Key Points
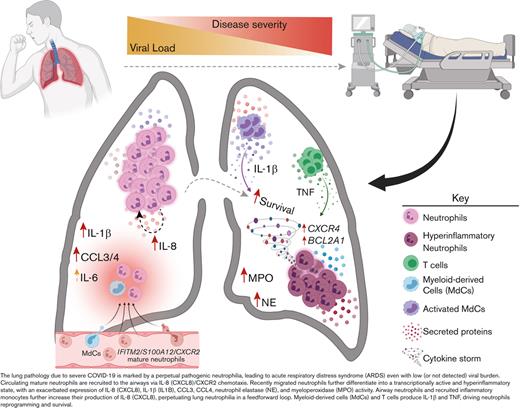
Blood neutrophils are transcriptionally reprogrammed within the airways of patients with severe COVID-19, exacerbating acute respiratory distress syndrome despite low viral load.
Persistent and self-sustaining pathogenic neutrophilia intensifies the cytokine storm in the airways of patients of African ancestry with COVID-19.
Abstract
Troubling disparities in COVID-19–associated mortality emerged early, with nearly 70% of deaths confined to Black/African American (AA) patients in some areas. However, targeted studies on this vulnerable population are scarce. Here, we applied multiomics single-cell analyses of immune profiles from matching airways and blood samples of Black/AA patients during acute SARS-CoV-2 infection. Transcriptional reprogramming of infiltrating IFITM2+/S100A12+ mature neutrophils, likely recruited via the IL-8/CXCR2 axis, leads to persistent and self-sustaining pulmonary neutrophilia with advanced features of acute respiratory distress syndrome (ARDS) despite low viral load in the airways. In addition, exacerbated neutrophil production of IL-8, IL-1β, IL-6, and CCL3/4, along with elevated levels of neutrophil elastase and myeloperoxidase, were the hallmarks of transcriptionally active and pathogenic airway neutrophilia. Although our analysis was limited to Black/AA patients and was not designed as a comparative study across different ethnicities, we present an unprecedented in-depth analysis of the immunopathology that leads to acute respiratory distress syndrome in a well-defined patient population disproportionally affected by severe COVID-19.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic, caused by SARS-CoV-2, is associated with high morbidity and mortality. The pandemic has caused over a million deaths in the United States, where historically marginalized groups and communities of color are disproportionately burdened with disease severity and mortality.1,2 Despite persistent health disparities in COVID-19 outcomes among Black/African Americans (AA)3,4 and the deployment of government health equity strategies through the Centers for Disease Control and Prevention, an in-depth understanding of the immunopathology of severe COVID-19 in marginalized groups is lacking. Hence, there is an urgent need to investigate the immune responses that lead to acute respiratory distress syndrome (ARDS) and death in the most vulnerable population, particularly in the Black/AA community that has been disproportionally affected by severe COVID-19.
A hallmark of COVID-19 pathogenesis is the vast array of clinical outcomes, ranging from asymptomatic or mild self-limiting disease to ARDS, multiorgan system failure, and death. Such diversity in COVID-19 pathogenesis poses challenges in identifying processes that dictate progression to severe disease. Early single-cell analyses of bronchoalveolar lavage fluid (BALF) samples from patients with severe disease implicated dysregulated monocyte and macrophage responses as central features in poor outcomes.5,6 As such, most early efforts have focused on characterizing and constraining aberrant monocyte/macrophage responses. Concurrently, reports of neutrophilia in the peripheral blood arose, and the neutrophil to lymphocyte ratio emerged as an independent risk factor for disease progression.7
The role of neutrophils in response to bacterial infections is well established,8 but neutrophils also have diverse functions in antiviral immunity and pathogenesis.9,10 Neutrophils are efficient scavengers and can directly phagocytize influenza A virus (IAV)11 and other respiratory viruses.10 Neutrophil degranulation is a primary effector function at the outset of infection, and potent proteases such as myeloperoxidase (MPO) and neutrophil elastase (NE) within primary granules can be released and catalyze the formation of reactive oxygen species that have antiviral properties.9 However, if neutrophil responses are overexuberant, this can lead to adverse effects during the antiviral response causing diffuse tissue damage and exacerbated disease pathogenesis. Indeed, a previous systems analysis of lethal IAV infection in mice highlighted that neutrophilia largely accounted for poor prognosis, whereby proinflammatory neutrophils established a chemokine-driven dysregulated hyperinflammatory feedforward loop,12 suggesting a self-potentiating pathogenic neutrophilia in severe viral respiratory infections.
Most studies implicating pathogenic neutrophilia in severe COVID-19 have been limited to blood specimens.13-17 More recent studies have expanded on these findings by evaluating neutrophil responses in the airways during COVID-19 pneumonitis.5,16,18-20 However, these studies have yielded inconclusive results regarding the role of lung neutrophils in disease severity.5,18,20-22 Most importantly, they have not investigated the role of pathogenic neutrophilia among the most vulnerable population, particularly Black/AA patients disproportionally affected by severe COVID-19.
Hence, to address this gap and better understand the clinical impact of exacerbated neutrophilia and the relationship between lung and blood immune responses associated with severe COVID-19 in the most vulnerable population, we used a systems immunology approach from the airways and matching blood of Black/AA patients we serve in the Atlanta area. We combined multiomics single-cell mRNA sequencing (scRNA-seq) with high-dimensional (Hi-D, 30-parameter) flow cytometry and multiplexed immunoassays, which revealed transcriptionally active and self-sustained pathogenic airway neutrophilia as a hallmark of severe disease within this cohort. Specifically, we identified mature IFITM2+/ S100A12+ circulating neutrophils as the source population in the blood, which are primarily recruited to the lungs via the CXCL8 (IL-8)/CXCR2 axis. Upon recruitment, neutrophils undergo transcriptional reprogramming in the airways to acquire a self-sustained hyperinflammatory phenotype characterized by exacerbated levels of IL-8, IL-1β, IL-6, and CCL3/4, along with copious amounts of NE and MPO.
Altogether, our findings indicate that therapeutic efforts to reduce pathogenic airway neutrophilia may constrain ARDS in severe COVID-19, particularly in our cohort of Black/AA patients. Although our study was not designed as a comparative analysis between different races/ethnicities, we provide an unprecedented in-depth analysis of the immunopathology that leads to severe COVID-19 and ARDS in a cohort of Black/AA patients. We expect our results and the large multiomics datasets made available here to inform future comparative studies aimed at reducing persistent health disparities among historically marginalized groups.
Methods
Ethics and biosafety
A total of 35 individuals were enrolled for this study (supplemental Tables 1 and 2). The 18 patients with severe COVID-19 were recruited from the Intensive Care Units of Emory University, Emory St. Joseph's, Emory Decatur, and Emory Midtown Hospitals. We also recruited 9 mild COVID-19–infected outpatients in the Emory Acute Respiratory Clinic and 8 healthy adults from the Emory University Hospital. All studies were approved by the Emory Institutional Review Board under protocol numbers IRB00058507, IRB00057983, and IRB00058271. Informed consent was obtained from the patients when they had decision-making ability, or from a legally authorized representative if the patient was unable to provide consent. Blood, sputum, and endotracheal aspirate (ETA) samples were obtained. Control blood samples were obtained from healthy adults and were matched by age and race. Study inclusion criteria were confirmed COVID-19 diagnosis by polymerase chain reaction amplification of SARS-CoV-2 viral RNA obtained from nasopharyngeal or oropharyngeal swabs, age ≥18 years and willingness to provide informed consent. Individuals with a confirmed history of COVID-19 were excluded from the healthy donor group. All work with infectious virus and respiratory samples from patients with COVID-19 was conducted inside a biosafety cabinet within the Emory Health and Safety Office and the US Department of Agriculture (USDA)-approved BSL3 containment facility in the Health Sciences Research Building at Emory University following protocols approved by the Institutional Biosafety Committee and Biosafety Officer.23
Patient sample collection and processing
Primary leukocytes from the airways of patients with COVID-19 requiring mechanical ventilator support were collected bedside via endotracheal aspiration, and whole blood was collected by standard venipuncture. Plasma from whole blood was isolated by centrifugation at 400 × g for 10 minutes at 4°C. To remove platelets, the isolated plasma was centrifuged at 4000 × g for 10 minutes at 4°C. Untouched circulating leukocytes were isolated using EasySep RBC Depletion Reagent (StemCell Technologies). ETA (from severe patients) or noninduced sputum (from mild patients) was mixed 1:1 with a 50mM EDTA solution (final concentration 25 mM EDTA) in custom RPMI-1640 medium deficient in biotin, L-glutamine, phenol red, riboflavin, and sodium bicarbonate (defRPMI-1640) with 3% newborn calf serum (NBCS), and mechanically dissociated using a syringe to liberate leukocytes from mucins and other respiratory secretions. Supernatants were collected for further analysis, and then the cells underwent an additional mechanical dissociation step using 1 to 3 mL of 10 mM EDTA in defRPMI-1640 + 3% NBCS and a P1000 pipettor. Cells were then washed with 10 mL defRPMI-1640 + 3% NBCS, passed through a 70 μm nylon strainer, and pelleted through a 2 mL 100% NBCS layer before counting and downstream processing.
High-dimensional (Hi-D) 30-parameter flow cytometry
Cells (up to 107 total) were resuspended in defRPMI-1640 with 3% newborn calf serum and Benzonase (fluorescence-activated cell sorter [FACS] buffer) in 5 mL FACS tubes and preincubated with GolgiStop (BD Biosciences) for ∼20 minutes at 4°C. Human TruStain FcX was then added, followed by a 10 minutes incubation at room temperature. The 24-color extracellular staining master mix (supplemental Table 3) was prepared 2× in BD Horizon Brilliant Stain Buffer to prevent staining artifacts from BD Horizon Brilliant dye interactions and was added 1:1 to the cells, then incubated for 30 minutes at 4°C. After staining (and total 1 hour exposure to GolgiStop), cells were washed with ∼4 mL FACS buffer. Next, the cells were resuspended in 200 μL of BD Cytofix/Cytoperm fixation/permeabilization solution and incubated at 4°C for 30 minutes, followed by washing with ∼4 mL of BD Perm/Wash Buffer. The 4-color intracellular staining (supplemental Table 3) was prepared in BD Perm/Wash Buffer, and the cells were stained for 30 minutes at 4°C. Cells were washed with ∼4 mL BD Perm/Wash Buffer, resuspended for a final 20 minutes incubation in 4% PFA, and transported out of the BSL3 containment facility. Cells were washed in ∼4 mL FACS buffer, then resuspended in 200 to 1000 μL FACS buffer for acquisition using BD FACSDiva Software on the Emory Pediatric/Winship Flow Cytometry Core BD FACSymphony A5. To distinguish autofluorescent cells from cells expressing low levels of a particular surface marker, we established upper thresholds for autofluorescence by staining samples with fluorescence minus one (FMO) control stain sets, in which a reagent for a channel of interest is omitted. The data were analyzed using FlowJo v10.8 (FlowJo LLC).
Cell-surface antibody-derived tag (ADT) staining, single-cell encapsulation, and library generation
Leukocytes from whole blood and ETA samples were incubated with oligo-conjugated Ig-A/D/G/M for 10 minutes at 4°C, followed by the addition of Human TruStain FcX (BioLegend) and 10 minutes incubation at room temperature. The cells were then surface stained with oligo-conjugated monoclonal antibody panel (total 89 antibodies; supplemental Table 4) for 30 minutes at 4°C, followed by 2 washes in defRPMI-1640/0.04% BSA. Cells were resuspended at a concentration of 1200 to 1500 cells/μL in defRPMI-1640/0.04% BSA and passed through a 20 or 40 μm cell strainer before loading onto a Chromium Controller (10× Genomics, Pleasanton, CA). The cells were loaded into a target encapsulation of 10 000 cells. Gene expression (GEX) and ADT libraries were generated using the Chromium Single Cell 5′ Library & Gel Bead Kit v1.1 with feature barcoding following the manufacturer's instructions. GEX libraries were pooled and sequenced at a depth of approximately 540 000 000 reads per sample in a single S4 flow cell and ADT libraries at a depth of approximately 79 000 000 reads per sample in a single lane of an S4 flow cell on a NovaSeq 6000 (Illumina, San Diego, CA; supplemental Table 5).
Multiomics single-cell RNA sequencing (scRNA-seq) analysis
Single-cell 5′ unique molecular identifier (UMI) counting and barcode de-multiplexing were performed using the Cell Ranger Software (v.5.0.0). To detect SARS-CoV-2 vRNA reads, we built a custom reference genome from human GRCh38 and SARS-CoV-2 references (severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome, GenBank MN908947.3). The splicing-aware aligner STAR24 was implemented to align the FASTQ inputs to the reference genome, and the resulting files were automatically filtered using Cell Ranger to include only cell barcodes representing real cells. This determination is based on the distribution of the UMI counts. The ADT reads were aligned to a feature reference file containing antibody-specific barcode sequences. To recover neutrophils, we applied our SUPERR pipeline as previously described.25 Briefly, we recovered neutrophils from Cell Ranger unfiltered count matrices by plotting surface CD16 ADT and CD66b ADT using the "FeatureScatter" function in Seurat v4.026 (R version 4.0.2). The double-positive cell barcodes were then extracted and further evaluated using GEX to confirm the identity of the viable neutrophil. A threshold for mitochondrial content per barcode was determined for each sample independently and applied as a cutoff to remove dead or dying cells (supplemental Table 8). Most samples showed high cell viability with a minimal proportion of dead cells.
The UMI counts of the GEX data were log-normalized by the "NormalizeData" function in Seurat before downstream analysis, following the optimal workflow we previously described for sample normalization and data integration.27 Center log-ratio transform in Seurat was performed on ADT UMIs when recovering neutrophils from the unfiltered matrices. For surface protein visualization to classify major lineages using our SUPERR workflow,25 ADT UMIs were normalized using the R package Denoised and Scaled by Background28 (DSB) to remove ambient UMI counts (ie, background) before manual sequential gating by surface expression (supplemental Figure 2) in SeqGeq v1.7 (FlowJo, LLC). DSB used empty droplets to calculate the background expression, which was manually selected according to the distribution of the total ADT per cell in the raw count matrices (supplemental Table 6). To minimize the influence of noninformative empty droplets, we removed cell barcodes with less than 100 ADT UMIs before plotting the ADT distribution.
Before integrating the multiple datasets, we first classified major lineages in individual samples based on a combination of gene transcripts and surface protein markers (SUPERR workflow25), for samples in which the ADT library was of sufficient quality to allow manual gating (supplemental Figure 2). Cell barcodes within each major lineage that coexpressed markers exclusive to other major lineages were considered cell doublets and were removed (supplemental Figure 4). In addition, we removed cell barcodes with extremely high total ADT UMIs, which we considered aggregated cells. To efficiently integrate the replicate samples, we concatenated the major lineages derived from the same tissue from different donors. To minimize batch effects and optimize data integration, we followed the data normalization and merging strategies described previously.27 Briefly, samples were first treated individually, log-normalized count matrices were scaled/Z-transformed, and the "vst" method of the Seurat function "FindVariableFeatures" was used to select the top 1000 highly variable genes (HVGs) of each sample. HVGs shared between replicate samples were used to perform principal component analysis. To visualize the data, we performed UMAP reduction of the first 30 PCs, and cell clustering was generated using the Leiden community detection algorithm at a resolution of 0.8. UMAP visualizations for integrated blood and integrated ETA were generated using the Seurat v4 data integration workflow. Data for SARS-CoV-2-infected Calu-3 cells were previously published and were analyzed as previously described.23
Receptor-ligand interaction analyses
Clustered cells from the lung and blood samples from each patient were investigated for evidence of intercellular communication using CellChat.29 The clustered cell populations from the lung samples were combined with blood neutrophils to determine which lung cell populations could recruit circulating neutrophils. We utilized NicheNet30 to determine which ligand-receptor pairs could be responsible for the different transcriptional states of neutrophil populations in lung and blood samples. We focused on certain neutrophil clusters as receiver populations, considering the remaining neutrophils and other lung cell populations as senders, and thus potential interactors. The target set of genes was determined using Seurat::FindMarkers(min.pct = 0.1), keeping only those genes with an adjusted P-value <0.05 and an average log2-fold change of more than 0.2. To address changes in gene expression due to lung infiltration, we ran an algorithm using blood neutrophil cluster 2 as the most likely candidate for lung infiltration. The genes considered were those differentially expressed between blood cluster 2 and lung neutrophils, and genes differentially expressed between blood cluster 2 and those cells that progressed along "Trajectory 2" in the lung (Figure 3).
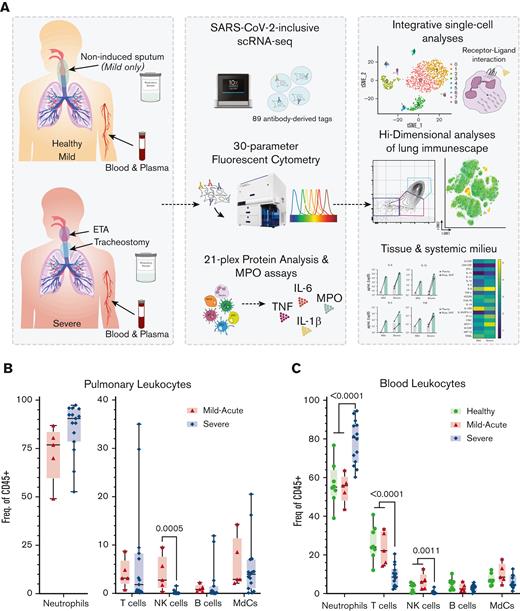
Experimental design for the systems immunology approach (integrated multiomics single-cell assays) to study patients with COVID-19 and identify robust neutrophilia in the lungs. (A) Respiratory samples (sputum or endotracheal aspirates) and matching blood from all subjects were collected for 21-plex Mesoscale analysis, high-dimensional (Hi-D) 30-parameter flow cytometry, and multiomics scRNA-seq. Cells from endotracheal aspirates (ETA) and blood of patients with severe COVID-19 along with blood from healthy individuals were surface-stained with a panel of 89 oligo-conjugated monoclonal antibodies before single-cell encapsulation, and analyses were performed with a custom human reference genome that included the SARS-CoV-2 genome to simultaneously detect viral mRNA transcripts. Integrative multiomics analyses were performed on the resulting datasets. (B) Box plots showing the distribution of leukocytes isolated from endotracheal aspirates (ETA). (C) Box plots showing the distribution of leukocytes isolated from the whole blood of severe patients. For comparisons across the 3 patient groups (ie, healthy, MA, and severe), ordinary one-way ANOVA (if equal variance) or Brown-Forsythe and Welch ANOVA (if unequal variance) tests were performed for data with a normal distribution. Data with a lognormal distribution were analyzed using the Kruskal-Wallis test.
Experimental design for the systems immunology approach (integrated multiomics single-cell assays) to study patients with COVID-19 and identify robust neutrophilia in the lungs. (A) Respiratory samples (sputum or endotracheal aspirates) and matching blood from all subjects were collected for 21-plex Mesoscale analysis, high-dimensional (Hi-D) 30-parameter flow cytometry, and multiomics scRNA-seq. Cells from endotracheal aspirates (ETA) and blood of patients with severe COVID-19 along with blood from healthy individuals were surface-stained with a panel of 89 oligo-conjugated monoclonal antibodies before single-cell encapsulation, and analyses were performed with a custom human reference genome that included the SARS-CoV-2 genome to simultaneously detect viral mRNA transcripts. Integrative multiomics analyses were performed on the resulting datasets. (B) Box plots showing the distribution of leukocytes isolated from endotracheal aspirates (ETA). (C) Box plots showing the distribution of leukocytes isolated from the whole blood of severe patients. For comparisons across the 3 patient groups (ie, healthy, MA, and severe), ordinary one-way ANOVA (if equal variance) or Brown-Forsythe and Welch ANOVA (if unequal variance) tests were performed for data with a normal distribution. Data with a lognormal distribution were analyzed using the Kruskal-Wallis test.
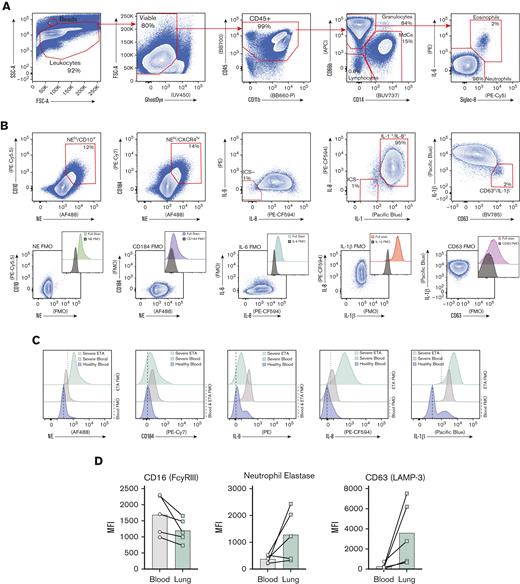
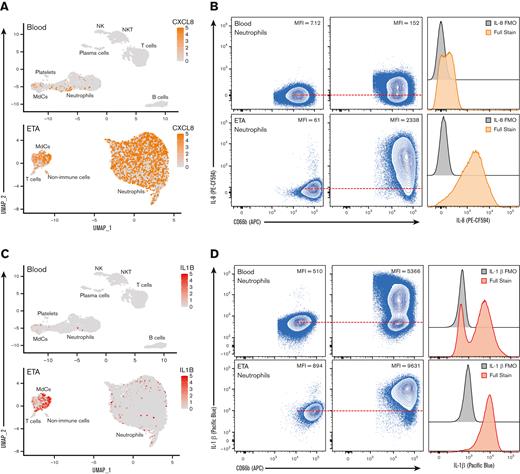
Exacerbated neutrophilia in the airways and matching blood in patients with severe COVID-19. (A) Representative gating strategy for all samples (supplemental Figure 1 for full gating strategy). (B) Representative plots demonstrating the inflammatory profile of pulmonary neutrophils, including NE, CD184 (CXCR4), and intracellular staining of IL-6, IL-8, and IL-1β, including the full stain and FMO controls. (C) Representative histograms showing the median fluorescence intensity (MFI) of key markers in healthy blood (blue), severe blood (gray), and severe ETA (green) samples. (D) MFI of CD16 (FCγRIII), NE, and CD63 (LAMP-3) reveal a GRIM-like phenotype in neutrophils from paired blood (gray circles) and lung (green squares) samples.
Exacerbated neutrophilia in the airways and matching blood in patients with severe COVID-19. (A) Representative gating strategy for all samples (supplemental Figure 1 for full gating strategy). (B) Representative plots demonstrating the inflammatory profile of pulmonary neutrophils, including NE, CD184 (CXCR4), and intracellular staining of IL-6, IL-8, and IL-1β, including the full stain and FMO controls. (C) Representative histograms showing the median fluorescence intensity (MFI) of key markers in healthy blood (blue), severe blood (gray), and severe ETA (green) samples. (D) MFI of CD16 (FCγRIII), NE, and CD63 (LAMP-3) reveal a GRIM-like phenotype in neutrophils from paired blood (gray circles) and lung (green squares) samples.
Cell trajectory analyses
The Python toolkit scVelo31 was used to infer the trajectories using biological data from ETA neutrophils. Input data for scVelo analysis were intron, exon, and spanning count matrices estimated using the dropEST tool,33 then filtered with previously identified neutrophil cell barcodes in R studio. Intron, exon, and spanning matrices were compared with identify missing rows (genes) and were added to each matrix to equalize the dimensions. The exon matrix contained the spliced matrix and the sum of the intron and spanning matrices constituted the unspliced matrix. Spliced and unspliced matrices were imported using an anndata library, and the pandas library was used to import the gene names and cell barcodes. Raw count matrices were added to anndata object layers as spliced and unspliced. Gene names and cell barcodes were then attached to the variables and observations of the anndata object, respectively. The anndata object was transposed, followed by a regular scVelo analysis. The default parameters of plotting velocity streams include vkey=velocity, colorbar=True, alpha=0.3, sort_order=True, and legend_loc=on data.
Pathway and process enrichment analyses
Mesoscale U-PLEX assays
U-PLEX Biomarker Group 1 Human Multiplex Assays (Meso Scale Discovery) were used to evaluate the levels of 21 analytes following the manufacturer's protocol (supplemental Table 7) in plasma and UVC-inactivated respiratory supernatants (see ref23). Samples were diluted 1:5 for all assays, except for IL-8, MCP-1, and IL-1RA, which were above the upper limit of detection for the assay and diluted 1:200 to acquire measurements within the assay range. The final values were obtained by multiplying the measurements with their respective dilution factors.
Myeloperoxidase (MPO) content and activity
The abundance and activity of MPO were quantified as previously described.34 MPO activity and protein concentration were measured sequentially, following immunocapture. On average, across 6 96-well plate assays, lower limits of quantification were 4.0 ng/mL (activity) and 0.84 ng/mL (protein). Samples above the lower limit of detection but below the lower limit of quantification were imputed as half of the latter, and those detected above the highest standard of 50 ng/mL (ie, above the upper limit of detection) at all dilutions were imputed as twice the standard concentration (supplemental Table 7).
SARS-CoV-2 quantitative reverse transcription PCR (RT-qPCR)
Viral (v)RNA was extracted from the respiratory secretions of patients with COVID-19 using the Quick-RNA Viral Kit (Zymo Research), following the manufacturer's protocol, and complementary (c)DNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's instructions and then diluted 1:5 in nuclease-free water. Ten μL diluted cDNA was used with the NEB Luna Universal Probe qPCR Master Mix (New England BioLabs Inc) following the manufacturer's protocol, and was performed in 384-well plates using a QuantStudio 5 Real-Time PCR System (Applied Biosystems). The primer/probe pairs were: AGAAGATTGGTTAGATGATGATAGT (forward), TTCCATCTCTAATTGAGGTTGAACC (reverse), and /56-FAM/TCCTCACTGCCGTCTTGTTGACCA/3IABkFQ/ (probe), which were designed from sequences previously described35 (Integrated DNA Technologies; IDT). To generate a standard curve for the quantification of SARS-CoV-2 genome copies a gBlock from IDT with the following sequence was used as a standard: AATTAAGAACACGTCACCGCAAGAAGAAGATTGGTTAGATGATGATAGTCAACAAACTGTTGGTCAACAAGACGGCAGTGAGGACAATCAGACAACTACTATTCAAACAATTGTTGAGGTTCAACCTCAATTAGAGATGGAACTTACAGTTTCAGTGTTCAATTAA.
Statistical analyses
Statistical analyses were performed using GraphPad Prism 9.4.0. Data were independently analyzed for distribution (normal [Gaussian] vs lognormal) using the D'Agostino and Pearson test for normality in the untransformed and Log10-transformed data. In cases where the sample size (N) was too small for D'Agostino and Pearson normality tests, the Shapiro-Wilk test was used to assess the distribution. When the data passed both distribution tests, the likelihood of each distribution (normal vs lognormal) was computed, and quantile-quantile (QQ)-plots were generated. When Log10 transformed data had a higher likelihood of a normal distribution (passing normal distribution test) and/or failed lognormal distribution test, paired t-tests were performed to compare matching blood and respiratory supernatant samples within a single group. If the data showed unequal variance (as determined using an F-test), a ratio paired t test was performed. All instances where lognormal distribution was likely a nonparametric Wilcoxon matched-pairs sign ranked test were performed. For instances when only lung supernatants (ie, mild-acute [MA] sputum and severe ETA) were compared, Mann-Whitney U tests were performed. For comparisons across the 3 patient groups (ie, healthy, MA, and severe), ordinary 1-way ANOVA (if equal variance) or Brown-Forsythe and Welch ANOVA (if unequal variance) tests were performed for data with a normal distribution. Alternatively, data with a lognormal distribution were analyzed using the Kruskal-Wallis test.
Results
Study cohort
From June 2020 through February 2021, before vaccine accessibility, most patients admitted to the intensive care unit (ICU) of Emory-affiliated hospitals were of African ancestry. Therefore, we focused our study on this demographic, which has been disproportionately affected by severe COVID-19. We collected airway (endotracheal aspirate, ETA) and matching blood samples from a cohort of 35 Black/AA patients with COVID-19 who presented to Emory University Hospitals (severe) or the Emory Acute Respiratory Clinic (MA) in Atlanta, GA (Figure 1A and supplemental Table 1), including 8 matched healthy adults as controls. Of the 27 patients confirmed as positive by PCR from nasopharyngeal swabs, 18 had an NIH severity score of critical (https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/), referred to as severe herein, and were admitted to the ICU requiring mechanical ventilator support. The remaining 9 were mild (MA) outpatients. All patients were unvaccinated. All severe patients in our cohort received corticosteroids (dexamethasone or equivalent), and approximately half received 1 or more doses of the antiviral medication remdesivir, with an average ICU stay of 26 days (supplemental Tables 1 and 2).
Exacerbated neutrophilia in the airways and matching blood of patients with severe COVID-19
We first characterized the major immune lineages in the ETA and matching blood samples by Hi-D flow cytometry and observed pronounced circulating neutrophilia and lymphopenia (notably T and NK cells), which were similarly reflected in the airways (Figure 1B,C; supplemental Figure 1). This is in line with recent reports showing that lymphopenia is associated with significant alterations in the myeloid compartment.16,36 Strikingly, in most cases of severe disease, ≥85% of all pulmonary leukocytes were neutrophils (Figure 1B). This contrasts with other studies that have reported much more heterogeneity in neutrophil frequency in patients' blood and lungs.21,37 In addition, circulating T cells and NK cells decline with increased disease severity (Figure 1C), concomitant with a notable decrease in pulmonary NK cells (Figure 1B), which has recently been reported as a feature of COVID-19 pneumonia.38 However, there were only subtle differences in the B-cell compartment (which were detected at very low levels in the airways) and myeloid-derived cells (MdCs) compartment compared with other reports,5,39,40 highlighting the importance of neutrophilia and neutrophil-to-lymphocyte ratio in our cohort.
Recruited airway neutrophils are mature, transcriptionally active, and further differentiate into a highly inflammatory state
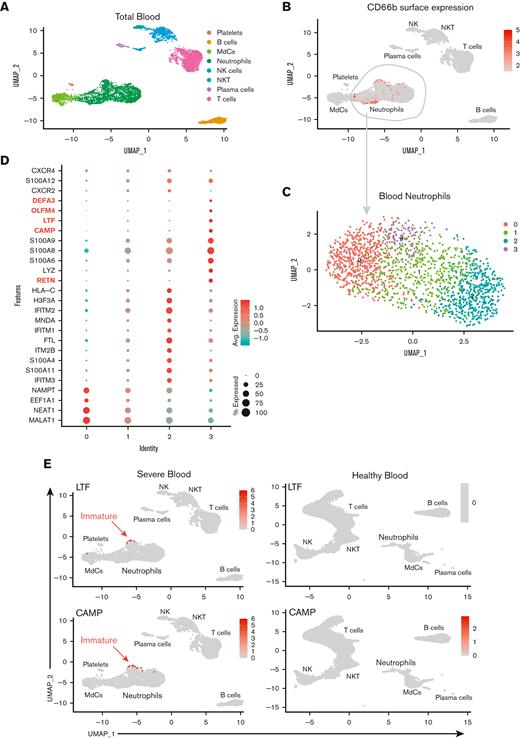
To gain insight into the cellular states and transcriptional regulation in patients with severe COVID-19, we performed multiomics scRNA-seq on cells from whole blood and ETA samples of severe patients and whole blood from demographic-matched healthy controls (Figure 1A). We assessed the immune features in the data integrated from 4 or more patients from the same cohort (ie, healthy vs severe; see Methods). First, we manually gated the major lineages (supplemental Figure 2A,C) from the blood (Figure 2A) and ETA (Figure 2F) using the ADT data for cell-surface protein expression (supplemental Figure 2), and then used the gene expression (GEX) data to generate clusters for each major lineage identified based on their cell-surface markers (see our previously published SUPERR pipeline25; supplemental Figure 2). For example, the total neutrophils identified by CD66b and CD16 surface ADT in the blood (Figure 2B) and ETA (Figure 2G) were clustered independently using GEX data (Figure 2C,H) to identify further heterogeneity.
A previous study reported an increase in immature neutrophils in the lung concomitant with increased circulating immature neutrophils from emergency hematopoiesis in patients with severe COVID-19,36 which is believed to be further increased upon administration of dexamethasone.20 Therefore, we assessed the immature neutrophil phenotype in the blood (Figure 2A-E) and lung (Figure 2F-J) by scRNA-seq. We readily identified a cluster of neutrophils expressing CAMP, LTF, RETN, OLFM4, DEFA3, CD24, and MMP8 (Figure 2E; supplemental Figure 3) in the blood of severe patients (cluster 3 in blood neutrophils) that also had high expression of calprotectin (S100A8/9) and other calgranulins (Figure 2C-E). This is consistent with the immature neutrophil phenotypes described by others16,18 and was notably absent in neutrophils from healthy blood, confirming emergency hematopoiesis/granulopoiesis in our patient cohort. However, in contrast to previous studies,20,36 we did not observe any signature of immature neutrophils in the lungs of severe patients (Figure 2I,J) despite pronounced neutrophilia.
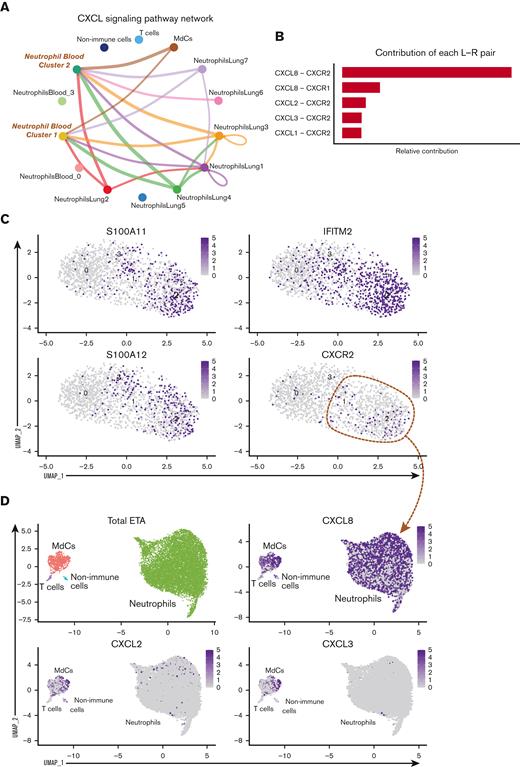
To determine which neutrophil subset(s) in the blood can infiltrate the lung and further differentiate into a pathogenic state, we explored cell-cell communication (CellChat29) of clustered cells from blood (Figure 2C) and lung (Figure 2F) samples. We found significant communication through the CXCL pathway between lung neutrophil, myeloid, and non-immune populations (ie, epithelial/stromal cells) and CXCR2-expressing neutrophils from the blood (blood clusters 1 and 2; Figure 3A). The CXCL8 (IL-8)/CXCR2 pathway was identified as the primary recruitment axis for circulating CXCR2+ neutrophils (Figure 3B). Virtually all lung neutrophils, myeloid cells, and nonimmune cells showed potential to recruit circulating neutrophils (Figure 3A,D), indicating a robust and redundant mechanism of neutrophil recruitment to the airways via the CXCL8 (IL-8)/CXCR2 axis. In the blood, a defined subset of mature neutrophils expressing high levels of CXCR2, along with interferon-induced IFITM2 and S100A11/12, identified cluster 2, and, to a lesser extent, cluster 1, as putative neutrophil subsets that can infiltrate the lung via IL-8 (Figure 2D and 3C,D; supplemental Figure 4a). In contrast, immature neutrophils (blood cluster 3) lacked CXCR2 (Figure 3C), suggesting that they were unlikely to infiltrate the lungs via IL-8.
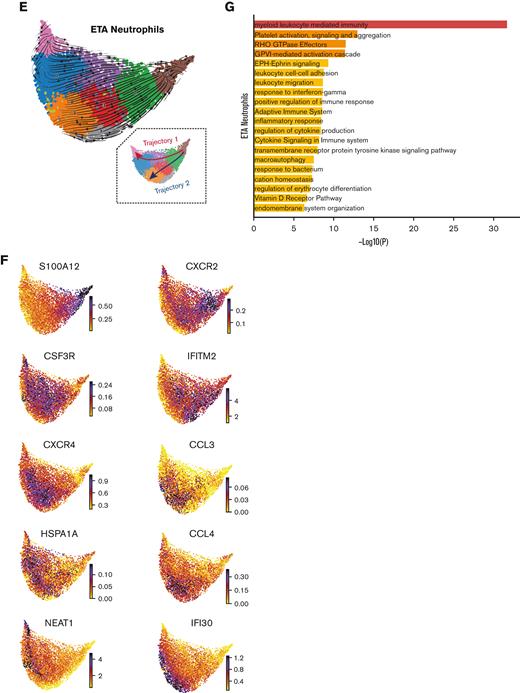
After identifying blood neutrophil clusters 1 and 2 as putative sources of lung-recruited neutrophils, we sought to identify their cell trajectory/differentiation once they entered inflamed airways. Cell trajectory analyses (scVelo31) revealed 2 differentiation pathways (Trajectories 1 and 2) for the recently infiltrated neutrophils (Figure 3E). Indeed, airway neutrophils expressing the highest levels of CXCR2 and S100A11/12 were observed at the beginning of the trajectory, further supporting this as a putative emigrant population from the blood (Figure 3F). Notably, neutrophils recruited to the lung and differentiated along Trajectory 2 experienced transcriptional reprogramming to acquire a heightened inflammatory phenotype (Figure 3E-G; supplemental Figure 4b). This is consistent with recent reports of neutrophil transmigration in other respiratory illnesses, such as cystic fibrosis,41 in which neutrophils undergo lung-specific adaptations42 instead of the canonical rapid and transient effector function, resulting in cell death. Indeed, we observed increased expression of the interferon-stimulated gene (ISG) IFI30, along with increased expression of CCL3 (MIP-1α) and CCL4 (MIP-1β) (Figure 3F), which in turn can promote the recruitment of inflammatory monocytes and/or proinflammatory macrophage phenotypes in the airways. Importantly, most neutrophils increased the expression of CXCL8 (IL-8) when recruited to the airways (Figure 3D), perpetuating neutrophil recruitment to the lungs. Furthermore, upon airway infiltration, neutrophils increased the expression of CXCR4 (Figure 3F). Interestingly, we found that CXCR4 expression, previously attributed to immature neutrophils,36 was increased in mature lung-recruited neutrophils (Figure 3F). Taken together, the progressive increase in IFI30 and CCL3/4, along with abundant de novo CXCL8 and CXCR4 mRNA transcripts, reveals a transcriptionally active state in neutrophils that sustains a hyperinflammatory milieu in the lungs.
Pulmonary TNF and IL-1β promote neutrophil reprogramming in the lungs
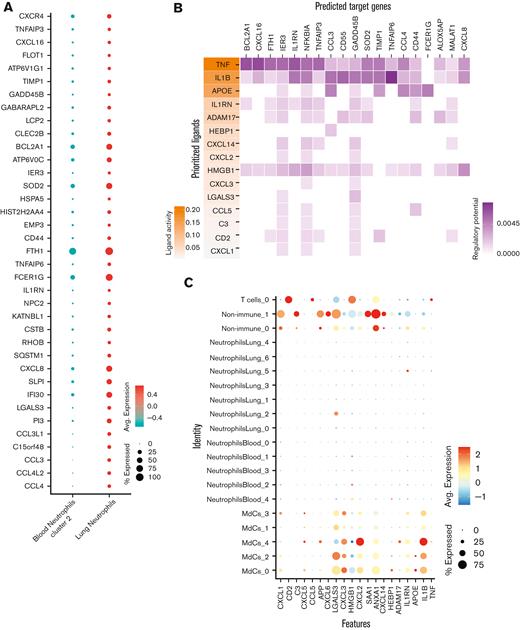
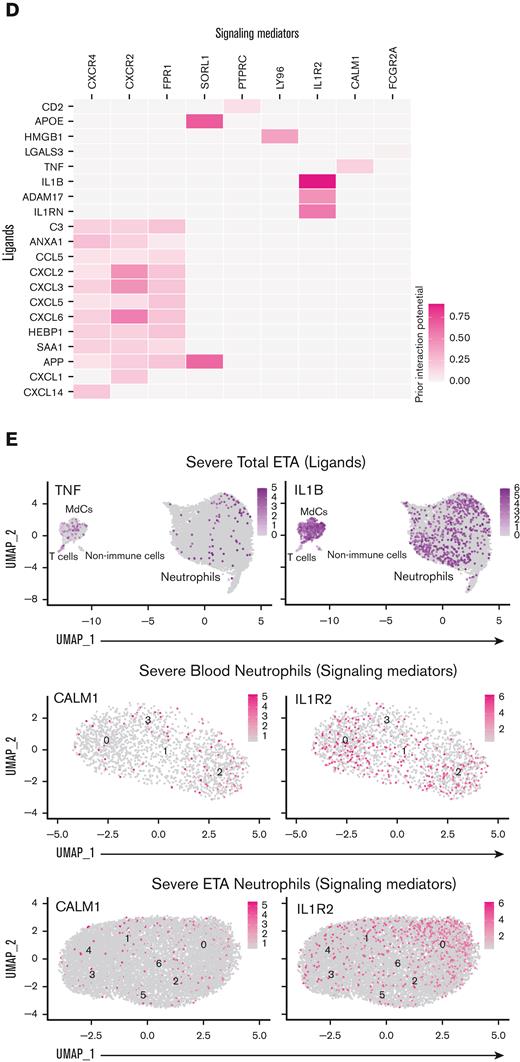
To ascertain which ligand-receptor pairs are potentially responsible for the transcriptional reprogramming of neutrophils in the lung, we first performed a DGE analysis between blood neutrophil cluster 2 and lung neutrophils to identify genes that are upregulated in lung-recruited neutrophils (Figure 4A). Next, we used the computational method NicheNet30 to identify potential (prioritized) ligands that could induce the upregulation of genes identified by DGE analysis. TNF, IL1B, and APOE were the highest prioritized ligands with high ligand activity, and their signaling axes have the regulatory potential to drive the gene expression profiles observed in lung-recruited neutrophils (Figure 4B). NicheNet analysis revealed that TNF as the ligand predicted to increase the expression of BCL2A1, CCL4 (MIP-1β), and CXCL16 in lung neutrophils, which are known antiapoptotic factors and monocyte chemoattractants, respectively. Furthermore, both TNF and IL1B are ligands predicted to induce NFKBIA and CXCL8 (IL-8), and IL1B has the greatest potential to induce the expression of the monocyte chemotactic factor CCL3 (MIP-1α), whereas APOE is predicted to upregulate the expression of FCER1G. Interestingly, TNF, IL1B, and HMGB1 are ligands with the widest range of regulatory potential to induce neutrophil reprogramming by upregulating most genes that we identified as differentially expressed in lung-recruited neutrophils (Figure 4A,B).
To investigate the cellular source of these regulatory ligands, we explored the transcriptome data and found that pulmonary T cells represent the primary source of TNF and that MdCs expressed the highest levels of IL1B and APOE transcripts (Figure 4C,E). From the transcriptome data, ligand-receptor pair analyses further identified putative signaling mediators for the top 15 prioritized ligands (Figure 4D). TNF is predicted to signal through a receptor associated with CALM1 (Figure 4D). Intriguingly, it has been previously reported that CALM1 can bind to other transmembrane proteins, including ACE-2,43 and regulate their cell surface expression.44IL1B is predicted to signal through the canonical IL1B/IL1R2 pathway and APOE through the APOE/SORL1 lipid pathway. HMGB1 is predicted to signal through LY96 (MD-2), which is commonly associated with TLR4 (Figure 4D).45-47 In agreement with our predicted NicheNet analysis, IL1R2 and CALM1 transcripts were abundant in neutrophils from blood cluster 2, and their expression was sustained upon recruitment to the airways (Figure 4E).
Collectively, we showed that TNF- and IL-1β-mediated transcriptional reprogramming of lung infiltrating neutrophils leads to the induction of proinflammatory NFKBIA, CCL3 (MIP-1α), and CCL4 (MIP-1β), along with TNFAIP3/6 and CXCL8 (Figure 3 and 4). Notably, neutrophil-derived CCL3 and CCL4 attract inflammatory monocytes from circulation to the airways. Interestingly, lung-infiltrating monocytes also produced elevated levels of CXCL8, potentiating the recruitment of circulating neutrophils through the CXCL8 (IL-8)/CXCR2 axis (Figure 3A,D). Hence, both neutrophils and inflammatory monocytes in the airways exacerbate pathogenic neutrophilia in the lungs of severe patients in a hyperinflammatory feedforward loop.
Upon recruitment, airway neutrophils acquire a distinct phenotype and produce highly inflammatory molecules, including NE, IL-1β, IL-6, and IL-8
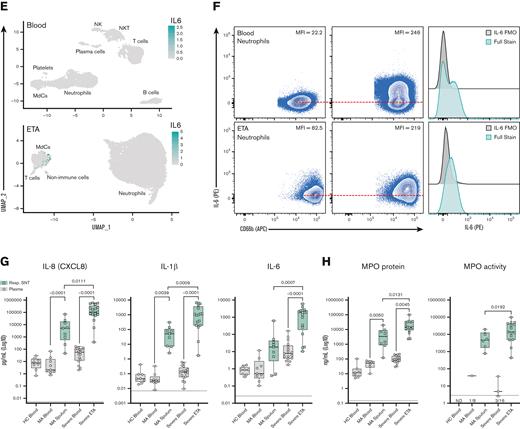
Since we identified prominent neutrophilia in our cohort, we developed a Hi-D, 30-parameter flow cytometry panel to reveal additional inflammatory neutrophil phenotype and validate our findings from the multiomics scRNA-seq data (see "CoV-Neutrophil" panel in supplemental Table 3). We used extracellular staining of NE and CD63 (LAMP-3) to assess primary granule release,48 coupled with intracellular staining for key effectors implicated in cytokine storm (IL-1β, IL-6, and IL-8), which appeared to be upregulated in our single-cell GEX data. We observed that nearly all neutrophils recruited to the lung developed an inflammatory profile characterized by exacerbated levels of NE and elevated production of IL-1β and IL-8 (protein levels) with a dramatic increase in primary granule release (Figure 5A-D), validating our finding that CXCL8 (IL-8) mRNA transcript was dramatically increased in neutrophils by scRNA-seq (Figure 3D). Most neutrophils in the airways were positive for all 3 intracellular cytokines and expressed CD63 on the cell surface (Figure 5C-D), suggesting that neutrophils are likely to release cytokines upon degranulation. In contrast, only a small proportion of blood neutrophils underwent granule release and instead expressed a cytokine signature limited mainly to IL-1β production, with much lower levels of IL-6 and IL-8 (Figure 5C). We then explored cell-surface markers to identify the potential pathogenic state of lung-recruited neutrophils. Of note, we found a subset of neutrophils in the lung that was characterized by high levels of surface CD10, CD184 (CXCR4), and NE (Figure 5B,C). We also observed a partial reduction in surface CD16 (FcγRIII) expression concomitant with increased NE and CD63, suggesting a GRIM-like phenotype in transcriptionally active pulmonary neutrophils, as previously described.32 In addition, neutrophils in the lung had lower levels of FcγRII (CD32) than those in the blood (supplemental Figure 4d). Together, the Hi-D flow cytometry results confirmed that upon recruitment to the lungs, neutrophils produced exacerbated levels of potentially damaging enzymes (eg, NE) and inflammatory cytokines (IL-1β/6/8) while undergoing pronounced primary granule release and phenotypic changes.
Neutrophil-secreted inflammatory molecules, not viral load, in the airways potentiate ARDS in severe patients
To better understand the inflammatory signaling milieu and relate the intracellular cytokine data to protein secretion, we measured 21 total analytes in the airways and plasma using the Mesoscale U-PLEX platform (supplemental Table 7). We found that IL-8 was the most abundant chemokine in the airways of severe patients and was secreted at very high levels (Figure 6). These findings further validated our Hi-D flow cytometry and single-cell GEX analysis identifying IL-8 as the most abundant neutrophil-derived chemokine (and neutrophil chemotactic factor) in the airways of patients with severe COVID-19 (Figure 6A,B,G). Exacerbated levels of IL-8 were accompanied by high levels of IL-1β and IL-6 (Figure 6A-G). Strikingly, IL-8 levels were ∼100-fold higher in the airways of severe vs mild patients and that of IL-1β and IL-6 levels in severe patients (Figure 6G). These findings are in stark contrast to an early report49 that showed no differences between circulating and pulmonary IL-6 and IL-8 secretions in the <100 pg/mL range. However, more recent studies on both ETA50 and BALF51 samples have noted similar findings, which corroborate our findings. Therefore, these data implicate the IL-8/CXCR2 signaling axis in the persistent pathogenic neutrophilia observed in the airways of severe patients in our Black/AA cohort.
We noticed only minor differences in circulating (plasma) levels of many targets between healthy individuals and those affected by severe COVID-19, suggesting an uncoupling of local and systemic inflammatory responses,50 particularly for IL-8 and IL-1β, but also IL-6 to a lesser extent (Figure 6G; supplemental Figure 5). Conversely, we observed an increase in circulating IL-1RA levels (supplemental Figure 5) with increasing disease severity, similar to other reports.52 However, we observed an opposite trend in the airways, where IL-1RA levels appeared to be decreased in severe patients (supplemental Figure 5). This is in stark contrast to IL-1β levels, which were not markedly different in the circulation of mild vs severe patients but were significantly increased in the airways with disease severity (Figure 6G). Levels of TNF were also significantly increased in the airways with disease severity (supplemental Figure 5), further supporting our NicheNet analyses that pulmonary TNF and IL-1β are inflammatory mediators that drive the transcriptional reprogramming of neutrophils recruited to the lungs via IL-8 (Figure 4).
We also measured the concentration of neutrophil-derived MPO and its enzymatic activity across patient groups. MPO is an important neutrophil effector molecule (originating from primary granules, such as NE) implicated in respiratory illnesses such as cystic fibrosis,34 and can contribute to lung tissue damage during neutrophilic pneumonitis. MPO protein concentrations increased stepwise with disease severity in plasma and respiratory supernatants (Figure 6H). Importantly, MPO activity was mostly detected in the airways and increased with increasing disease severity (Figure 6H).
To understand the viral burden in the airways and how viral load might influence neutrophils and other pulmonary cells, we performed SARS-CoV-2-specific real time qualitative polymerase chain reaction (supplemental Figure 6a), which revealed that viral load was decreased in the upper airways of severe patients admitted to the ICU compared with MA patients (supplemental Figure 6b). However, we did note a gene signature in neutrophils associated with response to IFN-γ (Figure 3G), including a pronounced increase in IFITM2 (Figure 3C,F; supplemental Figure 4a), which has been shown to promote SARS-CoV-2 infection in human lung cells.53 By including the SARS-CoV-2 genome sequence into our human reference transcriptome in the multiomics single-cell analysis, we were able to assess viral mRNA (vRNA) transcripts at a single-cell level (supplemental Figure 6C-E) in recruited neutrophils or other cell types (ie, myeloid, lymphoid, and nonimmune cells) from severe patients. Notably, we did not detect vRNA in any cell type in the blood or airways of severe patients (supplemental Figure 6C,D). In contrast, copious amounts of vRNA were detected in infected Calu-3 cells that were subjected to the same experimental procedure (supplemental Figure 6E), as described previously.23 Together, these data indicate that the viral load was low (or not detected) in severe patients at the sample timepoints, implicating the inflammatory milieu rather than the viral burden in neutrophil transcriptional reprogramming.
Discussion
The COVID-19 pandemic further highlighted some of the socio-economic and behavioral inequalities that may have contributed to troubling disparities in COVID-19–associated morbidity and mortality,54 with almost 70% being Black/AA patients in some areas.1,2 Although socioeconomic and behavioral differences indeed contribute to health disparities among demographics, a systematic investigation to determine the immunological features that characterize disease severity in Black/AA patients is lacking. Our systematic immunology approach represents the first comprehensive study to address this knowledge gap by analyzing matching airway and blood samples from Black/AA patients. Our analysis revealed new therapeutic targets to inhibit neutrophil migration, retention, and/or survival in the lung as potentially effective interventions for individuals with severe disease that have been disproportionally affected by COVID-19.
Previous studies have reported increased neutrophilia in severe COVID-19, particularly in the circulation.55-58 In contrast, reports on exacerbated airway neutrophilia and the implication of lung neutrophils as the main cell type driving ARDS in severe patients have yielded inconclusive results.5,18,20-22 In SARS-CoV-2-infected rhesus macaques59,60 and mice,61,62 where conditions and sample collection are more controlled, exacerbated neutrophilia has been identified as the key immunological feature associated with disease severity. Notably, NE is a potent serine protease that is released from degranulating neutrophils in the lungs, and has the potential to stimulate production of TNF, IL-1β, and IL-8,63,64 and also abrogates the protective effector functions of T cells and MdCs in the lung by cleaving cell surface receptors such as TLRs and Fc-receptors.65,66 Indeed, we demonstrated NE staining on the surface of T cells and MdCs in the lung in addition to neutrophils, which may explain, in part, the reduction of FcγRII (CD32) expression on pulmonary neutrophils. However, reduction in CD32 expression may prevent IgG-mediated suppression of ISG induction, as previously reported58 in pulmonary neutrophils. Indeed, we demonstrated pronounced expression of IFITM1, IFITM2, IFITM3, and IFI30 in pulmonary neutrophils, suggesting a persistent ISG-gene signature. Similarly, intracellular showed that pulmonary neutrophils produce exacerbated levels of IL-8 and IL-1β proteins that are likely released upon degranulation along with IL-6, as indicated by their high concentration in the ETA. Intriguingly, this was coupled with the production and secretion of TNF by pulmonary T cells, which our analyses suggest drives the transcriptional reprogramming of circulating neutrophils to attain a hyperinflammatory state within the lung microenvironment, providing compelling evidence of a self-potentiating pathogenic neutrophilia in severe COVID-19.
In addition to neutrophils, other immune and nonimmune cell populations in the lungs may contribute to COVID-19 pathogenesis. Nonimmune cells (eg, stromal and epithelial cells) are known targets of SARS-CoV-2 in the lung67 and are posited to influence immune cell dynamics, especially at the outset of infection.68 Indeed, our data indicate that nonimmune cells play a role in granulocyte activation and therefore may initiate neutrophilia early in SARS-CoV-2 pathogenesis.69 Similarly, the myeloid lineage is also abundant in the airways and may influence disease progression. For example, we and others36 have shown that infiltrating neutrophils sustain local production of calgranulins (S100A8/9/11/12), which can signal through and activate myeloid cells via TLRs and RAGE receptors,70-72 compounding the already hyperinflammatory lung milieu. Interestingly, inflammatory monocytes that are likely recruited to the lungs via neutrophil-secreted CCL3/4 also show elevated levels of CXCL8 (IL-8), which helps sustain the recruitment of pathogenic neutrophils in a positive feedback loop. Furthermore, T-cell–derived TNF and HMGB1, along with myeloid-derived IL-1β, can induce the inflammatory gene signatures observed in lung-recruited neutrophils. Strikingly, TNF is a ligand predicted to increase BCL2A1 expression in pulmonary neutrophils, which is an antiapoptotic factor known to regulate neutrophil survival.73,74 Further studies should investigate other effector functions of myeloid and lymphoid subsets and how these cells interact with neutrophils to promote protection or pathology in the lungs of patients with severe COVID-19.
Modulating inflammation through corticosteroids (particularly dexamethasone) has shown clinical efficacy and is now the standard of care for patients progressing to severe COVID-19.75 However, the pleiotropic effects of glucocorticoids and their propensity to cause neutrophilia are well documented in asthma and chronic obstructive pulmonary disease.76 Furthermore, previous studies have documented differences in leukocyte sensitivity to glucocorticoids across racial/ethnic backgrounds, which drive variations in transcriptomic signatures.77 In a recent study, Sinah et al proposed that dexamethasone reduced the ISG- and prostaglandin-gene signatures characteristic of aberrant neutrophils specific to COVID-19 ARDS20 concomitant with the expansion of immunosuppressive immature neutrophils. However, despite all patients in our cohort receiving dexamethasone or its equivalent, we noted that circulating neutrophils recruited to the airways already had an established ISG signature (increased IFITM2 expression) that was further upregulated upon recruitment and transcriptional reprogramming in the inflamed airways. Furthermore, we did not identify immature neutrophils within the airways, and this phenotype was restricted to blood.
Another well-established mechanism-of-action for dexamethasone is via transcriptional repression of proinflammatory cytokines, including IL-1β, IL-8, and TNF.76,78 However, we observed very high CXCL8 and IL1B transcripts concomitant with elevated pulmonary IL-8 and IL-1β proteins in the airway fluid with prominent signatures by intracellular staining, particularly in pulmonary neutrophils. In contrast, IL-6 levels were lower than that of IL-8 and IL-1β in our patient cohort, and the overly abundant neutrophils were not producing as much IL-6 comparatively, which may explain, in part, why anti-IL-6/IL-6R studies failed to meet primary endpoints.79 Here, we additionally provide evidence of an uncoupled cytokine profile in airway fluids vs plasma where the lung microenvironment exhibits features of cytokine-induced ARDS driven largely by a proinflammatory, neutrophilic feedforward loop. Beyond COVID-19, it has been shown that resolution of neutrophilia in ARDS has substantial prognostic benefit.80 Hence, taken together with other recent reports on neutrophilia, our analyses suggest that patients of African ancestry may have either decreased sensitivity to glucocorticoids as previously described in GWAS studies77 and/or divergent mechanism of action, highlighting a critical gap in our knowledge warranting further investigation.
Although lung pathology in COVID-19 is initiated by a viral infection, ICU patients with severe disease no longer show signs of uncontrolled viral replication. In fact, not only did we not detect viral transcripts by scRNA-seq, but we also noted a decreased (or undetectable) viral burden in severe patients in the ICU vs MA patients seen in the outpatient clinic. This is further supported by our previous study where we performed plaque assays on the respiratory secretions from severe patients and revealed significantly diminished, if any, viral plaques from the ETA23 along with postmortem tissue analyses from patients who succumbed to COVID-19 disease.81,82 This may explain, in part, why antiviral drugs such as remdesivir are not able to prevent death when administered to severe patients in the ICU.83
It is important to consider the limitations in our study. Although a major strength of our study is the uniformity of sample collection in a single disease state, our airway samples were limited to ETA, in contrast to the bronchoalveolar lavage fluid (BALF) used in other studies.5,16,18-21 Although the relative abundance of immune cells may vary across the upper and lower airways, previous studies have observed a correlation between paired ETA and BALF samples. In addition, these studies show that ETA samples are not inherently neutrophilic,84 although neutrophilia may be a shared feature of non–COVID-19 and COVID-19 ARDS.80,85 In addition, although our systems immunology approach provides a comprehensive assessment of molecular disease signatures, such approaches rely on mathematical and bioinformatic methods that are inherently predictive and, in some cases, (such as CellChat and NicheNet) infer functional relationships from the transcriptional states of cells. However, we were able to validate some of these findings using our complementary Hi-D flow cytometry and secreted protein assays. Finally, our study was limited in scope to 35 subjects and single collection time points, limiting our ability to interrogate correlations with clinical outcomes and warranting further longitudinal studies in larger patient cohorts. Finally, our study was not designed to compare immune responses across different races/ethnicities, and future studies should reveal whether the immune signatures uncovered here are shared across different demographics. In fact, other studies have noted similar features of COVID-19 disease severity,16,18,36 suggesting that the findings in our target demographic may be broadly applicable to other groups, while also highlighting some important differences.
In conclusion, our findings implicate airway neutrophilia in the immunopathophysiology of severe COVID-19, where perpetual, transcriptionally active, and highly inflammatory pulmonary neutrophils drive ARDS despite a low viral burden. Thus, therapeutic interventions targeting neutrophil recruitment/retention and/or survival/reprogramming, particularly via blocking IL-8, CXCR2, and CXCR4, have the potential to constrain ARDS in severe patients, especially those most vulnerable to succumb to COVID-19. We expect these findings and the datasets we make available here to lay the groundwork for future comparative studies and inform efforts to mitigate health disparities in the ongoing COVID-19 pandemic.
Acknowledgments
This study was supported by the National Institutes of Health (NIH) National Institute of Allergy and Infectious Diseases R01AI123126 and R01AI123126-05S1 (E.E.B.G.), NIH National Heart, Lung, and Blood Institute T32-HL116271 to 07 (R.P.R.), the Program for Breakthrough Biomedical Research (E.E.B.G.), and the Lowance Center for Human Immunology (E.E.B.G.). D.J.E. was partially supported by Emory's Laney Graduate School Fellowship, and J.D.C. was supported in part by CF@LANTA, a component of Emory University and Children's Healthcare of Atlanta. The graphical abstract and supplemental Figure 6A were generated, in part, using BioRender. The authors thank Keivan Zandi, Ann Chahroudi, Nils Schoof, Kira Moresco, and Stacy Heilman of the Department of Pediatrics (Emory University), along with the Emory Biosafety Officers Kalpana Rengarajan and Esmeralda Meyer for their assistance in setting up the BSL3 facility and kindly providing SARS-CoV-2 viral stocks. The authors also thank Nadia Roan (Gladstone Institutes/University of California, San Francisco) and Sulggi Lee (University of California, San Francisco) for their helpful discussions. Flow cytometry data were collected at Emory's Pediatrics/Winship Flow Cytometry Core (access supported in part by Children's Healthcare of Atlanta). The authors acknowledge the Genomic Cores at the Yerkes Non-Human Primate Research Center at Emory University (NIH Office of The Director, NIH P51 OD011132 and S10 OD026799), Baylor College of Medicine, and Parker H. Petit Institute for Bioengineering and Bioscience at the Georgia Institute of Technology for single-cell library sequencing. The authors are grateful for the efforts of Sang N. Le, John Varghese, Anum Jalal, Saeyun Lee, and Rahul Patel, who also contributed to the patient recruitment. The authors also thank the nurses, staff, and providers in the 71 ICU at Emory University Hospital Midtown, the medical ICU in Emory Decatur Hospital, the 5G/6G ICU in Emory University Hospital, and the ICU in Emory Saint Joseph's Hospital for their dedication and commitment during the COVID-19 pandemic. The authors thank all the healthy individuals, patients, and their families for their participation in this study, without whom the work would not have been possible.
Authorship
Contribution: D.J.E. contributed to conceptualization, methodology, investigation, formal analysis, data curation, visualization, and wrote the original draft and reviewed and edited the manuscript; J.Y. contributed to methodology, formal analysis, data curation, visualization, and reviewed and edited the manuscript; A.K. contributed to methodology, investigation, data curation, and reviewed and edited the manuscript; V.D.G. contributed to methodology, formal analysis, investigation, data curation, and reviewed and edited the manuscript; X.P-G. contributed to methodology, formal analysis, data curation, visualization, and reviewed and edited the manuscript; J.D.C. contributed to methodology, formal analysis, investigation, data curation, and reviewed and edited the manuscript; J.E. contributed to formal analysis and visualization; B.R.B. contributed to investigation and data curation; B.S.D. contributed to investigation, data curation, and formal analysis; M.R.H. contributed to resources; F.A. and G.L.C. contributed to investigation; R.P.R. contributed to resources, data curation, and reviewed and edited the manuscript; D.Y.O. contributed to methodology and data curation; I.S. contributed to resources; C.M. contributed to methodology, and formal analysis; F.E.H.L. contributed to methodology, data curation, resources, and reviewed and edited the manuscript; R.M.T. contributed to methodology, formal analysis, data curation, resources, and reviewed and edited the manuscript; E.E.B.G. contributed to conceptualization, methodology, formal analysis, resources, data curation, wrote the original draft, and contributed to visualization, supervision, project administration, and funding acquisition; and all authors discussed the results and read and approved the final manuscript.
Conflict-of-interest disclosure: F.E.L. is the founder of MicroB-plex, Inc.; serves on the SAB of Be Biopharma Inc.; receives grants from BMGF and Genentech; and receives royalties from BLI, Inc. C.M., D.Y.O., and X.P. are employees of Genentech Inc. D.Y.O. and C.M. own Roche stocks. The remaining authors declare no competing financial interests.
The current affiliation for R.P.R. is Division of Pulmonary, Department of Medicine, Allergy and Critical Care Medicine, University of Pittsburgh School of Medicine, Pittsburgh, PA.
Correspondence: Eliver E. B. Ghosn, Lowance Center for Human Immunology, Health Sciences Research Building, 1760 Haygood Dr NE, E240, Atlanta, GA 30322; e-mail: eliver.ghosn@emory.edu.
References
Author notes
Single-cell sequencing datasets presented here are available through NCBI GEO, accession number GSE186267. Contact the corresponding author for other forms of data sharing (eliver.ghosn@emory.edu).
The full-text version of this article contains a data supplement.
J.Y. and A.K. contributed equally to this study.