TO THE EDITOR:
Coagulation factor VIII (FVIII) is essential for hemostasis,1-3 and elevated FVIII is a risk factor for venous thromboembolism (VTE).2-6 The liver is a major source of FVIII, and a subset of liver endothelial cells (ECs), marked by CD32+, produce high levels of FVIII.7-12
FVIII is secreted by vesicle trafficking from the endoplasmic reticulum (ER) to the Golgi and finally to the plasma membrane in a pathway that includes the proteins COPII, lectin mannose binding 1 (LMAN1), and the ER cargo receptor complex MCFD2.13,14 Lipopolysaccharide stimulates EC release of FVIII,15,16 but little is known about other agonists that induce FVIII release from ECs.
Genome-wide association studies of FVIII have identified candidate genes that regulate FVIII levels in humans. A genome-wide association studies and whole-exome sequencing association study found 13 loci associated with plasma FVIII activity, of which 1 locus includes the CD36 gene.17-19 The lead genetic variant of CD36 associated with FVIII, designated rs3211938, is a loss-of-function variant.20 The minor allele frequency of this variant is 8% in individuals of African ancestry, and is not detected in individuals of European ancestry.17 These data identify CD36 as a candidate protein that regulates FVIII levels in humans.
CD36 is a cell surface protein expressed in diverse cells, including ECs.21,22 CD36 is a scavenger receptor that interacts with ligands including long-chain fatty acids and oxidized low-density lipoprotein (oxLDL).21,22 We hypothesized that CD36 regulates FVIII levels by controlling its release from the ECs.
We purified CD32+ human liver ECs (HLEC) from a commercial source of liver ECs, cultured HLEC in vitro, and measured FVIII release by enzyme-linked immunosorbent assay. We silenced CD36 in HLEC, added oxidized LDL and an inhibitor of p38, and measured FVIII release. Detailed methods are provided in supplemental Material.
We first identified HLEC that express FVIII.10-12 We reanalyzed publicly available single cell RNA sequencing (scRNA-Seq) data of the human liver (GSE115469)10 and, consistent with previous reports, identified 3 subpopulations of ECs in the human liver (Figure 1A).10,11 A set of differentially expressed genes separated these 3 subpopulations (Figure 1B). One cluster of ECs (cluster 11) expressed high levels of CD36 and F8 (Figure 1C). This subpopulation also expressed high levels of cell surface protein CD32 (encoded by FCGR2B) (Figure 1C). We refer to these CD32-expressing HLEC as CD32+ HLEC.
A CD32+ subpopulation of human liver ECs (ECs and HLEC) expresses FVIII. A data set for scRNA-seq of human liver cells was analyzed and a subpopulation of ECs was purified and studied. (A) Human liver cells contained 3 subpopulations of vascular ECs. (B) Differentially expressed genes identify subpopulations of ECs, including CD32 (FCGR2B), which marks endothelial cluster 11. Differentially expressed genes were plotted for all-liver cell clusters; the size of the circle indicates the percentage of cells in each population expressing each gene, and the intensity of the color indicates the level of expression. (C) scRNA-Seq demonstrates that CD32+ HLECs express higher levels of F8 and CD36 but lower levels of VWF. The expression of selected genes in each endothelial cluster and hepatocytes is plotted. (D) Expression of CD32 in CD32+ HLEC. HLEC were sorted by CD32+ expression, and CD32 mRNA expression was measured by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (N = 3, mean ± standard deviation [SD], ∗∗P < .005). (E) Localization of CD32 in CD32+ HLEC. CD32+ HLEC were isolated, hybridized with antibody to CD32, and imaged by confocal microscope (green = CD32, blue = DNA). Representative images are shown using the left 20× objective and the right 63× immersion objective (scale bar, 50 μM). (F) CD32+ HLEC express F8. CD32+ mRNA was purified from HLEC and HUVEC, and F8 mRNA expression was measured by qRT-PCR (N = 3, mean ± SD, ∗∗∗P = .0001). (G) CD32+ HLEC release FVIII into the media. Media were collected from CD32+ HLEC at 2, 4, 6, 8, and 24 hours. Top: FVIII antigen was measured by enzyme-linked immunosorbent assay (ELISA) (N = 3, mean ± SD). Bottom: FVIII activity was measured using a chromogenic FVIII activity assay (N = 6, mean ± SD). (H) FVIII was localized in a punctate pattern in CD32+ HLEC. CD32+ HLEC were hybridized with antibody to FVIII and imaged using a confocal microscope. Representative images are shown using the 20× objective and the 63X immersion objective (scale bar, 100 μM and 50 μM, respectively). (I) FVIII and COPII are colocalized in CD32+ HLEC. CD32+ HLEC were hybridized with antibodies to FVIII and COPII and imaged using confocal microscope using the 20× objective and the 63X immersion objective (scale bar, 100 μM and 50 μM, respectively). The Pearson correlation coefficient = 0.62 ± 0.08. (J) CD32+ HLEC express minimal levels of VWF and release minimal levels of VWF into the media. mRNA was isolated from CD32+ HLEC and HUVEC and analyzed by qRT-PCR (top) (N = 6, mean ± SD, ∗∗∗∗P < .0001). Ten thousand HUVEC and HLEC CD32+ cells were seeded in p96-collagen coated plates, cultured for 3 days, the media was replaced, cells were treated with media or 10 μM histamine for 1 hour, and the concentrations of VWF released into the media were measured by ELISA (bottom). (N = 8, mean ± SD, ∗∗∗∗P < .0001).
A CD32+ subpopulation of human liver ECs (ECs and HLEC) expresses FVIII. A data set for scRNA-seq of human liver cells was analyzed and a subpopulation of ECs was purified and studied. (A) Human liver cells contained 3 subpopulations of vascular ECs. (B) Differentially expressed genes identify subpopulations of ECs, including CD32 (FCGR2B), which marks endothelial cluster 11. Differentially expressed genes were plotted for all-liver cell clusters; the size of the circle indicates the percentage of cells in each population expressing each gene, and the intensity of the color indicates the level of expression. (C) scRNA-Seq demonstrates that CD32+ HLECs express higher levels of F8 and CD36 but lower levels of VWF. The expression of selected genes in each endothelial cluster and hepatocytes is plotted. (D) Expression of CD32 in CD32+ HLEC. HLEC were sorted by CD32+ expression, and CD32 mRNA expression was measured by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (N = 3, mean ± standard deviation [SD], ∗∗P < .005). (E) Localization of CD32 in CD32+ HLEC. CD32+ HLEC were isolated, hybridized with antibody to CD32, and imaged by confocal microscope (green = CD32, blue = DNA). Representative images are shown using the left 20× objective and the right 63× immersion objective (scale bar, 50 μM). (F) CD32+ HLEC express F8. CD32+ mRNA was purified from HLEC and HUVEC, and F8 mRNA expression was measured by qRT-PCR (N = 3, mean ± SD, ∗∗∗P = .0001). (G) CD32+ HLEC release FVIII into the media. Media were collected from CD32+ HLEC at 2, 4, 6, 8, and 24 hours. Top: FVIII antigen was measured by enzyme-linked immunosorbent assay (ELISA) (N = 3, mean ± SD). Bottom: FVIII activity was measured using a chromogenic FVIII activity assay (N = 6, mean ± SD). (H) FVIII was localized in a punctate pattern in CD32+ HLEC. CD32+ HLEC were hybridized with antibody to FVIII and imaged using a confocal microscope. Representative images are shown using the 20× objective and the 63X immersion objective (scale bar, 100 μM and 50 μM, respectively). (I) FVIII and COPII are colocalized in CD32+ HLEC. CD32+ HLEC were hybridized with antibodies to FVIII and COPII and imaged using confocal microscope using the 20× objective and the 63X immersion objective (scale bar, 100 μM and 50 μM, respectively). The Pearson correlation coefficient = 0.62 ± 0.08. (J) CD32+ HLEC express minimal levels of VWF and release minimal levels of VWF into the media. mRNA was isolated from CD32+ HLEC and HUVEC and analyzed by qRT-PCR (top) (N = 6, mean ± SD, ∗∗∗∗P < .0001). Ten thousand HUVEC and HLEC CD32+ cells were seeded in p96-collagen coated plates, cultured for 3 days, the media was replaced, cells were treated with media or 10 μM histamine for 1 hour, and the concentrations of VWF released into the media were measured by ELISA (bottom). (N = 8, mean ± SD, ∗∗∗∗P < .0001).
To study the regulation of FVIII release, we isolated CD32+ HLEC from a commercial source by cell sorting (detailed methods are provided in the supplemental Material). CD32+ HLEC expressed CD32 messenger RNA (mRNA) (Figure 1D) and protein (Figure 1E) as detected by quantitative reverse transcription polymerase chain reaction and immunofluorescence microscopy, respectively. Compared with CD32- HLEC, CD32+ HLEC expressed higher levels of CD36 and F8 mRNA as detected by quantitative reverse transcription polymerase chain reaction (supplemental Figure 1A-B), in concordance with results from scRNA-Seq of the human liver.
We next examined CD32+ HLEC expression of F8 and the release of FVIII. CD32+ HLEC expressed F8 mRNA at higher levels than human umbilical vein EC (HUVEC) (Figure 1F) and constitutively released FVIII into the media over time (Figure 1G). FVIII localized in a punctate pattern within HLEC (Figure 1H). FVIII was expressed in CD32+ HLEC but not in other types of ECs (eg, HUVEC or EA.hy926) or hepatoblastoma (HepG2) cells (supplemental Figure 1C). In CD32+ HLEC, FVIII did not colocalize with Weibel-Palade Body markers (CD63 or RAB27, supplemental Figure 1D). CD32+ HLEC did not express Weibel-Palade Body cargo such as von Willebrand factor (VWF) or angiopoietin 2 but HUVEC expressed both VWF and angiopoietin 2 in a punctate pattern (supplemental Figure 1E). FVIII colocalized with COPII, a protein that mediates vesicle transport of cargo from the ER to the Golgi apparatus (Figure 1I).23 Taken together, our data show that CD32+ HLEC express F8, store FVIII in vesicles, and constitutively release FVIII.
Because VWF serves as a carrier for FVIII in the blood, we also compared VWF expression in CD32+ HLEC and HUVEC. We found that CD32+ HLEC did not express VWF mRNA but HUVEC expressed VWF mRNA (Figure 1J, top). Furthermore, HLEC did not constitutively release VWF into the media, whereas HUVEC released more VWF after stimulation with histamine (Figure 1J, bottom).
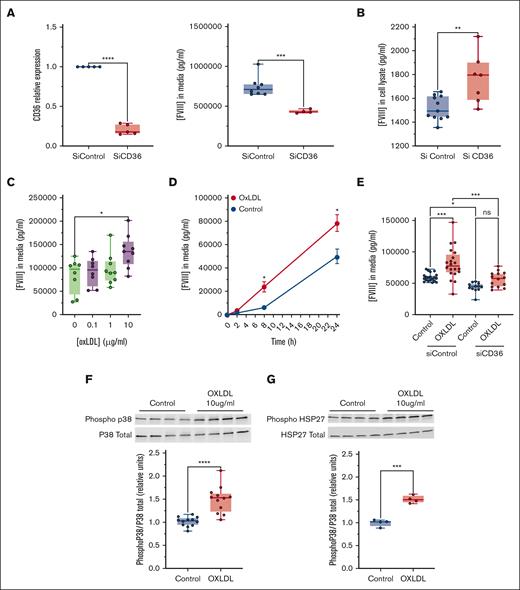
To explore the role of CD36 in FVIII release from ECs, we silenced CD36 in CD32+ HLEC and measured FVIII release. Silencing of CD36 decreased CD32+ HLEC release of FVIII into the media (Figure 2A, right) and increased intracellular FVIII (Figure 2B). These data suggest that CD36 promotes the basal release of FVIII, and in the absence of CD36, FVIII accumulates inside CD32+ HLEC.
The rs3211938 variant changes the CD36 amino acid 325 from tyrosine to a stop codon. Inspection of this variant in the GTEX portal indicated that the arterial expression of CD36 mRNA was reduced in heterozygous carriers (supplemental Figure 2A). To test the effect of this variant on CD36 expression in vitro, we constructed 2 plasmids: 1 encoding mCherry-CD36(wild-type) and 1 encoding mCherry-CD36(Y325X). Although Telo-HAEC cells transfected with mCherry-CD36(wild-type) expressed CD36, cells transfected with mCherry-CD36(Y325X) did not (supplemental Figure 2B). These results suggest that the rs3211938 truncating variant decreases CD36 expression in ECs.
CD36 agonists include long-chain fatty acids and oxLDL.21,22 Therefore, we determined the effects of several CD36 agonists on FVIII release. Oleic acid, 1-(palmitoyl)-2-(5-keto-6-octene-dioyl) phosphatidylcholine, and oxLDL each increased FVIII release from CD32+ HLEC (supplemental Figure 2C; Figure 2C-D). To confirm that CD36 mediates the effects of oxLDL on FVIII release, we treated CD32+ HLEC with oxLDL before or after silencing CD36 and found that CD36 silencing blunted the effect of oxLDL on FVIII release from ECs (Figure 2E). We also tested the ability of CD36 to mediate endocytosis of oxLDL. After silencing CD36 in CD32+ HLEC, oxLDL uptake was impaired compared with that in the siControl (supplemental Figure 2D).
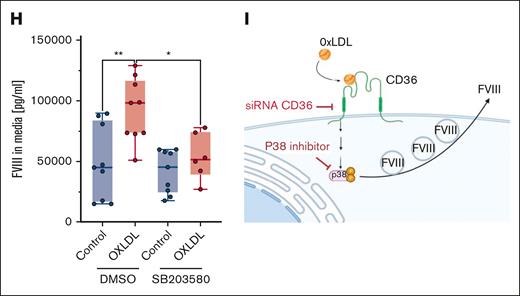
Because the binding of oxLDL to CD36 activates an intracellular signaling cascade,21,22 we next tested whether p38 mediates the effects of CD36 upon FVIII release. First, we treated CD32+ HLEC with vehicle or oxLDL and measured active phospho-p38. OxLDL increased the relative levels of phospho-p38 (Figure 2F) as well as phosphorylation of a downstream target of p38, heat shock protein 27 (HSP27) (Figure 2G). Finally, we tested the effect of blocking p38 on CD36 activation and FVIII release. Oxidized LDL increased the endothelial release of FVIII in control cells but failed to boost the release of FVIII from ECs treated with the p38 inhibitor SB203580 (Figure 2H). Taken together, these data suggest that oxLDL promotes FVIII release from CD32+ HLEC by engaging CD36 and triggering downstream p38 MAPK signaling (Figure 2I).
Despite its central role in hemostasis and thrombosis, FVIII trafficking within ECs is not fully understood. Our findings advance this field in several ways. First, our data are consistent with reports that FVIII secretion is mediated by COPII (Figure 1I)13,14,24 but refute the premise that FVIII release depends on the presence of Weibel-Palade bodies.25 This observation has implications for understanding cellular machinery and is not required to support FVIII production in gene therapy approaches. Second, our findings provide functional validation and define an intracellular mechanism mediating the association between a CD36 variant and a 19% decrease in FVIII levels.17 Characterization of the effect of oxLDL-CD36 engagement on FVIII release offers a potential explanation for elevated FVIII levels in diseases associated with increased oxLDL (eg, atherosclerosis and diabetes) and a hypothesis linking these diseases with VTE risk. We propose that the binding of oxLDL to CD36 increases circulating FVIII and consequently enhances procoagulant activity and thrombotic risk. Finally, if borne out, our findings offer a novel potential strategy for mitigating thrombosis risk related to elevated FVIII levels. All current antithrombotic strategies reduce the levels and/or activity of procoagulant proteins and are accompanied by an increased bleeding risk. CD36 antagonists may represent a different method to decrease FVIII and VTE risk in selected patients, particularly in those with high oxLDL levels.
Appendix
The members of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium Hemostasis Working Group are:
Airwave:
Abbas Dehghan (a.dehghan@imperial.ac.uk)
Dipender Gill (dipender.gill@imperial.ac.uk)
Paul Elliot (p.elliott@imperial.ac.uk)
ARIC:
Nathan Pankratz (pankr018@umn.edu)
Weihong Tang (tang0097@umn.edu)
Peng Wei (Peng.Wei@uth.tmc.edu)
Paul deVries (paul.s.devries@uth.tmc.edu)
Michael Brown (michael.r.brown@uth.tmc.edu)
Jan Bressler (Jan.Bressler@uth.tmc.edu)
Julie Hahn (Julie.M.Hahn@uth.tmc.edu)
Rachel K. Friedman (Rachel.K.Friedman@uth.tmc.edu)
Adam Heath (Adam.S.Heath@uth.tmc.edu)
Alanna Morrison (Alanna.C.Morrison@uth.tmc.edu)
B58C:
David Strachan (sgjd950@sgul.ac.uk)
CARDIA:
Alex P. Reiner (apreiner@u.washington.edu)
CHS:
Matt Conomos (mconomos@uw.edu)
Bruce M. Psaty (psaty@u.washington.edu)
Jen Brody (jeco@u.washington.edu)
Joshua C. Bis (joshbis@u.washington.edu)
Kerri L. Wiggins (kwiggins@uw.edu)
Nicholas L. Smith (nlsmith@u.washington.edu)
CRO/GenScot:
Caroline Hayward (caroline.hayward@igmm.ed.ac.uk)
Anne Richmond (Anne.Richmond@igmm.ed.ac.uk)
FHS:
Chris O’Donnell (Christopher.ODonnell@va.gov)
Jenny Huffman (jennifer.huffman@nih.gov)
Ming-Huei Chen (ming-huei.chen@nih.gov)
Qiong Yang (qyang@bu.edu)
Florian Thibord (florian.thibord@nih.gov)
Melissa Liu (melissa.liu3@nih.gov)
Andrew Johnson (johnsonad2@nhlbi.nih.gov)
GABC:
Karl Desch (kdesch@med.umich.edu)
GAIT2:
Maria Sabater-Lleal (msabater@santpau.cat)
Gerard Temprano (gtemprano@santpau.cat)
Laia Díez (ldieza@santpau.cat)
GeneStar:
Lisa Yanek (lryanek@jhmi.edu)
Genoa:
Patricia Peyser (ppeyser@umich.edu)
Mina Jhun (minajhun@umich.edu)
Gutenberg HS:
Philipp Wild (Philipp.Wild@unimedizin-mainz.de)
JHS:
Alex Reiner (apreiner@u.washington.edu)
Laura Raffield (laura_raffield@unc.edu)
Herman Taylor (htaylor@umc.edu)
Jayna Nicholas (jnicholas@unc.edu)
Kim Youkhana (mkimbe@email.unc.edu)
KORA:
Annette Peters (peters@helmholtz-muenchen.de)
Martina Mueller (martina.mueller@helmholtz-muenchen.de)
Cavin Ward-Caviness (cavin.wardcaviness@gmail.com)
Wolfgang Koenig (koenig@dhm.mhn.de)
Lothian:
Simon Cox (Simon.cox@ed.ac.uk)
Gail Davies (gail.davies@ed.ac.uk)
Riccardo Marioni (riccardo.marioni@ed.ac.uk)
LURIC:
Marcus Kleber (marcus.kleber@medma.uni-heidelberg.de)
MARTHA:
David Tregouet (david.tregouet@upmc.fr)
Pierre Morange (Pierre.MORANGE@ap-hm.fr)
MEGA:
Frits R. Rosendaal (f.r.rosendaal@lumc.nl)
Astrid van Hylckama Vlieg (A.van_Hylckama_Vlieg@lumc.nl)
MESA:
Jerry Rotter (jrotter@labiomed.org)
Jie Yao (jyao@labiomed.org)
Xiuqing Guo (xguo@labiomed.org)
MVP:
Chris O’Donnell (Christopher.ODonnell@va.gov)
Scott Damrauer (Scott.Damrauer@uphs.upenn.edu)
NEO:
Ruifang Li (R.Li@lumc.nl)
Dennis Mook (D.O.Mook@lumc.nl)
Frits R. Rosendaal (f.r.rosendaal@lumc.nl)
Orcades:
Harry Campbell (Harry.Campbell@ed.ac.uk)
PREVEND:
Pim van der Harst (P.vanderHarst@umcutrecht.nl)
Folkert Asselbergs (F.W.Asselbergs@umcutrecht.nl)
PROCARDIS:
Maria Sabater-Lleal (msabater@santpau.cat)
PROSPER:
Wouter Jukema (J.W.Jukema@lumc.nl)
Stella Trompet (S.Trompet@lumc.nl)
RETROVE:
Maria Sabater-Lleal (msabater@santpau.cat)
RS:
Maryam Kavousi (m.kavousi@erasmusmc.nl)
Frank Leebeek (f.leebeek@erasmusmc.nl)
Moniek de Maat (m.demaat@erasmusmc.nl)
SHIP:
Stephan Felix (felix@uni-greifswald.de)
Uwe Voelker (voelker@uni-greifswald.de)
Georg Homuth (georg.homuth@uni-greifswald.de)
TwinsUK:
Massimo Mangino (massimo.mangino@kcl.ac.uk)
WHI:
Alex P. Reiner (apreiner@u.washington.edu)
WGHS:
Daniel Chasman (DCHASMAN@research.bwh.harvard.edu)
Acknowledgments: This work was supported by grants from the National Institutes of Health HL134894 and R33 HL141791 (C.J.L., N.L.S., and A.S.W.), HL126974 (A.S.W.), HL166690 (M.A.), and HL139553 (P.S.d.V., A.C.M., N.L.S., C.J.L., and W.O.). M.S-L. is supported by a Miguel Servet contract from the ISCIII Spanish Health Institute (CPII22/00007) and cofinanced by the European Social Fund. P.R. is supported by the American Heart Association (AHA) 23POST1021300. M.A. is supported by NIH HL166690 and AHA 940488. C.J.L. is supported by the Michel Mirowski MD Professorship in Cardiology. The infrastructure for the Cohorts for Heart and Aging in Genomic Epidemiology Consortium is supported in part by the National Heart, Lung, and Blood Institute grant R01HL105756. Figure 2I was created with BioRender.com.
Contribution: P.R., P.T.-S., N.A., and M.V. performed the experiments; P.R., M.A., W.O., and C.J.L. analyzed the results; M.A. and C.J.L. designed the study; and P.R., A.C.M., M.S-L., A.S.W., P.S.d.V., N.L.S., W.O., M.A., and C.J.L. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the Cohorts for Heart and Aging in Genomic Epidemiology Hemostasis Working Group appears in “Appendix.”
Correspondence: Charles J. Lowenstein, Division of Cardiology, Department of Medicine, The Johns Hopkins University School of Medicine, 600 N. Wolfe St, Blalock 910, Baltimore, MD 21287; email: clowens1@jhmi.edu.
References
Author notes
Data are available upon reasonable request from the corresponding author, Charles J. Lowenstein (clowens1@jhmi.edu).
The full-text version of this article contains a data supplement.

![A CD32+ subpopulation of human liver ECs (ECs and HLEC) expresses FVIII. A data set for scRNA-seq of human liver cells was analyzed and a subpopulation of ECs was purified and studied. (A) Human liver cells contained 3 subpopulations of vascular ECs. (B) Differentially expressed genes identify subpopulations of ECs, including CD32 (FCGR2B), which marks endothelial cluster 11. Differentially expressed genes were plotted for all-liver cell clusters; the size of the circle indicates the percentage of cells in each population expressing each gene, and the intensity of the color indicates the level of expression. (C) scRNA-Seq demonstrates that CD32+ HLECs express higher levels of F8 and CD36 but lower levels of VWF. The expression of selected genes in each endothelial cluster and hepatocytes is plotted. (D) Expression of CD32 in CD32+ HLEC. HLEC were sorted by CD32+ expression, and CD32 mRNA expression was measured by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (N = 3, mean ± standard deviation [SD], ∗∗P < .005). (E) Localization of CD32 in CD32+ HLEC. CD32+ HLEC were isolated, hybridized with antibody to CD32, and imaged by confocal microscope (green = CD32, blue = DNA). Representative images are shown using the left 20× objective and the right 63× immersion objective (scale bar, 50 μM). (F) CD32+ HLEC express F8. CD32+ mRNA was purified from HLEC and HUVEC, and F8 mRNA expression was measured by qRT-PCR (N = 3, mean ± SD, ∗∗∗P = .0001). (G) CD32+ HLEC release FVIII into the media. Media were collected from CD32+ HLEC at 2, 4, 6, 8, and 24 hours. Top: FVIII antigen was measured by enzyme-linked immunosorbent assay (ELISA) (N = 3, mean ± SD). Bottom: FVIII activity was measured using a chromogenic FVIII activity assay (N = 6, mean ± SD). (H) FVIII was localized in a punctate pattern in CD32+ HLEC. CD32+ HLEC were hybridized with antibody to FVIII and imaged using a confocal microscope. Representative images are shown using the 20× objective and the 63X immersion objective (scale bar, 100 μM and 50 μM, respectively). (I) FVIII and COPII are colocalized in CD32+ HLEC. CD32+ HLEC were hybridized with antibodies to FVIII and COPII and imaged using confocal microscope using the 20× objective and the 63X immersion objective (scale bar, 100 μM and 50 μM, respectively). The Pearson correlation coefficient = 0.62 ± 0.08. (J) CD32+ HLEC express minimal levels of VWF and release minimal levels of VWF into the media. mRNA was isolated from CD32+ HLEC and HUVEC and analyzed by qRT-PCR (top) (N = 6, mean ± SD, ∗∗∗∗P < .0001). Ten thousand HUVEC and HLEC CD32+ cells were seeded in p96-collagen coated plates, cultured for 3 days, the media was replaced, cells were treated with media or 10 μM histamine for 1 hour, and the concentrations of VWF released into the media were measured by ELISA (bottom). (N = 8, mean ± SD, ∗∗∗∗P < .0001).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/1/10.1182_bloodadvances.2023010023/2/m_blooda_adv-2023-010023-gr1.jpeg?Expires=1768030180&Signature=3e~wQJa~YmzkczopVqI~rtc2WBYu-xc5fTYgBZ7NkapyJuqv7M5ox5z7onvTqeZtIdn-B2h~BG~1WdwgDxWr5sy2OqEn4WvIxgToFptUR~M8-h5tiiy1aFsLnndSXY4PydKwOoJPDfKTBcDOlLqkFaRf3zDk~d3QuNq4Fw7JlkeqTxD6EDuB8IOpBXybjRTvBJ~WzURchK-2E7vFrihRXV-mkZ4Z1voFdXE3IhPkgFi33dLSirpPQl-CDa-FqBLQKs6Cdf8sxDlky-O4z8YwLawdGp-803qlT~335JsqqMnnwYs~1H4ZuMp1c~MfBshFljqKP1Al8YnRI6PPuxK5Gg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![A CD32+ subpopulation of human liver ECs (ECs and HLEC) expresses FVIII. A data set for scRNA-seq of human liver cells was analyzed and a subpopulation of ECs was purified and studied. (A) Human liver cells contained 3 subpopulations of vascular ECs. (B) Differentially expressed genes identify subpopulations of ECs, including CD32 (FCGR2B), which marks endothelial cluster 11. Differentially expressed genes were plotted for all-liver cell clusters; the size of the circle indicates the percentage of cells in each population expressing each gene, and the intensity of the color indicates the level of expression. (C) scRNA-Seq demonstrates that CD32+ HLECs express higher levels of F8 and CD36 but lower levels of VWF. The expression of selected genes in each endothelial cluster and hepatocytes is plotted. (D) Expression of CD32 in CD32+ HLEC. HLEC were sorted by CD32+ expression, and CD32 mRNA expression was measured by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (N = 3, mean ± standard deviation [SD], ∗∗P < .005). (E) Localization of CD32 in CD32+ HLEC. CD32+ HLEC were isolated, hybridized with antibody to CD32, and imaged by confocal microscope (green = CD32, blue = DNA). Representative images are shown using the left 20× objective and the right 63× immersion objective (scale bar, 50 μM). (F) CD32+ HLEC express F8. CD32+ mRNA was purified from HLEC and HUVEC, and F8 mRNA expression was measured by qRT-PCR (N = 3, mean ± SD, ∗∗∗P = .0001). (G) CD32+ HLEC release FVIII into the media. Media were collected from CD32+ HLEC at 2, 4, 6, 8, and 24 hours. Top: FVIII antigen was measured by enzyme-linked immunosorbent assay (ELISA) (N = 3, mean ± SD). Bottom: FVIII activity was measured using a chromogenic FVIII activity assay (N = 6, mean ± SD). (H) FVIII was localized in a punctate pattern in CD32+ HLEC. CD32+ HLEC were hybridized with antibody to FVIII and imaged using a confocal microscope. Representative images are shown using the 20× objective and the 63X immersion objective (scale bar, 100 μM and 50 μM, respectively). (I) FVIII and COPII are colocalized in CD32+ HLEC. CD32+ HLEC were hybridized with antibodies to FVIII and COPII and imaged using confocal microscope using the 20× objective and the 63X immersion objective (scale bar, 100 μM and 50 μM, respectively). The Pearson correlation coefficient = 0.62 ± 0.08. (J) CD32+ HLEC express minimal levels of VWF and release minimal levels of VWF into the media. mRNA was isolated from CD32+ HLEC and HUVEC and analyzed by qRT-PCR (top) (N = 6, mean ± SD, ∗∗∗∗P < .0001). Ten thousand HUVEC and HLEC CD32+ cells were seeded in p96-collagen coated plates, cultured for 3 days, the media was replaced, cells were treated with media or 10 μM histamine for 1 hour, and the concentrations of VWF released into the media were measured by ELISA (bottom). (N = 8, mean ± SD, ∗∗∗∗P < .0001).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/1/10.1182_bloodadvances.2023010023/2/m_blooda_adv-2023-010023-gr1.jpeg?Expires=1768030181&Signature=oFo06w8lUww1Bgv5MbzWiG1hAli9X0~eotxIw5-UhRvUlzs1Xc4PTssX~VNXnQVmktd4qkJ290ryCjtaubAsgCxA3jiopiuoU6rjLNcHZ28~hZHs08O5qxCORcK0GwWzdWtl4mNMFJJrrK9oQ5wa~oA-~zllM-Z3uGdI7F6nJVd0~~8H3BOu-BZAC6Vmz~1Yrip9PrD4l7LErgIT8HrLF7y9sLhBhS6Nq7ARrb5HrBYYbWfhMIF4qncwAXeKmwlSd~tCqL1m155ifSEu1iLVjlrAaCIT2lITQy0RTEuNGArrOI1TF9O9QoGRJKoiS1kccU9Rnv~unAJ1bn1pO5eBWw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)