocyte count on day 1 of mavorixafor treatment (maximum fold change of 2.55, 2.52, and 2.64 respectively; X4, unpublished data, 20 September 2022; Figure 1H). Thus, we propose that mavorixafor may be considered a potential leukopenia treatment in CXCR4D84H patients.
The average CXCR4 (c.250G>C) allele frequency suggests 7.5 per 100 000 individuals may harbor CXCR4D84H (supplemental Table 3). The relatively high allele frequency, P4’s later symptom onset age, and P2’s healthy mother harboring CXCR4D84H suggest incomplete penetrance and/or variable expressivity of the CXCR4D84H phenotype.
Our observations from CXCR4D84H patients and results of the functional characterization of PBMCs and cells expressing the D84H variant highlight several novel findings, expanding the current understanding of genotype-phenotype correlations within the CXCR4 landscape.
Acknowledgments: The authors thank patients and their families for their participation and acknowledge the contributions of Richard Pollison to the chronic neutropenic disorders clinical trial. Molecular dynamics simulations were performed by Jian Yin and Junkun Lei at WuXi AppTec. Biostatistical consulting was provided by Precision for Medicine, Gladstone, New Jersey, and medical writing assistance was provided by Jane Nguyen of PRECISIONscientia in Yardley, Pennsylvania, which was supported financially by X4 Pharmaceuticals, Inc, in compliance with international Good Publication Practice guidelines.
The biorepository for processing and storing blood specimens was supported by a grant from the Jeffrey Modell Foundation. B.G. is funded by the Deutsche Forschungsgemeinschaft (GR1617/14-1/iPAD; SFB1160/2_B5; RESIST-EXC 2155 [project ID 390874280]; and the EU-H2020-MSCA-COFUND EURIdoc programme [number 101034170]) and the Bundesministerium fur Bildung and Forschung (GAIN 01GM1910A).
Contribution: K.Z., S.P., C.N., and A.B. designed the study; S.O.S., C.B.G., K.C., J.W.S., S.A., E.P., I.S., K.W., B.G., M.P., J.R.B., and T.K.T. provided patient data and samples; K.Z., S.P., C.B.G., I.W., C.N., B.U., H.M., S.M.-M., K.C., B.G., M.P., S.G., and J.L.N. acquired and generated data; K.Z., S.P., C.B.G., C.N., M.E., J.E.W., A.B., J.L.N., N.S., and T.K.T. analyzed and interpreted the results; K.Z., S.P., C.B.G., J.E.W., A.G.T., and T.K.T. assisted in the writing and editing of the manuscript; and all authors read and approved the submitted version of the manuscript.
Conflict-of-interest disclosure: X4 Pharmaceuticals funded the development of this manuscript. T.K.T. is a principal investigator in the phase 3 trial of mavorixafor for WHIM syndrome and consultant to X4 Pharmaceuticals. N.S. is a consultant to X4 Pharmaceuticals. K.Z., C.N., H.M., K.C., and A.G.T. are employees, and K.Z., C.N., H.M., and A.G.T. hold shares of X4 Pharmaceuticals. A.B., S.P., S.M.-M., and I.W. hold shares of X4 Pharmaceuticals. J.E.W. has received funding for investigator initiated and sponsored clinical trials from X4 Pharmaceuticals; served as consultant to X4 Pharmaceuticals in the past; and is a principal investigator of investigator-initiated and sponsored studies funded by X4 Pharmaceuticals. J.L.N. serves as a speaker for EUSA Pharma; an adviser for EUSA Pharma and AbbVie; and a consultant for Guidepoint Global. The remaining authors declare no competing financial interests.
Correspondence: Katarina Zmajkovicova, X4 Pharmaceuticals (Austria) GmbH, Helmut-Qualtinger-Gasse 2, 1030 Vienna, Austria; email: katarina.zmajkovicova@x4pharma.com; and Teresa K. Tarrant, Duke University Medical Center 3874, 200 Trent Dr, Durham, NC 27710; email: teresa.tarrant@duke.edu.
References
Author notes
K.Z. and S.P. contributed equally to this study.
The data that support the in vitro experimental findings and molecular dynamics simulations in this study are available from the corresponding author upon reasonable request. Qualified scientific and medical researchers may make requests for clinical trial data that underlie the results (text, tables, figures) reported in this article, after de-identification, at medicalinfo@x4pharma.com. Methodologically sound proposals for such data will be evaluated and approved by X4 Pharmaceuticals, Inc in its sole discretion. All approved researchers must sign a data access agreement prior to accessing the data. Data will be available as soon as possible but no later than within 1 year of the acceptance of the article for publication, and for 3 years after article publication. X4 Pharmaceuticals, Inc. will not share identified participant data or a data dictionary.
The full-text version of this article contains a data supplement.