Key Points
Responses to inotuzumab were observed in all leukemic subtypes, genomic alterations, and risk groups.
Inotuzumab demonstrated superior efficacy vs standard-of-care in patients with high-risk BCR::ABL1–like subtype.
Visual Abstract
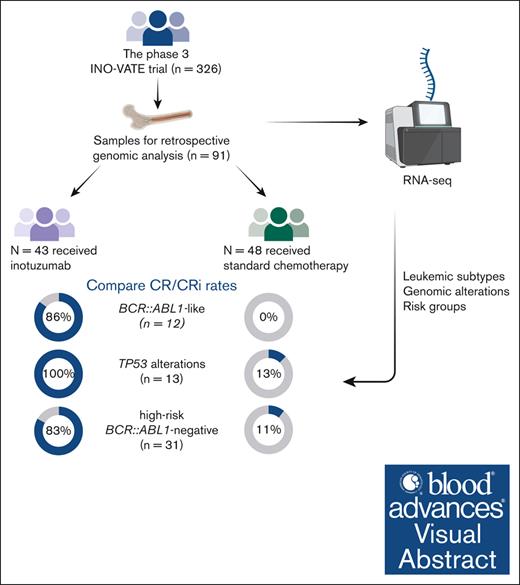
The phase 3 INO-VATE trial demonstrated higher rates of remission, measurable residual disease negativity, and improved overall survival for patients with relapsed/refractory (R/R) acute lymphoblastic leukemia (ALL) who received inotuzumab ozogamicin (InO) vs standard-of-care chemotherapy (SC). Here, we examined associations between genomic alterations and the efficacy of InO. Of 326 randomized patients, 91 (InO, n = 43; SC, n = 48) had samples evaluable for genomic analysis. The spectrum of gene fusions and other genomic alterations observed was comparable with prior studies of adult ALL. Responses to InO were observed in all leukemic subtypes, genomic alterations, and risk groups. Significantly higher rates of complete remission (CR)/CR with incomplete count recovery were observed with InO vs SC in patients with BCR::ABL1–like ALL (85.7% [6/7] vs 0% [0/5]; P = .0076), with TP53 alterations (100% [5/5] vs 12.5% [1/8]; P = .0047), and in the high-risk BCR::ABL1– (BCR::ABL1–like, low-hypodiploid, KMT2A-rearranged) group (83.3% [10/12] vs 10.5% [2/19]; P < .0001). This retrospective, exploratory analysis of the INO-VATE trial demonstrated potential for benefit with InO for patients with R/R ALL across leukemic subtypes, including BCR::ABL1–like ALL, and for those bearing diverse genomic alterations. Further confirmation of the efficacy of InO in patients with R/R ALL exhibiting the BCR::ABL1–like subtype or harboring TP53 alterations is warranted. This trial was registered at www.ClinicalTrials.gov as #NCT01564784.
Introduction
The advent of genomics has transformed the landscape of acute lymphoblastic leukemia (ALL), with better-defined subtypes that are associated with prognosis and that drive risk stratification with conventional therapies.1,2 The associations between ALL subtypes and outcomes with novel therapies are poorly understood; however, greater understanding of these associations may lead to the identification of new therapeutic approaches and improvement of existing therapies.
Inotuzumab ozogamicin (InO) is a calicheamicin-conjugated antibody that targets cluster of differentiation 22 (CD22) on leukemic blast cells and has demonstrated clinical activity in patients with relapsed/refractory (R/R) ALL.3-7 In the phase 3 INO-VATE trial, treatment with InO vs standard-of-care chemotherapy (SC) significantly improved rates of remission (80.7% vs 29.4%; P < .001) and measurable residual disease (MRD) negativity in responding patients with R/R ALL.7 After ≥2 years of follow-up, the median overall survival (OS) with InO was 7.7 months vs 6.2 months with SC (hazard ratio [HR], 0.75; 97.5% confidence interval [CI], 0.57-0.99; 1-sided P = .0105), with more responding patients proceeding directly to hematopoietic stem cell transplantation (HSCT) with InO than with SC (39.6% vs 10.5%; P < .0001).8 INO-VATE exploratory analyses have demonstrated InO clinical benefit across cytogenetic subgroups, including patients with BCR::ABL1+ R/R ALL.9,10 In the first of these analyses, rates of complete remission (CR) and CR with incomplete hematologic recovery (CRi) were higher in patients treated with InO than in those treated with SC, in all cytogenetic subgroups (BCR::ABL1+ ALL, complex cytogenetics [≥5 abnormalities]; normal diploid karyotype; and other cytogenic abnormalities); however, analyses were based primarily on karyotype data, which limited the ALL subgroups that could be evaluated.9 In the BCR::ABL1+ leukemic subtype, CR/CRi and MRD negativity were higher with InO (CR/CRi, 73%; MRD, 81%) vs SC (CR/CRi, 56%; MRD, 33%); however, the study findings were limited by the small number of patients, limited availability of cytogenic and molecular data to fully classify leukemia subtypes, heterogeneity of prior therapy, and no stratification at randomization for BCR::ABL1 status.10
Here, we further characterize the molecular profile of patients with R/R ALL in the INO-VATE trial and assess the impact of leukemic subtypes and alterations, and risk groups on the efficacy of InO vs SC.
Methods
Study design, patients, and treatment
INO-VATE (ClinicalTrials.gov identifier: NCT01564784) was an open-label, randomized, phase 3 trial; methods have been previously described.7 Briefly, adult patients with R/R B-cell ALL (B-ALL; ≥5% bone marrow blasts based on local morphologic analysis), CD22+, BCR::ABL1+, or BCR::ABL1− ALL who were due to receive their first or second salvage-treatment were enrolled. Patients were randomized 1:1 to receive InO or an SC regimen of the investigator’s choice, consisting of fludarabine/cytarabine (Ara-C)/granulocyte colony–stimulating factor, Ara-C/mitoxantrone, or high-dose Ara-C. Stratification at randomization was based on duration of first remission (<12 vs ≥12 months), salvage-treatment phase (1 vs 2), and age (<55 vs ≥55 years).
The study was conducted in accordance with the Declaration of Helsinki. Patients provided written informed consent, and the protocol was approved by the independent ethics committee and/or institutional review board at each study site. Differences between treatment groups were tested using χ2 test or Fisher exact test. This post hoc analysis was based on the final data from 4 January 2017.
Laboratory investigations
Bone marrow samples were collected at screening from after protocol amendment 2 (24 June 2013). Total RNA was extracted from frozen bone marrow cell pellets using Direct-zol RNA Miniprep Kit (Zymo Research Corp, Irvine, CA) and checked for integrity (TapeStation, Agilent Technologies, Santa Clara, CA) and quantity (QUBIT RNA assay, Invitrogen, Waltham, MA). Total stranded transcriptome sequencing (RNA-sequencing) was performed using the TruSeq Stranded Total RNA library preparation kit and sequencing performed using NovaSeq 6000 platforms (Illumina Inc, San Diego, CA). Genetic subtypes were determined by integrating gene expression, rearrangement, and copy number, as previously described.11,12 Single-nucleotide variants (SNVs) and short inserts and deletions (indels) were called from RNA-sequencing data by following the best practice workflow from the Genome Analysis Toolkit.13 The following criteria were applied to filter mutations: (1) at least 3 reads supported the mutation, and the mutant allele frequency was >0.1; and (2) the mutation was not observed in the common single-nucleotide polymorphism database, dbSNP 150.
Outcomes
Outcomes were reported by leukemic subtype and genomic alterations at screening. Patients were assigned to risk groups based on a classification of leukemic subtypes previously described by Paietta et al2: high-risk BCR::ABL1+ (BCR::ABL1+); high-risk BCR::ABL1− (defined as BCR::ABL1–like, low-hypodiploid, and KMT2A-rearranged); standard to intermediate risk (defined as DUX4-rearranged, hyperdiploid, MEF2D-rearranged, PAX5alt, TCF3::PBX1, and ZNF384-rearranged); undetermined risk (defined as CDX2/UBTF, ZEB2/CEBP, and B-other [cases without an identified subtype-defining driver genetic alteration]); and tumors unable to be classified because of low-blast content (<30%) of samples.2 Although recently published data suggest CDX2/UBTF to be a high-risk leukemic subtype,12,14 the number of patients evaluated has been small, and in our analysis we used the classification scheme reported by Paietta et al that considered CDX2/UBTF to be in the undetermined risk group.2
Response and disease progression were assessed using modified Cheson criteria.15 MRD negativity was assessed as <0.01% bone marrow blasts centrally analyzed using multicolor, multiparameter flow cytometry. Progression-free survival (PFS) was defined as the time from randomization to first event of disease progression, starting new induction therapy, or poststudy HSCT without achieving CR or CRi, or death from any cause. OS was defined as the time from randomization to death from any cause.
Statistical analysis
Analyses were carried out on the intent-to-treat (ITT) population. Median PFS and OS were estimated using the Kaplan-Meier method, with the HR and corresponding 97.5% CIs calculated using a Cox proportional hazard regression model.16 Unstratified HR and corresponding CI are displayed, except for “all patients (stratified),” which used the same stratification factors for randomization. Genes with an overall alteration prevalence cutoff of ≥8 patients were selected for correlative analysis. No additional candidate genomic alterations associated with efficacy differences emerged based on a further exploratory analysis based on alteration prevalence of 5 to 7 patients (not shown). P values were not corrected for multiple testing.
Results
Patients
Of 326 patients in the INO-VATE ITT population, 91 patients (InO, n = 43; SC, n = 48) had samples evaluable for molecular-profiling. Baseline characteristics were generally similar between the molecular-profiling cohort and the overall ITT population and were also comparable between study arms in the molecular-profiling cohort, although there were some variations because of the small sample size (Table 1).8
Molecular alterations
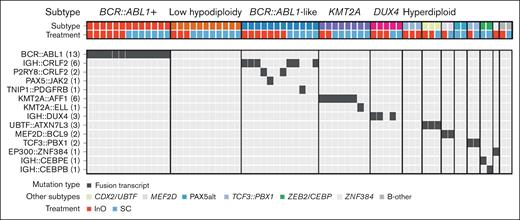
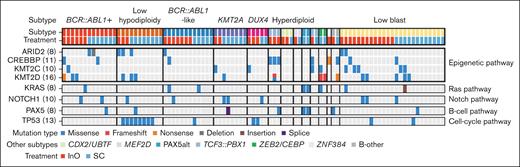
Thirteen different leukemic subtypes were identified (Table 2). The most common (≥5%) subtypes across treatment arms were BCR::ABL1+ (14.3%), BCR::ABL1–like (13.2%), low hypodiploidy (12.1%), KMT2A rearranged (8.8%), and DUX4 rearranged (5.5%); 25 of 91 (27.5%) patients could not be subtyped because of low-blast content of samples. The most common (≥5%) non–BCR::ABL1 rearrangements were IGH::CRLF2 (6.6%) and KMT2A::AFF1 (6.6%) detected in the BCR::ABL1–like and KTM2A-rearranged subtypes, respectively (Figure 1). The majority of patients with BCR::ABL1–like subtype had CRLF2/JAK pathway fusions (9/12; Figure 1), rather than ABL-class fusions (1 patient had TNIP1::PDGFRB). Among nonrearrangement genomic alterations, sequence mutations of chromatin-modifying genes were common (KMT2D, 17.6%; CREBBP, 12.1%; KMT2C, 11.0%; and ARID2, 8.8%; Figure 2; supplemental Table 1). Other common gene alterations included TP53 (14.3%), NOTCH1 (11.0%), and KRAS (8.8%). Most of the TP53 mutations (8/13 [62%]) were observed in low-hypodiploid B-ALL (Figure 2; supplemental Table 2).
Gene fusions of interest by ALL subtype. The most common subtypes across treatment arms were BCR::ABL1+, BCR::ABL1–like, low hypodiploidy, KMT2A rearranged, and DUX4 rearranged. The most common non–BCR::ABL1 rearrangements were IGH::CRLF2 detected in the BCR::ABL1–like subtype, and KMT2A::AFF1 detected in the KMT2A rearranged subtype. Oncoprint displays cases of both canonical and noncanonical fusion orientation. The numbers in parentheses denote the number of patients carrying that gene fusion.
Gene fusions of interest by ALL subtype. The most common subtypes across treatment arms were BCR::ABL1+, BCR::ABL1–like, low hypodiploidy, KMT2A rearranged, and DUX4 rearranged. The most common non–BCR::ABL1 rearrangements were IGH::CRLF2 detected in the BCR::ABL1–like subtype, and KMT2A::AFF1 detected in the KMT2A rearranged subtype. Oncoprint displays cases of both canonical and noncanonical fusion orientation. The numbers in parentheses denote the number of patients carrying that gene fusion.
Common nonfusion genomic alterations by ALL subtype and molecular pathway. Among nonrearrangement genomic alterations, sequence mutations of chromatin-modifying genes were common. Other common gene alterations included TP53, NOTCH1, and KRAS. The numbers in brackets denote the number of patients carrying that genomic alteration.
Common nonfusion genomic alterations by ALL subtype and molecular pathway. Among nonrearrangement genomic alterations, sequence mutations of chromatin-modifying genes were common. Other common gene alterations included TP53, NOTCH1, and KRAS. The numbers in brackets denote the number of patients carrying that genomic alteration.
Efficacy
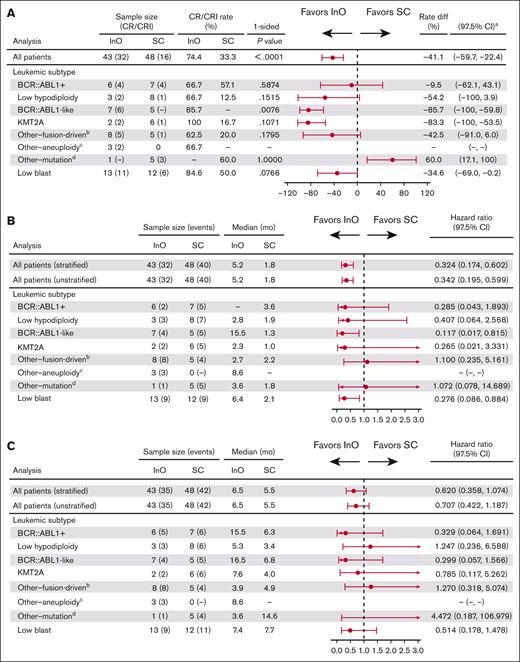
In 91 patients evaluated for response by molecular genetic subset, the CR/CRi rate was 74.4% in the InO group vs 33.3% in the SC group (P < .0001; Figure 3A). CR/CRi rates were higher with InO than with SC across leukemic subtypes, with the difference reaching statistical significance in the BCR::ABL1–like subtype (6/7 patients [85.7%] vs 0/5 patients [0%], respectively; P = .0076; the patient who did not respond to InO had IGH::CRLF2+ ALL). Among patients in the InO group who achieved CR/CRi, MRD negativity was typically achieved. MRD negativity was achieved by all 4 responding patients with the BCR::ABL1+ subtype and all 6 responding patients with the BCR::ABL1–like subtype, compared with 1 of 4 and 0 of 0 patients, respectively, treated with SC. A trend toward a PFS benefit with InO was observed across leukemic subtypes and a trend toward OS benefit with most subtypes (Figure 3B-C).
Efficacy outcomes with InO or SC by ALL subtype. (A) CR/CRi rates were higher with InO than with SC across leukemic subtypes, with the difference reaching statistical significance in the BCR::ABL1–like subtype. (B) A trend toward benefit with InO was observed across leukemic subtypes for PFS; and (C) across most subtypes for OS. aCIs were approximated using nominal distribution when ≥5 patients were in both the CR/CRi and non–CR/CRi subgroups. Where <5 patients were in either subgroup, exact method was used; bincluded DUX4, CDX2/UBTF, MEF2D, TCF3::PBX1, and ZNF384; cincluded hyperdiploid; dincluded B-other, PAX5alt, and ZEB2/CEBP.
Efficacy outcomes with InO or SC by ALL subtype. (A) CR/CRi rates were higher with InO than with SC across leukemic subtypes, with the difference reaching statistical significance in the BCR::ABL1–like subtype. (B) A trend toward benefit with InO was observed across leukemic subtypes for PFS; and (C) across most subtypes for OS. aCIs were approximated using nominal distribution when ≥5 patients were in both the CR/CRi and non–CR/CRi subgroups. Where <5 patients were in either subgroup, exact method was used; bincluded DUX4, CDX2/UBTF, MEF2D, TCF3::PBX1, and ZNF384; cincluded hyperdiploid; dincluded B-other, PAX5alt, and ZEB2/CEBP.
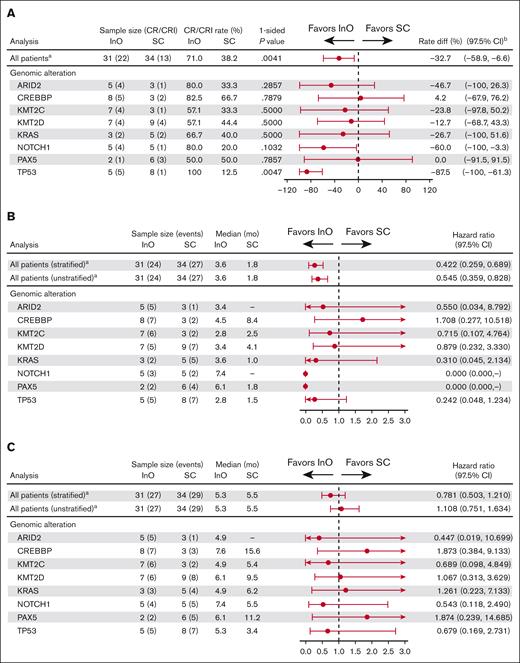
It was evident that across genomic alterations, there was a potential for response to InO treatment (Figure 4A). Of note, CR/CRi rates were significantly higher in patients with TP53 alterations who received InO (5/5 [100%]) than SC (1/8 [12.5%]; P = .0047). Although a trend toward a PFS benefit was observed across genomic alterations, no such trend was observed for OS (Figure 4B-C). Responses were observed in 1 of 3 (33.3%) patients with BCR::ABL1+ ALL and the ABL1 T315I mutation treated with InO compared with 3 of 6 (50%) patients treated with SC (P = not significant).
Efficacy outcomes with InO or SC by select genomic alterations. (A) Across genomic alterations, there was a potential for response to InO treatment, and CR/CRi rates were significantly higher in patients with TP53 alterations who received InO. (B) A trend toward a PFS benefit was observed across genomic alterations; however, (C) no such trend was observed for OS. aAll patients bearing an alteration in a gene with an overall alteration prevalence n of at least 8; bCIs were approximated using nominal distribution when ≥5 patients were in both the CR/CRi and non–CR/CRi subgroups. Where <5 patients were in either subgroup, exact method was used.
Efficacy outcomes with InO or SC by select genomic alterations. (A) Across genomic alterations, there was a potential for response to InO treatment, and CR/CRi rates were significantly higher in patients with TP53 alterations who received InO. (B) A trend toward a PFS benefit was observed across genomic alterations; however, (C) no such trend was observed for OS. aAll patients bearing an alteration in a gene with an overall alteration prevalence n of at least 8; bCIs were approximated using nominal distribution when ≥5 patients were in both the CR/CRi and non–CR/CRi subgroups. Where <5 patients were in either subgroup, exact method was used.
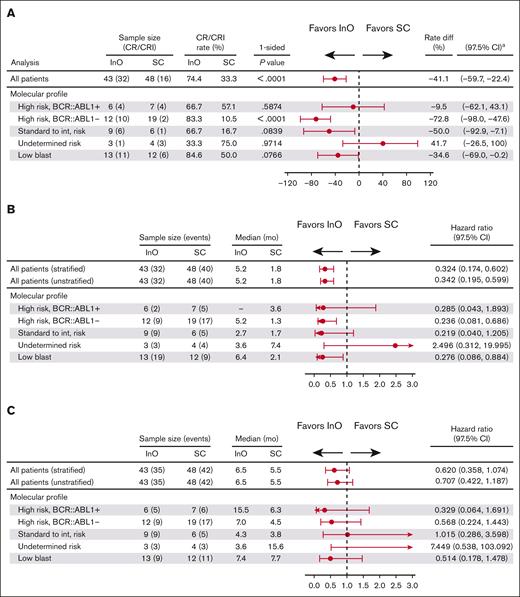
In an evaluation of efficacy outcomes according to risk group based on a classification of leukemic subtypes (high-risk BCR::ABL1+, high-risk BCR::ABL1−, standard to intermediate risk, undetermined risk, and low-blast; Table 2 for full definitions of risk groups), response to InO was observed across the risk groups (Figure 5A). In the high-risk BCR::ABL1− group, CR/CRi rates were significantly higher with InO (10/12 [83.3%]) compared with SC (2/19 [10.5%]; P < .0001). A trend toward a PFS benefit was evident across most risk groups, which was significant in the high-risk BCR::ABL1− group (HR, 0.236; 97.5% CI, 0.081-0.686) and the low-blast content group (HR, 0.276; 97.5% CI, 0.086-0.884; Figure 5B). Similarly, a trend toward OS benefit was observed across most risk groups (Figure 5C). In general, trends for the low-blast content group appeared most similar to the overall “all patients” group that combined all risk groups (Figures 3 and 5), likely reflecting that the low-blast population had a similar fraction of subtypes as the overall cohort.
Efficacy outcomes with InO or SC by leukemic subtype–based risk group. (A) Response to InO was observed across risk groups group based on a classification of leukemic subtypes. In the high-risk BCR::ABL1− group, CR/CRi rates were significantly higher with InO compared with SC. (B) A trend toward a benefit with InO was evident across most risk groups for PFS; (C) OS. aCIs were approximated using nominal distribution when ≥5 patients were in both the CR/CRi and non–CR/CRi subgroups. Where <5 patients were in either subgroup, exact method was used. int, intermediate.
Efficacy outcomes with InO or SC by leukemic subtype–based risk group. (A) Response to InO was observed across risk groups group based on a classification of leukemic subtypes. In the high-risk BCR::ABL1− group, CR/CRi rates were significantly higher with InO compared with SC. (B) A trend toward a benefit with InO was evident across most risk groups for PFS; (C) OS. aCIs were approximated using nominal distribution when ≥5 patients were in both the CR/CRi and non–CR/CRi subgroups. Where <5 patients were in either subgroup, exact method was used. int, intermediate.
Discussion
This retrospective analysis of the INO-VATE trial demonstrated potential for benefit with InO for patients with R/R ALL across leukemic subtypes and for patients bearing different genomic alterations. Although the number of cases of ALL for several subtypes was small, the spectrum of gene fusions and other genomic alterations observed was generally consistent with that previously reported for adult ALL.2,16
Similar to previously reported findings of the INO-VATE trial, response rates were higher with InO than with SC across leukemic subtypes.7-9 A notable finding was the low response rate observed in patients with BCR::ABL1–like ALL who were treated with SC (0/5 [0%]) vs InO (6/7 [85.7%]), with the difference reaching statistical significance. The lack of response in the SC group was not surprising given the small sample size and the poor prognosis of this patient group2; nevertheless, the results with InO for this high-risk subtype are encouraging.
The potential for response with InO was also evident across molecular alterations, including patients with ALL harboring TP53 mutations, for whom the CR/CRi rate was significantly higher for patients receiving InO (5/5 [100%]) than those receiving SC (1/8 [12.5%]). Higher response rates with InO in patients with TP53 alterations may reflect sensitization to calicheamicin-induced double-strand DNA breaks via abrogation of DNA repair and/or cell cycle checkpoints.17 In contrast, in cellular models, TP53 mutations have been reported to be associated with resistance to InO, which can be overcome via overexpression of wild-type TP53.18TP53 alterations were significantly associated with low-hypodiploid ALL, with 8 of 13 (61.5%) TP53 mutations occurring in this high-risk subtype, which is consistent with the literature.2,19,20TP53 alterations have been identified as a hallmark of low hypodiploidy in both adults and children with ALL, with many of the alterations in children present in the germ line.21 Our results warrant further evaluation of the interplay between TP53 mutation, low-hypodiploid subtype, and InO efficacy in a larger cohort of patients.
Although all patients with TP53 alterations treated with InO achieved a response, no significant PFS or OS benefit was observed, which may reflect the poor prognosis associated with TP53 alterations,19 especially in the R/R setting. In a study of patients with R/R disease with TP53 mutations, treatment with mini-hyper-CVD (referring to low-intensity therapy administered on a hyperfractionated schedule) and InO was associated with high morphologic response rate (16/16 [100%]) but a similarly short OS (median OS, 5 months). The authors suggested that the timing of the mutation (baseline or R/R) was important, with inherently more aggressive TP53 mutant clones expanding after chemotherapy.22
A previous analysis of patients with BCR::ABL1+ R/R ALL in the INO-VATE trial demonstrated a substantial improvement in responses and HSCT rates with InO vs SC.10 The authors discussed the potential ability of InO to target BCR::ABL1+ ALL regardless of ABL-kinase mutational status, and highlighted the need to examine concurring mutations in patients with R/R BCR::ABL1+ ALL (ABL- kinase mutation, particularly T315I). Our analysis identified the ABL1 T315I mutation in 9 of 13 (69.2%) patients with BCR::ABL1+ ALL (data not shown); this is a “gatekeeper” mutation in the ABL-kinase domain and commonly seen in relapse after treatment with first- and second-generation tyrosine kinase inhibitors.10,23,24
The number of patients with the KMT2A-rearranged subtype treated with InO was small (n = 2), which limits the interpretability of these data. Interestingly, both patients had a relatively high CD22 positivity (90% and 84%, respectively), which is atypical because KMT2A rearrangements have been associated with lower CD22 positivity, and both patients achieved CR/CRi.25 The percentage of CD22+ blasts fell to <10% after only 1 cycle of InO treatment, which was a similar but more rapid decline compared with that typically seen for CD22 positivity in the INO-VATE study.25 Regarding efficacy and progression in these patients, MRD in patient 1 was 90.8% at screening; 7.2% at cycle 1, day 20; and 1.4% at cycle 2, day 20, consistent with robust initial benefit. However, PFS was relatively short at 2.8 months. Likewise, for patient 2, MRD was 60.7% at screening; and 1.2% at cycle 1, day 21, with a PFS of 1.7 months.
The bispecific T-cell engager blinatumomab has also conferred activity in patients with high-risk BCR::ABL1+ or BCR::ABL1–like R/R ALL. In the phase 2 ALCANTARA study, 16 of 45 (36%) patients with BCR::ABL1+ R/R ALL achieved CR/CRi with blinatumomab, including 10 patients with the ABL1 T315I mutation, with a median OS of 7.1 months.26 In a post hoc analysis from the phase 3 TOWER study, 3 of 9 (33%) patients with BCR::ABL1–like R/R ALL treated with blinatumomab achieved CR/CRi. A retrospective, single-center study has reported a higher response rate of 70% (16/23) in patients with BCR::ABL1–like R/R ALL after treatment with blinatumomab.27,28 Acknowledging the small patient numbers, our results in BCR::ABL1+ or BCR::ABL1–like subtypes compare favorably with these response rates.
The limitations of this analysis include its ad hoc, retrospective nature; the small sample size in subgroups; sample collection being initiated midway through the trial; and some suboptimal samples. In addition, we prioritized leukemic subtype classification, and RNA-sequencing was used to report gene fusions and SNV-indels but not gene deletions. Without DNA-based profiling and gene deletions, some alterations may be underestimated; for example, our study reported IKZF1 SNVs/indels (4/91 [4.4%]), and a further investigation of IKZF1 splicing isoforms identified IK6 isoform (which lacks exons 4-7)29 at 13.2% (12/91) prevalence; because the most common IKZF1 alterations are IKZF1 deletions, this may explain the low rates of IKZF1 alterations reported in our study relative to those reported in the UKALLXII/ECOG-ACRIN E2993 study (31% [41/131]).2
In summary, our results suggest a treatment benefit with InO to patients with R/R ALL across leukemic subtypes, molecular alterations, and risk groups, including poor prognosis, high-risk subtypes. Further confirmation of the efficacy of InO in patients with R/R ALL exhibiting the BCR::ABL1–like subtype or harboring TP53 alterations is warranted.
Acknowledgments
The authors thank all patients who participated in the trial, and medical staff of participating centers. The authors thank Chunxu Qu, Wayne Peng, Vijaya Maroju, Shevali Kansal, and Pfizer Clinical Programming. Medical writing support was provided by Gemma Shay-Lowell and Anne Marie McGonigal of Engage Scientific Solutions, and was funded by Pfizer.
This study was funded by Pfizer. The study was supported by the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital. C.G.M is supported by a National Cancer Institute Outstanding Investigator Award (R35 CA197695) and G.N. reports grant support from the National Cancer Institute (P30 CA021765).
Authorship
Contribution: A.A., C.G.M., A.D.L., and E.V. conceived and designed the study; H.K., D.J.D., M.S., M.L., W.S., N.G., S.O., E.J., R.C.D., and A.A. provided study materials or recruited patients; Y.Z., K.G.R., R.Y., A.D.L., M.R.L., S.O., G.N., and C.G.M. performed experiments and/or assembled data; X.L. designed statistical analyses; and all authors analyzed and interpreted the data, wrote the manuscript, and approved the final manuscript.
Conflict-of-interest disclosure: H.K. received honoraria from AbbVie, Amgen, Daiichi-Sankyo, Pfizer, Bio Ascend, Novartis, Takeda, Adaptive Biotechnologies, Aptitude Health, Delta-Fly, Janssen Global, and Oxford Biomedical; research support from AbbVie, Amgen, Daiichi-Sankyo, and Pfizer; grants from Ascentage, Bristol Myers Squibb, Immunogen, Jazz Pharmaceuticals, and Sanofi; and serves on the advisory board of Actinium. D.J.D. received honoraria from Amgen, Autolus, Agios, Blueprint Pharmaceuticals, Forty Seven, Incyte, Jazz Pharmaceuticals, Kite Pharma, Novartis, Pfizer, Shire, and Takeda, and research support from AbbVie, GlycoMimetics, Novartis, and Blueprint Medicines. M.S. received research support and honoraria from Pfizer. M.L. received research support and honoraria from Pfizer and Amgen. W.S. received research support and/or honoraria from ADC Therapeutics, Agios, Amgen, Astellas, Gilead, Jazz, Kite, Pfizer, and Sigma-Tau. N.G. received research support and honoraria from Pfizer and Amgen. S.O. received research support and honoraria from Pfizer. E.J. received research support and consultancy fees from Amgen, Pfizer, Takeda, Adaptive Biotechnologies, AbbVie, Bristol Myers Squibb, Spectrum, and Genentech. R.D.C. reports consultancy for Amgen, Jazz, Kite/Gilead, and Pfizer; serves on boards and committees of PeproMene Bio and Autolus; received research support from Amgen, Kite/Gilead, Pfizer, Servier, and Vanda Pharmaceuticals; received honoraria from Kite/Gilead; and reports spouse employed by, and owns stocks in, Seagen. X.L. is a previous employee of Pfizer and is now an employee of Mirati Therapeutics Inc. C.G.M. reports research support from Pfizer and AbbVie; received honoraria from Amgen and Illumina; and reports equity in Amgen. A.A. reports consulting for Jazz Pharmaceuticals, Beam Theraputics, GlycoMimetics, Amgen, Pfizer, and Novartis, and received research support from Seattle Genetics, Immunogen, Glycomimetics, AbbVie, Pfizer, Amgen, Kite, and MacroGenics. A.D.L., R.Y., and E.V. are employees of, and own stock in, Pfizer. The remaining authors declare no competing financial interests.
Correspondence: Anjali Advani, Department of Hematology and Medical Oncology, Cleveland Clinic Taussig Cancer Institute, 9500 Euclid Ave, Cleveland, OH 44195; email: advania@ccf.org; and Charles G. Mullighan, St. Jude Children’s Research Hospital, 262 Danny Thomas Pl, Memphis, TN 38105; email: charles.mullighan@stjude.org.
References
Author notes
A.A. and C.G.M. contributed equally to this study.
Upon request, and subject to review, Pfizer will provide the data that support the findings of this study. Additionally, subject to certain criteria, conditions, and exceptions, Pfizer may also provide access to the related individual deidentified participant data. More information is available at https://www.pfizer.com/science/clinical-trials/trial-data-and-results.
The full-text version of this article contains a data supplement.