Mantle cell lymphoma (MCL) is an uncommon mature B-cell lymphoma that presents a clinical spectrum ranging from indolent to aggressive disease, with challenges in disease management and prognostication. MCL is characterized by significant genomic instability, affecting various cellular processes, including cell cycle regulation, cell survival, DNA damage response and telomere maintenance, NOTCH and NF-κB/ B-cell receptor pathways, and chromatin modification. Recent molecular and next-generation sequencing studies unveiled a broad genetic diversity among the 2 molecular subsets, conventional MCL (cMCL) and leukemic nonnodal MCL (nnMCL), which may partially explain their clinical heterogeneity. Some asymptomatic and genetically stable nnMCL not requiring treatment at diagnosis may eventually progress clinically. Overall, the high proliferation of tumor cells, blastoid morphology, TP53 and/or CDKN2A/B inactivation, and high genetic complexity influence treatment outcome in cases treated with standard regimens. Emerging targeted and immunotherapeutic strategies are promising for refractory or relapsed cases and a few genetic and nongenetic determinants of refractoriness have been reported. This review summarizes the recent advances in MCL biology, focusing on molecular insights, prognostic markers, and novel therapeutic approaches.
Introduction
Mantle cell lymphoma (MCL) is an infrequent B-cell neoplasm distinguished by the proliferation of mature B cells, commonly expressing CD5. It primarily affects elderly males, with a median age of ∼65 years.1 It is usually considered aggressive but it has a very heterogeneous clinical behavior. The translocation t(11;14)(q13;q32) and cyclin D1 overexpression are MCL pathognomonic features.1,2 Two distinct molecular subtypes with different clinicobiological features have been recognized: the most frequent conventional MCL (cMCL) and leukemic nonnodal MCL (nnMCL).3 There are 4 cytological variants, the most prevalent being the classic MCL with small to medium-sized cells, the less prevalent being the small cell variant, usually associated with low proliferation and mainly found in nnMCL, and the other 2 variants, the blastoid and pleomorphic, are characterized by larger cells and are associated with more aggressive clinical features. After the t(11;14), most MCL cells acquire a high number of secondary alterations affecting genes of several pathways, but only a small number impact the prognosis and response to treatment.3,4 In this review, we summarize recent data on MCL pathogenesis, including different molecular subtypes and variants, and prognostic/predictive factors under chemoimmunotherapy or current molecular-targeted approaches.
Primary genetic events in MCL: cyclin D1 as a key player
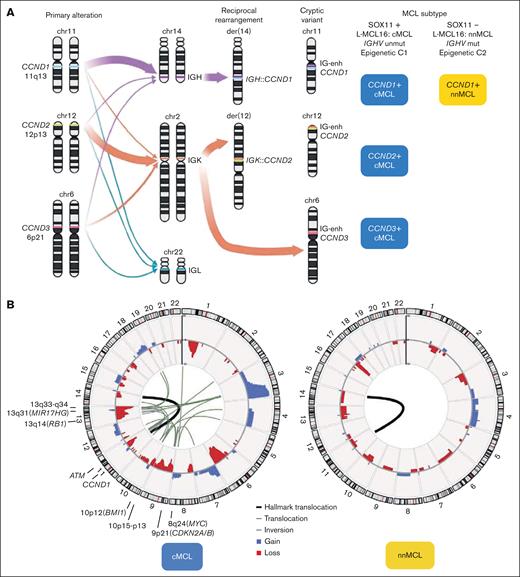
The primary genetic alteration in both molecular subtypes, cMCL and nnMCL, is the translocation t(11;14)(q13;q32), which juxtaposes the enhancer of the immunoglobulin heavy chain (IGH) gene near the CCND1 oncogene. In virtually all patients with MCL the translocation involves IGH::CCND1, but occasional variant translocations with immunoglobulin light chain genes have been reported, the t(2;11)(p11;q13) involving IGK::CCND1 and t(11;22)(q13;q11) involving IGL::CCND1 (Figure 1A).3 Fluorescence in situ hybridization (FISH) using a fusion IGH::CCND1 probe is widely used for diagnostic but the CCND1 break-apart probe is recommended to recognize these variant translocations. The molecular consequence of this rearrangement is a constitutive overexpression of the cyclin D1 protein. The main mechanism generating the translocation is aberrant V(D)J rearrangement, which takes place in the bone marrow precursors at the pre-B stage. Nevertheless, 5 cases have been reported in which the primary rearrangement was generated via aberrant class switch recombination (CSR) or somatic hypermutation (SHM), both processes considered to occur in the germinal center (GC).5 No clinicobiological differences had been identified in these cases.
Primary and secondary MCL chromosomal alterations. (A) The left panel is a schematic representation of CCND1, CCND2, and CCND3 genes and their rearrangements with IGH, IGK, and IGL genes. The size of the arrows represents their frequency. The right panel indicates the molecular subtype according to the CCND gene rearrangement and cMCL or nnMCL. Note that still there are no reported MCL cases with CCND2 or CCND3 rearrangements of the nnMCL subset. (B) Circular representation of copy number alterations (CNA) and structural variants (SV) in MCL with whole-genome sequencing (WGS) and pretreatment (45 cMCL and 16 nnMCL).5 In the inner side, the primary SV, t(11;14) found in all cases, was represented by a thicker black line, whereas other alterations affecting (or very near) driver genes are represented by black lines (translocations) or gray lines (insertions); in the outer side, the CNA are colored (gains in blue and losses in red). Driver genes or regions frequently targeted by SV (in addition to gains and losses) are indicated. Note that no recurrent rearrangements were found.
Primary and secondary MCL chromosomal alterations. (A) The left panel is a schematic representation of CCND1, CCND2, and CCND3 genes and their rearrangements with IGH, IGK, and IGL genes. The size of the arrows represents their frequency. The right panel indicates the molecular subtype according to the CCND gene rearrangement and cMCL or nnMCL. Note that still there are no reported MCL cases with CCND2 or CCND3 rearrangements of the nnMCL subset. (B) Circular representation of copy number alterations (CNA) and structural variants (SV) in MCL with whole-genome sequencing (WGS) and pretreatment (45 cMCL and 16 nnMCL).5 In the inner side, the primary SV, t(11;14) found in all cases, was represented by a thicker black line, whereas other alterations affecting (or very near) driver genes are represented by black lines (translocations) or gray lines (insertions); in the outer side, the CNA are colored (gains in blue and losses in red). Driver genes or regions frequently targeted by SV (in addition to gains and losses) are indicated. Note that no recurrent rearrangements were found.
Cyclin D1-negative MCL has been recognized,1 and in the largest reported series, 39 of 52 (75%) cases have CCND2 rearrangement with immunoglobulin genes (mainly IGK and IGL, and less frequently IGH), and were detectable by FISH using a CCND2 break-apart probe. The remaining 13 of 52 (25%) carry cryptic rearrangements, 4 cases with CCND2 and 9 with CCND3, involving the insertion of the IGK or IGL small enhancer region (∼27 kilobases). These rearrangements are undetectable by standard FISH approaches using break-apart probes (Figure 1A).6,7 A few cryptic immunoglobulin rearrangements with CCND1 have also been identified.8
Cyclin D1, although considered a weak oncogene on its own,9 plays a pivotal role in cell cycle regulation by facilitating the transition from G1 to S phase through CDK4-mediated phosphorylation of Rb1. Alongside the t(11;14), aggressive MCL cases often exhibit other CCND1 alterations, such as amplification of the translocated allele,5 and mutations or deletions in the 3' untranslated region (UTR) of CCND1 messenger RNA (mRNA),5,10,11 which generates a truncated protein without the 3'UTR regulatory region and microRNA target sites, with increased protein half-life. All of these alterations contribute to further potentiate cyclin D1’s oncogenic activity. CCND1 mutations in the 5' region and exon 1 are common in nnMCL with IGHV-mutated genes, suggesting their acquisition in the GC microenvironment by aberrant SHM,12 and several CCND1 coding mutations (E36K, Y44D, or C47S) have been associated with increased cyclin D1 protein levels in cell lines.13 Moreover, cyclin D1 functions as a transcriptional regulator, influencing the expression of genes involved in cell cycle progression and contributes to the DNA damage response (DDR) pathway.14
Differential diagnosis
The translocation t(11;14)(q13;q32) and/or cyclin D1 overexpression have been identified in other B-cell lymphomas, usually acquired as a secondary event at progression or relapse.15,16 High-grade B-cell lymphomas with CCND1 rearrangement as a secondary event usually harbor MYC and BCL2 and/or BCL6 rearrangements. Their immunophenotypic profile differs from MCL; they lack CD5 and SOX11 expression, and their genomic landscape is different from MCL and more similar to diffuse large B cell lymphoma, with BCL2, MYC, CDKN2A, KRAS, and TNFRSF14 mutations.17
MCL molecular subtypes
MCL comprises 2 molecular subtypes; cMCL and nnMCL.1,3 A leukemic presentation in the absence of lymphadenopathies, no expression of the oncogenic transcription factor SRY-related HMG-box 11 (SOX11), presence of IGHV somatic mutations, and low genomic complexity are clinical and biological features that differentiate nnMCL from cMCL. A gene expression signature identified in peripheral blood is able to distinguish these MCL subtypes and is based on the expression of 16 genes (L-MCL16).18 The L-MCL16 signature is stable over time. SOX11 detection by immunohistochemistry is useful for MCL diagnosis because it is expressed in MCL and also in the cyclin D1-negative MCL but is not expressed in other B-cell lymphomas, with the exception of some Burkitt and precursor cell lymphomas/leukemias.19,20 SOX11 oncogenic roles include alterations in several pathways and altogether seem to be responsible for the more aggressive behavior of cMCL compared with nnMCL.3,21 The primary cause of SOX11 expression is poorly understood and is related to epigenetic alterations.22,23 cMCL is characterized by low or no mutations in IGHV, whereas nnMCL has IGHV mutations, consistent with a different cell of origin, a similar scenario as in chronic lymphocytic leukemia (CLL). Although a clear cutoff is difficult to define, 97% identity with the germ line can predict survival.24 IGHV3-21 and IGHV4-34 are the most predominant families, and MCL with IGHV3-21 usage almost exclusively share the same light chain (IGLV3-19), with different copy number alterations (CNA) and a better prognosis, favoring the hypothesis that IGHV3-21 tumors may represent a distinct MCL subtype and that there is a possible role for antigens in MCL development.25-27 In the largest MCL study, IGHV3-21 was the most frequently rearranged gene (133/807 [17%]) and was found mainly in cases with unmutated IGHV (97% of cases with unmutated IGHV). However, in this study, there was no information regarding SOX11 or cMCL/nnMCL subtypes.26 The association between IGHV status and IGHV3-21 expression was also confirmed in another study, with 13 cases of IGHV3-21 and unmutated IGHV vs 3 with mutated IGHV.24 Moreover, by whole-genome sequencing (WGS), we found that MCL with IGHV3-21 are more frequent in the cMCL subset (6/44 [14%]) than in nnMCL (1/17 [6%]).5 The association of MCL IGHV3-21 with unmutated IGHV genes and better survival is interesting and further reinforces the hypothesis that MCL with IGHV3-21 may represent a distinct MCL subset among cMCL.
Similar to CLL, another layer of information supporting the 2 different MCL molecular subtypes came from DNA methylation studies.5,23 The 2 groups identified also reflect different cellular origin with cluster 1 MCL, the most frequent, characterized by an imprint of GC-unexperienced B cells, with few or no IGHV mutations, expressing SOX11, and corresponding to cMCL; and cluster 2, defined by an imprint of B cells that have experienced the GC and high levels of IGHV SHM, no expression of SOX11 and corresponding to nnMCL.23 This study also shows that all MCL (irrespective of the subtype) have a DNA methylation profile similar to that of antigen-experienced cells.
Secondary genetic events in MCL
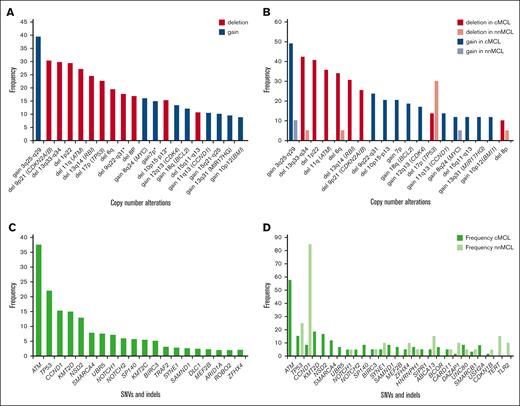
MCL is a lymphoma entity with the highest degree of genetic instability, only comparable to multiple myeloma.28,29 cMCL shows very frequent complex karyotypes, whereas nnMCL at diagnosis present usually with simple karyotypes with only t(11;14). Unexpectedly, a subset of cases also had 17p loss, frequently as isochromosome 17q.30,31 Subsequent studies using copy number arrays and WGS showed that >90% of cMCL and especially all blastoid variants, display highly altered genomes, whereas nnMCL at diagnosis are genetically very stable (Figure 1B; Table 1).5,32-34 We performed a review of CNA in diagnostic samples from 2 large studies (Figure 2A).5,32 All alterations are common in cMCL and very low or absent in nnMCL, except for deletion of 17p (TP53), which is more frequent in nnMCL (30%), whereas virtually exclusive alterations of cMCL are losses at 13q33-q34 (42%), 1p22 (41%), 11q22-q23 (36%), 6q (34%), 13q14 (RB1) (31%), 9p21 (CDKN2A/B) (25%), 9q22q31 (24%), and 10p15-p13 (20%), and gains at 3q25-q29 (49%), 18q21-q22 (BCL2) (19%), and 12q13 (CDK4) (17%) (Figures 1B and 2B). This profile of alterations, resulting from multiple intra- and interchromosomal translocations, is highly specific for MCL, including the cyclin D1-negative subset,6 and different from other B-cell lymphomas.
Recurrent genomic MCL alterations identified using WGS/WES analysis. The frequencies displayed correspond to the studies detailed in Table 1. (A) Illustrates the CNA observed (overall frequency ≥2%) in 202 patients with MCL. The frequency of genomic alterations highlighted with an asterisk “∗” is based only on the Nadeu et al5 publication. (B) CNA identified in Nadeu et al5 categorized by MCL subtype. (C) Single nucleotide variants (SNVs) and insertions and deletions (indels) with overall frequency ≥2%, identified in 432 patients with MCL. (D) SNVs and indels, identified in Nadeu et al5 and categorized by MCL subtype. Variants with an overall frequency ≥2% in at least 1 of the MCL subtypes are displayed.
Recurrent genomic MCL alterations identified using WGS/WES analysis. The frequencies displayed correspond to the studies detailed in Table 1. (A) Illustrates the CNA observed (overall frequency ≥2%) in 202 patients with MCL. The frequency of genomic alterations highlighted with an asterisk “∗” is based only on the Nadeu et al5 publication. (B) CNA identified in Nadeu et al5 categorized by MCL subtype. (C) Single nucleotide variants (SNVs) and insertions and deletions (indels) with overall frequency ≥2%, identified in 432 patients with MCL. (D) SNVs and indels, identified in Nadeu et al5 and categorized by MCL subtype. Variants with an overall frequency ≥2% in at least 1 of the MCL subtypes are displayed.
WGS studies unveiled that several genomic phenomena, such as enhancer hijacking, chromoplexy, chromothripsis, and breakage-fusion–bridge cycles, are relatively frequent in cMCL compared with nnMCL and could be responsible for the activation of several oncogenes, that is, BMI1 at 10p12, MIR17HG at 13q31, TERT at 5p15, and MYC at 8q24. In addition, several nonrecurrent structural variants affect the regions of driver genes (Figure 1B).5,MYC rearrangements with IGH and non-IG partners have been identified only occasionally in MCL, especially in pleomorphic variants.5,44 Other previously undetected alterations identified by WGS are the noncoding mutations in the TERT promoter.5 WGS studies also confirmed the high frequency of mono- and biallelic deletions of the 9p21.3 locus, including always the CDKN2A tumor suppressor gene, and very frequently (>90%) with codeletion of the neighboring genes CDKN2B and MTAP, which have essential roles in cell cycle/TP53 regulation and the methionine salvage pathway, respectively. CDKN2A deletions are especially enriched in blastoid variants (60%-92% of cases).5,45
In GC–derived mature B-cell lymphomas, there is a propensity for aberrant SHM (aSHM) and CSR potentially related to the increased genomic instability; however, in MCL, only the nnMCL subtype and a minority of cMCL with IGHV mutations have GC-experience and aSHM/CSR, and the only known off-target aSHM gene is CCND1. The relationship between CCND1 and genomic instability is still unclear. Transgenic mice with overexpression of cyclin D1 do not develop tumors or lymphomas9 but when cyclin D1 is expressed constitutively in the nucleus, they develop lymphomas that overexpress MYC and have alterations in TP53 or BCL2, 2 relevant genes for cell survival/apoptosis.46 One hypothesis to explain the high genomic instability of cMCL is the high prevalence of ATM and TP53 alterations. In fact, MCL are the tumors with the highest frequency of ATM inactivation (64% of cMCL have mutation and/or deletion), and the ATM gene is critically involved in DDR and maintenance of genome integrity. ATM alterations in cMCL are associated with increased chromosomal instability (8 CNA vs 3 CNA).47 Although not frequent, the downregulation of CHEK2 and CHEK1, also involved in signal transduction of DDR, may represent additional mechanisms leading to chromosomal instability.48 Additional evidence is that an animal model with early B-cell–specific ATM deficiency synergizes with ectopic cyclin D1 expression and promotes the development of tumors morphologically similar to human MCL.49 This suggests that the combination of high cyclin D1 and low ATM levels accelerates and increases the incidence of pre-GC lymphoma. Furthermore, these murine lymphomas have focal deletions of TP53, CDKN2A, KMT2D, and RB1 genes (genes frequently altered in human MCL). Besides ATM, cMCL also harbors TP53 alterations, which mediate several processes, including DDR pathways. Nevertheless, TP53 is not exclusively of cMCL, because nnMCL also has frequent TP53 alterations but generally has few or no genomic alterations. Thus, it is tempting to speculate that SOX11 expression (found in cMCL but not in nnMCL), a potent oncogene with multiple and diverse functions, may also contribute to the genetic instability characteristic of cMCL, although more research is needed to elucidate this.
Different mutational profile in cMCL, nnMCL, and blastoid subsets
The clinical heterogeneity observed in patients with MCL could be explained, in part, by the different distribution of genetic alterations in both molecular subsets (Table 1). nnMCL has a low number of driver genes per case (median, <2 per case), whereas cMCL accumulates alterations in several driver genes (median, 7 per case) affecting multiple pathways simultaneously.5 Virtually all cases (cMCL and nnMCL) had initial CCDN1 alteration and cell cycle deregulation, and additional inactivation of the cell cycle inhibitors CDKN2A/B, RB1, as well as MIR17HG, CDK4, and BMI1 oncogene amplifications. Besides cell cycle, the most altered pathway is DDR, with very frequent and early ATM inactivation. TP53, with multiple roles, especially in the cell cycle and DDR, is also frequently altered in MCL. Inactivation of ATM and TP53 genes is frequently biallelic, usually by mutation or deletion.5,32,50 Recent WGS, whole-exome sequencing (WES), and targeted approaches allowed the identification of new alterations in cell cycle genes (SAMHD1, CDKN1B), and alterations in new pathways such as telomere maintenance (TERT), chromatin remodelers (KMT2D/C, NSD2, SMARCA4, and SP140), NF-κB signaling pathways (BIRC3, CARD11, TRAF2, and TLR2), NOTCH pathway (NOTCH1/2), and other genes (HNRNPH1 and UBR5).5,32,33,35-42 In 42 blastoid MCL analyzed by WES, mutations of NOTCH2, NOTCH3, and UBR5 were exclusively found, whereas NOTCH1 mutations were enriched in blastoid compared with nonblastoid forms.33
We performed a systematic review of 11 recent WGS and WES studies of MCL5,32,33,35-42 at diagnosis, including 432 cases, considering only single nucleotide variants and insertions and deletions. The most frequently altered (>13%) genes were ATM, TP53, CCND1, KMT2D, and NSD2 (Figure 2C; Table 1). The distribution of mutations varied significantly among MCL subtypes, with nnMCL characterized by CCND1 mutations (mainly due to aSHM) found in 85% of cases and TP53 in 25%, whereas the most recurrently mutated genes in cMCL include ATM, KMT2D, NSD2, and TP53 (Figure 2D).
A recent study32 using WES coupled with RNA sequencing (RNA-seq) identified 4 distinct MCL clusters: C1 has IGHV mutations, CCND1 aSHM, and active B-cell receptor (BCR) signaling; C2 is enriched with ATM alterations and upregulation of NF-κB and DDR pathways; C3 with SP140, NOTCH1, and NSD2 mutations and downregulation of BCR signaling and MYC targets; and C4 with TP53, RB1, CDKN2A/B inactivation, active MYC pathway, and high proliferation. Besides the nnMCL subtype (cluster C1) with well-known different clinicobiological features, the other 3 clusters (C2, C3, and C4) may represent cMCL with different underlying biology, which may impact prognostic and help personalizing therapy. C2 is enriched in ATM alterations and very frequent and early alterations in MCL but with no prognostic impact, despite its relationship with genome instability. In contrast, C3 and C4 are enriched in multiple and concomitant cell cycle defects, together with MYC deregulation, which make the cells more difficult to eradicate. The differences of BCR functionality observed among the 4 clusters may also be important for the response to different targeted therapies.
Biological prognostic factors
The cMCL molecular subset exhibits a more aggressive clinical course, characterized by shorter overall survival (OS) and a significantly reduced time to treatment compared with nnMCL. This observation has been consistently demonstrated across various studies using diverse methodologies for stratification, including SOX11 immunohistochemistry and gene expression analyses, such as microarrays, RNA-seq, quantitative polymerase chain reaction, and the NanoString signature L-MCL16.5,18,32 Other measures of genomic complexity, such as complex karyotype predict strongly for inferior OS and poor response to therapy,51,52 increased complexity by microarray,18 WGS and also an increased number of driver genes/regions.5,32,33 The genomic complexity in clinical practice can be assessed by karyotype (≥3 alterations),51 SNP or comparative genomic hybridization array (≥6 alterations),18 or WGS (>7 alterations).5 Similar to CLL, the genomic complexity of MCL has an independent prognostic value over TP53 alterations.5,32,33 The presence of chromotripsis, chromoplexia, and breakage-fusion-bridge detected by WGS are frequent in cMCL, enriched in blastoid forms, and is associated with poor outcome.5 Patients with blastoid histology, high proliferation levels (Ki-67, ≥50%), and a particular mutational profile (CCND1, NOTCH1, TP53, SPEN, SMARCA4, KMT2C, RANBP2, and NOTCH2) had very dismal outcomes.33 The 4 clusters defined by WES/RNA-seq present also distinct outcomes, with C1 having good prognosis, C2 and C3 intermediate, and C4 very dismal prognosis.32
Regarding individual genes, the key prognostic genetic alterations are TP53 (mutations and deletions) detected in both MCL subtypes, and CDKN2A/B deletions detected mainly in cMCL. Both alterations are especially enriched in blastoid and highly proliferative cases.5,33 Simultaneous alterations in TP53 and CDKN2A/B seem to have an additive prognostic effect and a dismal outcome despite the high-dose cytarabine received.53,TP53 mutations are associated with inferior OS and time to treatment in both cMCL and nnMCL subtypes.34,54,55 Whether only TP53 mutations or also TP53 deletions without mutation are associated with the worse outcomes is still unclear.32,54,CCND1 3’ UTR truncations are associated with aggressive behavior,10 and several other genes, such as KMT2D, NOTCH1, NOTCH2, SMARCA4, TRAF2, and SP140, and the recently described HNRNPH1.5,32,36,55,56,MYC gains have also been associated with worse outcomes,5,57 whereas others report that MYC translocations, rather than gains, are independent MCL prognostic factor.58
In 2 WES studies with 1132 and 2535 patients with MCL samples before and after standard chemoimmunotherapy, the authors noted a very high frequency of acquired CDKN2A deletions, TP53 mutations/deletions, and less frequently other high-risk genetic alterations, such as KMT2D, NOTCH1, NOTCH2, SMARCA4, and SP140. The resistant MCL clones detected at relapse were already present at diagnosis and were selected for therapy.35
Clinical prognostic factors
A prognostic score that has been confirmed in numerous series, the MIPI (MCL International Prognostic Index), was established by implementing 4 independent prognostic factors: age, performance status, lactate dehydrogenase, and leukocyte count.59 To allow a simplified calculation, a simplified MIPI has been defined. Although these scores have been confirmed in numerous studies, it became obvious that patient age may strongly influence them; therefore, they are not appropriate to guide individual treatment strategies. Alternatively, the most important biological prognostic markers independent of clinical features are the cell proliferation rate, as measured by Ki-67 expression and alterations of p53, and may even allow a more individualized risk-based therapeutic approach.60 Accordingly, a combined MIPI score (MIPI-c) integrating the clinical and biological features has been subsequently established. In fact, immunohistochemical determination of p53 expression has been prospectively confirmed as a reliable prognostic marker for p53 mutation analysis,61 and this combined clinicobiological score, combining MIPI with p53 high expression and Ki-67>30%, together with blastoid morphology, was recently reported to define high-risk cases with significantly shorter failure-free and OS,62 and may identify patients who may benefit from more experimental therapeutic approaches.
Furthermore, a cell proliferation gene signature (MCL35) that distinguishes patient subsets differing by >5 years in median survival has been identified63 and validated in diagnostic material from patients treated in the prospective MCL Younger (NCT00209222) and MCL Elderly (NCT00209209) trials of the European MCL Network.64,65
Early relapses between the first 2 years (progression of disease ≤24 months [POD24]) are associated with a significantly worse outcome.66-69 Most interestingly, patients with high MIPI-c and no POD24 event had the same prolonged OS compared with patients with low or intermediate MIPI-c and no early progression (median OS not reached).66
Concerning the prognostic impact of minimal residual disease (MRD) status, several studies have provided evidence of its strong prognostic potential predicting improved subsequent progression-free survival (PFS) for patients who are MRD-negative at the end of induction and before high-dose consolidation.34,70 Furthermore, the lack of molecular remission after the end of the currently recommended standard treatment has been shown to be strongly predictive of early clinical relapse within 1 to 2 years.70,71 However, the use of MRD analysis in clinical routine is still limited and the impact of MRD monitoring in the context of new targeted treatments remains unclear.
Molecular-targeted therapies
Covalent BTKis
Constitutive activation of BCR signaling is a characteristic pathogenic feature of malignant B cells, including MCL. Targeting the BCR pathway with the covalent BTK inhibitor (BTKi) ibrutinib resulted in remarkable response rates, leading to its approval for relapsed MCL.72 A pooled analysis (n = 370) of the results of 3 different trials testing ibrutinib as monotherapy revealed an overall response rate (ORR) of 66% with median PFS and OS of 13 and 25 months, respectively, which was even higher when patients were treated with ibrutinib at the first relapse compared with treatment at later relapses (ORR: 1 prior line 78% vs >1 prior line 67%; median PFS: 1 prior line 25 months vs >1 prior line 10 months).73 Based on these results and the results of a randomized phase 3 trial comparing ibrutinib to temsirolimus monotherapy in relapsed MCL, reporting a significant improvement in PFS for patients treated with ibrutinib vs temsirolimus (15 months vs 6 months),74 treatment with BTKi monotherapy was implemented as the standard of care for patients at first relapse.
Second-generation BTKi acalabrutinib was approved in October 2017 by the Food and Drug Administration for patients with relapsed/refractory (R/R) MCL who had received at least 1 prior therapy based on promising results, especially regarding tolerability, in an open-label, multicenter, single-arm phase 2 study of acalabrutinib.75 The final analysis of this trial including 124 patients, reported ORR and complete response (CR) rates of 82% and 48%, respectively, and a median PFS of 22 months.76 Another next-generation BTKi, zanubrutinib, is a highly potent, selective, bioavailable, and irreversible BTKi with maximized BTK occupancy. It was approved in 2019 in the United States and China for the treatment of patients with R/R MCL, based on results from a phase 2 study enrolling 86 Chinese patients with R/R MCL reporting an ORR of 84% and a CR rate of 78%. The median PFS was 33 months, and in patients with mutated TP53, median PFS was 15 months.77,78
Treatment alternatives for patients failing covalent BTKi
Noncovalent BTKi
Pirtobrutinib, a highly selective, noncovalent BTKi, inhibits both wild-type and C481-mutant BTK. This compound is very well tolerated, with only 6% of patients stopping the continuous treatment due to side effects. In 90 evaluable patients with MCL previously treated with covalent BTKi, the ORR observed within the BRUIN phase 1/2 trial was 58%, including 20% CRs.79 The currently recruiting phase 3 BRUIN MCL-321 trial compares pirtobrutinib with the investigator’s choice of any approved covalent BTKi for relapsed MCL.80
Bcl-2 inhibitors
A monotherapy with the bcl2-inhibitor venetoclax might be a promising alternative, as a phase 1 trial showed response rates of 75% in patients with relapsed MCL.81 Recently, an analysis of a retrospective data collection of 20 patients with BTKi-resistance treated with venetoclax in a UK-wide compassionate use program showed an ORR of 53%; however, the median PFS was only 3 months.82 A multicenter analysis of patients with R/R MCL with high-risk disease features and a median of 3 prior treatments including BTKis in 91% reported similar results for venetoclax alone or in combination, with an ORR of 40% and a median PFS of 3.7 months.30 Another study evaluated the outcomes of 24 multiple relapsed patients who received venetoclax-based therapies. Among them, 67% had progressed on BTKi, the ORR was 50%, and the PFS was 8 months.83
Immunomodulators
Several studies have confirmed the benefit of the orally available immunomodulatory drug lenalidomide in relapsed MCL, with response rates of 35% to 50%.84-86 In a randomized phase 2 trial, this approach was superior to monochemotherapy (response rate 46% vs 23%).87 However, in BTKi-failures response rates at least of lenalidomide alone are only modest.88
CAR T cells
The autologous CD19 chimeric antigen receptor (CAR) T-cell construct (CD28 costimulatory domain) brexucabtagene autoleucel (TECARTUS, KTE-X19) was approved for R/R MCL after a previous BTKi treatment, based on the results of the ZUMA-2 study.89 After a median follow-up of 36 months, treatment with brexucabtagene autoleucel was reported to induce a durable ORR of 91% and PFS of 26 months in patients refractory or intolerant toward BTKi treatment.90 In a subgroup analysis of patients who relapsed between 24 months (POD24), median PFS was substantially shorter than that in the non-POD24 group (11 months vs 29 months), although ORR and CR rates were equivalent in both cohorts.90 Another CD19–directed CAR T-cell product (lisocabtagene maraleucel) with 4-1BB as a costimulatory domain was evaluated in a phase 2 study, TRANSCEND NHL 001 (NCT02631044), for R/R MCL. Primary analysis of 83 patients evaluated reported an ORR of 83% and a CR rate of 72%. The median duration of response was 16 months and the PFS was 15 months. The most common grade ≥3 treatment-emergent adverse events were neutropenia (56%), anemia (38%), and thrombocytopenia (25%). Cytokine release syndrome was reported in 61% of the patients.91 Preclinical data suggest a synergistic effect of ibrutinib on apheresis product fitness, CAR T-cell expansion and toxicity. The phase 2 TARMAC study investigated the combination of time-limited ibrutinib with tisagenlecleucel reporting a 12-months PFS of 75%.92 Overall, the results achieved with CAR T-cell treatment are very promising, with long-lasting remissions even after BTKi-treatment failure. Therefore, CAR T-cell therapy should, to date, be first choice for the group of high-risk patients refractory to BTKi.
Resistance mechanisms to targeted therapies
Ibrutinib
Nearly one-third of patients receiving ibrutinib have been reported to develop primary intrinsic resistance and the clinical outcome of these patients is usually poor, with OS rates between 6 and 8 months.93,94 The molecular mechanisms underlying resistance to ibrutinib have been extensively studied, and sustained activation of the phosphoinositide-3-kinase-AKT pathway or other genetic alterations providing alternative activation of BCR signaling seem to play an important role.95 Mutations in CARD1140 and CCND113 have been reported to mediate BTKi resistance and upregulated BIRC5/survivin expression due to 17q gain was recently reported to contribute to resistancy.96 Sequencing of 165 samples from patients with MCL identified recurrent mutations in TRAF2 or BIRC3 in 15% of the patients resistant to ibrutinib treatment.97 In line with this, it was reported that MCL cell lines resistant to BTKi treatment displayed activation of the alternative NF-κB pathway, associated with genetic lesions in this pathway.98 Another report confirmed mutations in NF-κB signaling pathways, both canonical (eg, TNFAIP3/A20) and noncanonical (eg, BIRC2) to be associated with ibrutinib resistance.99 The well-described point mutation within the kinase domain of BTK at the cysteine 481 position (BTKC481S) and gain-of-function mutations in the PLCG2 gene have been reported to confer secondary resistance to BTKi. However, unlike in CLL, BTKC481S mutation is infrequent in MCL and PLCG2 gene mutations have not been observed.100
Besides the genetic changes underlying ibrutinib resistance, metabolic reprogramming to oxidative phosphorylation and glutaminolysis were shown to be associated with therapeutic resistance to ibrutinib in MCL.101-103 RNA-seq analysis of surviving cells after ibrutinib treatment of a sensitive MCL cell line also revealed increased activity of oxidative phosphorylation and upregulated CD52 expression in resistant cells.104
As covalent BTKi will be prospectively applied as part of first-line therapy, the frequency of patients with BTKi-resistance will rise and the identification of optimal treatment alternatives for these patients is highly warranted.
Venetoclax
Venetoclax has proven to be an effective alternative treatment strategy for MCL. However, many initial responders will ultimately progress and deciphering the underlying mechanisms of venetoclax resistance is of great importance. WES was performed on 7 patients, including samples before the initiation of venetoclax and after progression to venetoclax. Alterations in SMARCA4 and BCL2 were only observed after progression. Interestingly, clonal evolution of novel SMARCA4 and KMT2C/D mutations was demonstrated in 2 patients with serial samples.83
Molecular profiling of 24 tumor samples from patients treated within the AIM trial evaluating the combination of ibrutinib and venetoclax detected chromosome 9p21.1–p24.3 (CDKN2A/B) loss and/or mutations in components of the switching defective/sucrose nonfermenting chromatin-remodeling complex in all patients with primary resistance and in two-thirds of patients with relapsed disease.38
Conclusion
Recent studies on MCL have provided novel insights into its pathogenesis, including (1) identification of 2 MCL subsets with distinct cell of origin and clinical behavior; (2) discovery of variant primary translocations beyond the hallmark t(11;14); (3) recognition of a variety of mutated and altered driver genes and regions distributed differently in cMCL and nnMCL subtypes, contributing to their distinct clinical behavior; (4) exploration of combined molecular and clinical prognostic factors associated with poor outcomes; (5) evaluation of the efficacy of molecular-targeted therapies, even in refractory MCL cases; and (6) initial investigations into genetic and nongenetic resistance mechanisms to targeted therapies. Overall, these findings represent significant advancements in our understanding of MCL pathogenesis with potential implications for improved clinical management and patient outcomes. Despite this huge progress, other areas related to the complex MCL biology are yet to be explored and warrant further investigation. These areas include the MCL tumor microenvironment, longitudinal clonal evolution, molecular heterogeneity across different compartments (ie, bone marrow, peripheral blood, lymph nodes, gastrointestinal tract, and plasma), and metabolic reprogramming.
Acknowledgments
This work was mainly developed at the Centre Esther Koplowitz, Barcelona, Spain. The authors are grateful to Marta Grau for her help with the literature review, the preparation of the figures, and reference formatting.
This study was supported by a grant from the Leukemia & Lymphoma Society (M.D. and S.B.), Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III, “Cofinanciado por la Unión Europea” and Fondo Europeo de Desarrollo Regional (FEDER) “Una manera de fer Europa” (PI22/00203 and INT23/00037 [S.B.]), Marató TV3-Cancer (201904-30 [S.B.]), Generalitat de Catalunya Suport Grups de Recerca, Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR) (2021-SGR-01293 [S.B.]), National Institutes of Health, National Cancer Institute (P01CA229100s, participation of S.B. and C.L.), Beatriu de Pinós grant from Secretaria d’Universitats i Recerca del Departament d’Empresa i Coneixement de la Generalitat de Catalunya, and Marie Sklodowska-Curie COFUND program from Horizon 2020 (2018-BP-00055 [C.L.]).
Authorship
Contribution: All authors wrote the review and prepared the table and figures.
Conflict-of-interest disclosure: M.D. has received research support from AbbVie, Bayer, Bristol Myers Squibb (BMS)/Celgene, Gilead/Kite, Johnson & Johnson Innovative Medicine, Lilly, and Roche; speakers honoraria from AstraZeneca, BeiGene, Gilead/Kite, Janssen, Lilly, Novartis, and Roche; and serves on scientific advisory board for AbbVie, AstraZeneca, BeiGene, BMS/Celgene, Gilead/Kite, Janssen, Lilly/Loxo, Novartis, and Roche. The remaining authors declare no competing financial interests.
Correspondence: Sílvia Beà, Molecular Pathology of Lymphoid Neoplasms, Institut d’Investigacions Biomèdiques August Pi i Sunyer and Hematopathology Section, Hospital Clínic of Barcelona, Villarroel 170, 08036 Barcelona, Spain; email: sbea@clinic.cat.