Key Points
TP53 mutations are independently associated with inferior PFS, even in the presence of other high-risk features.
CLMA can reliably and rapidly identify genetic mutations, which may inform patient management.
Abstract
Genetic subgroups of diffuse large B-cell lymphoma (DLBCL) have been identified through comprehensive genomic analysis; however, it is unclear whether this can be applied in clinical practice. We assessed whether mutations detected by clinical laboratory mutation analysis (CLMA) were predictive of outcomes in patients with newly diagnosed DLBCL/high-grade B-cell lymphoma (HGBL). Patients diagnosed from 2018 to 2022 whose biopsy samples were subjected to CLMA and who received rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone or rituximab plus etoposide, prednisolone, vincristine, cyclophosphamide, and doxorubicin were analyzed for overall/complete response rate (ORR/CRR) and estimated progression-free/overall survival (PFS/OS). CLMA was successfully performed in 117 of 122 patient samples (96%), with a median turnaround time of 17 days. Median duration of follow-up was 31.3 months. Of the mutations detected in ≥10% of the samples, only TP53 was associated with both progression and death at 2 years. TP53 mutations were detected in 36% of tumors, and patients with TP53 mutations experienced significantly lower ORR (71% vs 90%; P = .009), CRR (55% vs 77%; P = .01), 2-year PFS (57% vs 77%; P = .006), 2-year OS (70% vs 91%; P = .001), and median OS after relapse (6.1 months vs not yet reached; P = .001) as than those without TP53 mutations. Furthermore, patients with TP53 loss-of-function (LOF) mutations experienced lower rates of 2-year PFS/OS than those with non-LOF mutations and inferior or near-inferior 2-year PFS if harboring high-risk clinicopathologic features. TP53 mutations identified through CLMA can predict for inferior outcomes in patients with newly diagnosed DLBCL/HGBL. Results of CLMA can be used in real time to inform prognosis and/or identify candidates for clinical trials.
Introduction
Although approximately two-thirds of patients diagnosed with diffuse large B-cell lymphoma (DLBCL) are cured after receipt of firstline immunochemotherapy, efforts to predict those at risk of developing relapsed/refractory disease are needed, given that the majority of patients who are not cured will die of complications from lymphoma. Molecular testing performed on biopsy samples from patients with newly diagnosed DLBCL has identified subgroups of patients with inferior survival after receipt of rituximab with cyclophosphamide, doxorubicin, vincristine and prednisone (R-CHOP), including those with activated B-cell cell of origin as per gene expression profiling1 and those with MYC rearrangements with or without concomitant BCL2 and/or BCL6 rearrangements as per fluorescence in situ hybridization (FISH).2-6
More recently, comprehensive genomic analyses performed on biopsy specimen from patients with newly diagnosed DLBCL have classified tumors into multiple genetic subgroups, some of which are associated with inferior progression-free survival (PFS) and overall survival (OS) after receipt of R-CHOP.7,8 Although comprehensive genomic analysis is not currently feasible to perform in clinical practice, mutation analysis can be performed in clinical laboratories through massively parallel sequencing, the result of which may aid in assignment of a genetic subgroup to some tumors through the LymphGen platform.9 One analysis of targeted mutational analysis performed on biopsy specimen from a large cohort of patients with newly diagnosed DLBCL identified genetic subgroups with similarities to those characterized through comprehensive genomic analyses; however, when restricted to patients treated with R-CHOP, a poor-risk genetic subgroup was not clearly identified.10
Additionally, given the heterogeneity of mutations detected within genetic subgroups, it is unclear that a common signaling pathway is altered in all tumors within a genetic subgroup or that it would be practical to combine a specific targeted therapy with firstline immunochemotherapy for all patients with DLBCL whose tumors are classified within a genetic subgroup. However, a subset analysis of the PHOENIX trial revealed that patients with DLBCL aged ≤60 years whose tumors were classified into MCD (9%) or N1 (4%) genetic subgroups experience significantly improved PFS and OS if treated with R-CHOP plus ibrutinib as compared with those treated with R-CHOP alone.11
Furthermore, many mutations are commonly found within multiple genetic subgroups, and combining a specific targeted therapy with firstline immunochemotherapy for patients with DLBCL whose tumors are classified in the genetic subgroup characterized by a given mutation may disregard those who harbor the mutation but are classified in other subgroups. For example, although nearly 90% of A53 tumors harbor a TP53 mutation, this mutation is also detected in ∼20% to 50% of tumors classified in other genetic subgroups, and given that the prevalence of the A53 genetic subtype is only 6.6%,9 restricting an experimental therapy targeting TP53 only to patients whose tumors are classified as A53 would mean that the vast majority of patients whose tumors harbor TP53 mutations would be excluded from such therapy.
We aimed to determine whether any individual recurring genetic mutations detected in tumors from patients with newly diagnosed DLBCL and high-grade B-cell lymphoma (HGBL) through clinical laboratory mutation analysis (CLMA) were predictive of response and survival outcomes after treatment with firstline immunochemotherapy.
Methods
Inclusion criteria for this analysis were patients diagnosed with DLBCL/HGBL (either de novo or transformed indolent lymphoma with non–small cell lymphocytic histology without receipt of prior cytotoxic chemotherapy) between January 2018 and July 2022 who received either standard-dose R-CHOP or rituximab with etoposide, prednisone, vincristine, cyclophosphamide, and doxorubicin (R-EPOCH) as firstline therapy and had mutation analysis performed on their initial diagnostic biopsy sample with 1 of 2 lymphoma-focused gene sequencing panels at the Penn Center for Personalized Diagnostics at the University of Pennsylvania (lymphoma sequencing panel [LSP] from 2018 to 2020 and PennSeq lymphoma panel [PSLP] from 2020 to 2022), details of which are provided in the supplemental Materials. Exclusion criteria included death due to nonlymphoma causes during firstline therapy and loss to follow-up <1 year after diagnosis if in remission.
Institutional standards for pathologic evaluation and diagnosis of tumor biopsy specimen included immunohistochemical staining for CD10, BCL6, and MUM1, among other makers, with the cell of origin assigned by Hans algorithm12 as well as FISH for MYC rearrangement, with reflex testing for BCL2 and BCL6 rearrangement if tested positive. Therapy was administered at the discretion of the treating physician. Disease response by computed tomography with or without positron emission tomography was determined using Lugano classification.13 PFS was defined as the interval between diagnosis of DLBCL/HGBL and relapse of DLBCL/HGBL or last follow-up in remission. OS was defined as the interval between diagnosis of DLBCL/HGBL and death due to any cause or last follow-up while alive. Survival curves were plotted using Kaplan-Meier estimates, and survival analysis was performed using the log-rank test. Univariate and multivariate analysis were performed using Cox proportional-hazards regression. Categorical variables were analyzed using Fisher exact test. Statistical significance was defined as a 2-tailed P value < .05. All statistical analyses were performed using Stata version 13 (StataCorp, College Station, TX). Data were censored on 1 July 2023. This protocol was approved by the institutional review board of the University of Pennsylvania.
Results
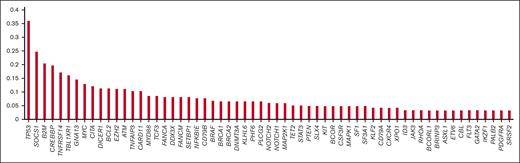
In total, 137 patients were initially included, with 15 subsequently excluded (8 because of death from nonlymphoma causes during firstline therapy, and 7 because of becoming lost to follow-up <1 year after diagnosis when in remission), and 117 of 122 patient biopsy samples underwent successful CLMA (LSP for 55 and PSLP for 62), with a success rate of 96%. Median time from receipt of biopsy in the genomics laboratory to report release (turnaround time) was 17 days. Analyzed samples of successful assays were paraffin-embedded tissue for 102, bone marrow for 12, and cytology for 3. Request for performing mutation analysis as per the review of the patient’s medical record for successful assays was made by the interpreting hematopathologist for 107 patients (101 as part of routine evaluation and 6 specifically to aid in rendering a diagnosis) and the treating clinician for 10 patients. A histogram depicting the frequency of mutations detected in ≥3% of cases tested is shown in Figure 1, and variants detected in each tumor are listed in supplemental Table 1.
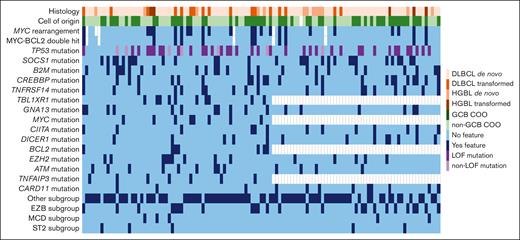
Baseline characteristics, including mutations occurring in ≥10% of cases tested as well as subgroup classification by LymphGen 2.0 based on available molecular data, are listed in Table 1. Additionally, individual patient tumor characteristics are shown in Figure 2. R-CHOP was received by 73 patients (62%), and R-EPOCH by 44 patients (38%).
Individual patient tumor characteristics. COO, cell of origin; GCB, Germinal center B.
Individual patient tumor characteristics. COO, cell of origin; GCB, Germinal center B.
For all patients, the overall response rate (ORR) was 84% (69% complete response rate [CRR] and 15% partial response rate). With a median follow-up of 31.3 months, the estimated 2-year PFS was 70% (95% confidence interval [CI], 60-78), estimated 2-year OS was 83% (95% CI, 74-89), and estimated median OS after relapse was 21.4 months (95% CI, 6.7 to not yet reached). Univariate Cox regression analysis of characteristics listed in Table 1 identified HGBL classification, MYC rearrangement, and TP53 mutation as significantly predictive of progression at 2 years; however, of these characteristics, only TP53 mutation remained predictive on multivariate analysis (hazard ratio [HR], 2.3; 95% CI, 1.1-4.8; P = .03). Additionally, univariate analysis identified International Prognostic Index (IPI) score ≥ 3, HGBL classification, MYC rearrangement, MYC-BCL2 double-hit lymphoma, TP53 mutation, and TNFRSF14 mutation as significantly predictive of death at 2 years; however, of these characteristics, only HGBL classification (HR, 5.0; 95% CI, 1.1-22.7; P = .04) and MYC rearrangement (HR, 4.4; 95% CI, 1.0-18.0; P = .04) remained predictive on multivariate analysis.
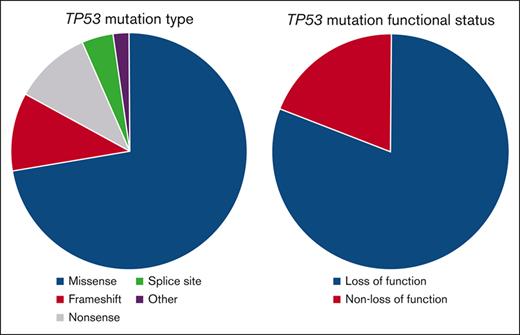
Given that TP53 mutations predicted for both progression and death at 2 years, we explored the characteristics of these mutations. Forty-nine TP53 mutations were detected in 42 patient samples (36% of patients) and were classified as missense in 34, frameshift in 6, nonsense in 6, splice site in 2, and other in 1. Forty-two (86%) TP53 mutations were deemed loss-of-function (LOF) mutations, either as a missense mutation designated as nonfunctional and/or LOF via query of The TP53 Database or any nonsense, frameshift, or splice site mutation as per ClinVar and/or PVS1 criteria for predicted LOF variants14 as well as existing literature.15 Details of each TP53 mutation are listed in Table 2.
Comparison of baseline characteristics of patients with (n = 42) vs without (n = 75) any TP53 mutation is listed in Table 3. The proportion of patients receiving R-CHOP/R-EPOCH was similar for patients with vs without TP53 mutations (61%/39% vs 64%/36%; P = .84).
For patients with vs without TP53 mutations, the ORR was 71% (55% CRR) vs 90% (77% CRR; P = .009 for ORR and P = .01 for CRR); estimated 2-year PFS was 57% (95% CI, 40-70) vs 77% (95% CI, 65-86; P = .006), as depicted in Figure 3A; estimated 2-year OS was 70% (95% CI, 53-82) vs 91% (95% CI, 80-96; P = .001), as depicted in Figure 3B; and estimated median OS after relapse was 6.1 months (95% CI, 4.2-11.5) vs not yet reached (95% CI, 16.3 to not yet reached; P = .01), as depicted in Figure 3C. Additionally, for patients with TP53 mutations treated with R-CHOP vs R-EPOCH, the estimated 2-year PFS was 70% (95% CI, 49-84), vs 33% (95% CI, 12-56; P = .01) and estimated 2-year OS was 85% (95% CI, 64-94), vs 43% (95% CI, 17-67; P = .001), as depicted in supplemental Figure 1. This finding is likely because of the fact that patients with TP53 mutations who were treated with R-EPOCH (n = 15) were more likely to have unfavorable baseline characteristics, specifically an IPI score ≥ 3 (80% vs 44%; P = .049), HGBL classification (73% vs 7%; P = .049), MYC rearrangement (87% vs 0%; P = .049), and MYC-BCL2 double-hit lymphoma (40% vs 0%; P = .049), than those with TP53 mutations treated with R-CHOP (n = 27).
Survival outcomes based upon TP53 mutation status. (A) PFS, (B) OS, and (C) OS after relapse based upon TP53 mutation status; (D) PFS and (E) OS based upon TP53 LOF and non-LOF mutation status. Mut, mutated.
Survival outcomes based upon TP53 mutation status. (A) PFS, (B) OS, and (C) OS after relapse based upon TP53 mutation status; (D) PFS and (E) OS based upon TP53 LOF and non-LOF mutation status. Mut, mutated.
In terms of TP53 variant allele frequency (VAF), we analyzed survival outcomes based on quartiles, with 25th, 50th, and 75th percentiles of 21%, 35%, and 50% VAF, respectively. There was no significant difference in the estimated 2-year PFS for patients with TP53 VAF falling within quartile 1 (70%; 95% CI, 33-89) vs quartile 2 (45%; 95% CI, 17-71) vs quartile 3 (48%; 95% CI, 16-75) vs quartile 4 (64%; 95% CI, 30-85; P = .53). Similarly, there was no significant difference in the estimated 2-year OS for patients with TP53 VAF falling within quartile 1 (80%; 95% CI, 41-95) vs quartile 2 (61%; 95% CI, 25-83) vs quartile 3 (70%; 95% CI, 33-89) vs quartile 4 (73%; 95% CI, 37-90; P = .78). Additionally,7 patients had biallelic TP53 mutations, 6 of whom developed progressive disease.
For patients with at least 1 TP53 LOF mutation (n = 35) vs only TP53 non-LOF mutation (n = 7) vs without TP53 mutation (n = 75), the estimated 2-year PFS was 51% (95% CI, 34-66) vs 86% (95% CI, 33-98) vs 77% (95% CI, 63-84; P = .002), as depicted in Figure 3D, and the estimated 2-year OS was 64% (95% CI, 46-78) vs 100% (95% CI, not calculable) vs 91% (95% CI, 80-96; P = .004), as depicted in Figure 3E.
For the subset of patients with TP53 mutations, HGBL classification (HR, 3.5; 95% CI, 1.4-8.8; P = .01) was significantly predictive of progression at 2 years on univariate Cox regression analysis, whereas HGBL classification (HR, 3.9; 95% CI, 1.2-12.2; P = .02) and MYC rearrangement (HR, 3.9; 95% CI, 1.2-12.2; P = .02) were significantly predictive of death at 2 years on univariate analysis; however neither remained predictive of death at 2 years on multivariate analysis. Similarly, for the subset of patients with TP53 LOF mutations, HGBL classification (HR, 3.3; 95% CI, 1.3-8.7; P = .01) was significantly predictive of progression at 2 years on univariate analysis whereas HGBL classification (HR, 3.2; 95% CI, 1.0-10.1; P = .046) and MYC rearrangement (HR, 3.2; 95% CI, 1.0-10.1; P = .046) were significantly predictive of death at 2 years on univariate analysis; however neither remained predictive of death at 2 years on multivariate analysis.
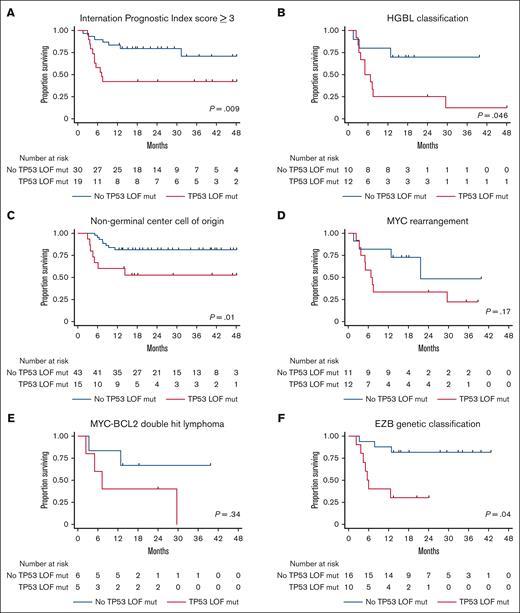
Because TP53 mutations were predictive of inferior estimated 2-year PFS on multivariate regression and patients with TP53 LOF mutations experienced a lower rate of estimated 2-year PFS than those with non-LOF mutations, an exploratory analysis was performed to evaluate whether TP53 LOF mutations were predictive of estimated 2-year PFS for subsets of patients with high-risk clinicopathologic features. Estimated 2-year PFS for patients with and without TP53 LOF mutations was 42% (95% CI, 20-62) vs 80% (95% CI, 61-90; P = .009), respectively, for the subset with an IPI score ≥ 3, as depicted in Figure 4A; 25% (95% CI, 6-50) vs 70% (95% CI, 33-89; P = .046), respectively, for the subset with HGBL classification, as depicted in Figure 4B; 53% (95% CI, 25-71) vs 81% (95% CI, 66-90; P = .01), respectively, for the subset with non-GCB cell of origin, as depicted in Figure 4C; 33% (95% CI, 10-59) vs 48% (95% CI, 9-81; P = .17), respectively, for the subset with MYC rearrangement, as depicted in Figure 4D; 40% (95% CI, 5-75) vs 67% (95% CI, 19-90; P = .34), respectively, for the subset with MYC-BCL2 double-hit lymphoma, as depicted in Figure 4E, and 30% (95% CI, 7-58) vs 72% (95% CI, 51-85; P = .04), respectively, for the subset with EZB genetic classification, as depicted in Figure 4F.
Survival outcomes based upon TP53 mutation and high-risk feature status. PFS for patients with (A) an IPI score ≥ 3, (B) HGBL classification, (C) non–germinal center cell of origin, (D) MYC rearrangement, (E) MYC-BCL2 double-hit lymphoma, and (F) EZB genetic classification based upon TP53 LOF mutation status.
Survival outcomes based upon TP53 mutation and high-risk feature status. PFS for patients with (A) an IPI score ≥ 3, (B) HGBL classification, (C) non–germinal center cell of origin, (D) MYC rearrangement, (E) MYC-BCL2 double-hit lymphoma, and (F) EZB genetic classification based upon TP53 LOF mutation status.
Although an external validation cohort was not available, analysis of survival outcomes by TP53 LOF mutation status for patients whose tumors were analyzed by LSP vs PSLP was conducted, as depicted in supplemental Figure 2. For patients whose tumors underwent LSP who had a median length of follow-up of 42.6 months, the estimated 2-year PFS for those with at least 1 TP53 LOF mutation (n = 15) vs no TP53 LOF mutation (n = 40) was 47% (95% CI, 21-69) vs 80% (95% CI, 64-89; P = .008), respectively, and the estimated 2-year OS was 67% (95% CI, 38-85) vs 90% (95% CI, 76-96; P = .03), respectively. Among patients whose tumor samples underwent PSLP who had a median duration of follow-up of 20 months, the estimated 2-year PFS for those with at least 1 TP53 LOF mutation (n = 20) vs no TP53 LOF mutation (n = 42) was 55% (95% CI, 31-73) vs 74% (95% CI, 53-87; P = .03), respectively, and the estimated 2-year OS was 65% (95% CI, 40-81) vs 92% (95% CI, 68-98; P = .009), respectively,.
Discussion
TP53 mutations have been historically reported to be predictive of inferior survival in patients with newly diagnosed DLBCL whose tumors harbor these mutations. Most notably, >100 patients with newly diagnosed DLBCL with tumors harboring TP53 mutations experienced inferior PFS and OS as compared with those without TP53 mutations when treated with R-CHOP or R-CHOP–like therapy, as identified through the International DLBCL Rituximab-CHOP Consortium Program.15 A similar finding was reported through an analysis of patients with newly diagnosed DLBCL included in the RICOVER-60 trial, which included ∼60 patients with tumors harboring TP53 mutations.16 Smaller studies of patients with newly diagnosed DLBCL have demonstrated similar results.17-20 Interestingly, the aforementioned report of targeted mutational analysis performed on biopsy samples from a large cohort of patients with newly diagnosed DLBCL reported an inferior OS for patients whose tumors were classified into some of their proposed genetic subgroups if demonstrating TP53 mutation.10 Another report of genetic classification of DLBCL tumors using a 38-gene LymphPlex algorithm defined 1 genetic subgroup characterized solely based upon the presence of TP53 mutation, which was associated with a lower rate of PFS than other subgroups.21
TP53 and p53 protein have been described as “undruggable” because of failure of targeted agents to be clinically effective in patients with TP53-mutated cancers and because that the desirable therapeutic outcome would be to restore the normal conformation of mutated p53 protein, which is overexpressed in TP53-mutated tumors, which is not a typical mechanism of action for small-molecule inhibitors.22 However, a strong association between increased expression of both p53 and EZH2 protein in tumors of patients with DLBCL has been reported,23 and p53-induced suppression of the EZH2 promoter leading to cell senescence does not occur in cells harboring TP53 mutations in vitro,24 suggesting that EZH2 inhibition may be an effective therapeutic strategy for patients with TP53-mutated DLBCL/HGBL. Furthermore, gene expression profiling of TP53-mutated tumors of patients with DLBCL identified activation of phosphoinositide 3-kinase signaling,21 which could support investigation of phosphoinositide 3-kinase inhibitors for patients with TP53-mutated DLBCL/HGBL. Finally, RNA sequencing demonstrated downregulation of interferon and apoptosis pathways as well as CD8+ T-cell infiltration in DLBCL tumors with TP53 mutations as compared with tumors without,25 and interestingly, a small number of patients with newly diagnosed DLBCL harboring TP53 mutations experienced a high overall response rate as well as increased expression of CD4+ and CD8+ T cells as well as serum interferon gamma levels after treatment with R-CHOP plus decitabine,26 suggesting that treatment with DNA methyltransferase inhibitors may benefit patients with TP53-mutated DLBCL/HGBL.
Classification of TP53 mutations as LOF vs non-LOF is notable as an effort to identify patients whose tumors are likely to develop as a result of dysregulation of TP53. The presence of a high-risk genetic alteration in a DLBCL/HGBL tumor does not necessarily equate with abnormal protein synthesis, and it is not clear that all patients with such an alteration in their tumors experience inferior survival if treated with standard-of-care therapy. One example of this is patients diagnosed with double-hit lymphoma harboring MYC and BCL2 rearrangements (MYC-BCL2 double-hit lymphoma), who are typically treated with R-EPOCH or other intensive therapies in the firstline setting; however, patients with MYC-BCL2 double-hit lymphoma whose tumors demonstrate non–MYC-IG rearrangement have a similar prognosis to those without MYC-BCL2 double-hit lymphoma when treated with R-CHOP.6
Additional strengths of our analysis are inclusion of patients with HGBL tumors as well as those treated with R-EPOCH, reflecting current practice in the diagnosis and treatment of aggressive B-cell lymphomas. Furthermore, analyses of subgroups of patients with high-risk features revealed that the presence of TP53 LOF mutations may further discriminate PFS, supporting exploration of efficacy outcomes for patients with TP53 LOF mutations enrolled on clinical trials investigating novel firstline treatment regimens for high-risk DLBCL/HGBL. Finally, we demonstrate the feasibility of performing CLMA, given the high success rate and relatively short turnaround time, and it is plausible that the result of CLMA could be factored into a decision regarding treatment modification, such as the addition of a small-molecule inhibitor to firstline immunochemotherapy at the start of cycle 2 in the experimental setting. Ongoing randomized clinical trials evaluating the benefit of venetoclax added to R-CHOP (NCT03984448) and acalabrutinib added to R-CHOP (NCT04529772) for patients with newly diagnosed DLBCL/HGBL allow 1 cycle of R-CHOP to be administered while confirming pathologic testing before study arm assignment at the start of cycle 2.
Weaknesses of our analysis include a smaller sample size of patients as well as the use of more limited gene panels as compared with some other published studies, which might have limited our ability to identify other mutations that predict for inferior clinical outcomes in this patient population. Additionally, although we did not analyze an external validation cohort of patients, it is notable that patients with TP53 LOF mutations experienced inferior estimated 2-year PFS and OS as compared with those without TP53 LOF mutations when analyzed as 2 separate cohorts based upon sequencing panel performed. Finally, it is likely that some patient tumors in our series were misclassified as “other” as per LymphGen because of our inability to detect all mutations as well as rearrangements and copy number changes included in this predictive algorithm.
Although it would be ideal if all clinical laboratories used identical algorithms for characterizing TP53 LOF mutations detected in DLBCL/HGBL tumors, this is unlikely to be the case in current practice, as suggested by reporting of different algorithms used for characterization of TP53 mutations in tumors from patients diagnosed with myelodysplastic syndrome and acute myeloid leukemia.27-29 We feel it is relevant to offer a framework for doing so, mainly using searchable online databases in hopes that this could contribute to development of a standardized and reproducible reporting algorithm for classification of TP53 mutations in DLBCL/HGBL tumors. Additional efforts to analyze p53 expression by immunohistochemical staining, and del17p status by FISH may also be informative in this effort.
In conclusion, TP53 mutations as detected by CLMA predict for inferior outcomes, most notably inferior estimated 2-year PFS on multivariable analysis, in patients with newly diagnosed DLBCL/HGBL treated with R-CHOP or R-EPOCH. Additionally, TP53 LOF mutations predict for inferior estimated 2-year PFS in high-risk subsets of these patients, including those with EZB genetic classification by LymphGen. Patients with newly diagnosed DLBCL/HGBL with tumors harboring TP53 mutation can be reliably and rapidly identified through CLMA, which can inform prognosis for those treated with R-CHOP/R-EPOCH as well as reveal candidates for treatment on protocols investigating therapies that target derangements caused by TP53 mutations. The use of CLMA in other treatment settings for patients with DLBCL/HGBL should be explored.
Authorship
Contribution: D.J.L. designed research, treated patients, analyzed data, and wrote and edited the manuscript; J.J.D.M. and A.B. performed research and edited the manuscript; and S.D.N., S.K.B., S.J.S., J.S., and E.A.C. treated patients and edited the manuscript.
Conflict-of-interest disclosure: D.J.L. declares a consultancy or advisory role with MorphoSys, Epizyme, Calithera, ADC Therapeutics, and Karyopharm; research funding from Curis and Calithera; and travel grants from Novartis. S.D.N. declares research funding from Pharmacyclics, Roche, Rafael, and FortySeven. S.K.B. declares a consultancy or advisory role with Daiichi Sankyo, Kyowa Kirin, Janssen, and Affimed and honoraria from Acrotech, Seagen, and Kyowa Kirin. S.J.S. declares a consultancy or advisory role with Novartis, Regeneron, Nordic, MorphoSys, Mustang Bio, Incyte, Genentech/Roche, Janssen, Legend Biotech, Loxo, Acerta, BeiGene, Celgene, and Nanovecter; and research funding from Novartis, Pharmacyclics, Merck, DTRM, Juno Therapeutics, AbbVie, Adaptive Biotechnologies, Incyte, Genentech/Roche, Celgene, and TG Therapeutics. J.S. declares a consultancy or advisory role with Seagen, Pharmacyclics, Incyte, Genmab, Bristol Myers Squibb, Atara, AstraZeneca, Adaptive, and ADCT; research funding from TG Therapeutics, Seagen, Pharmacyclics, Merck, Incyte, Bristol Myers Squibb, and AstraZeneca. E.A.C. received honoraria from Juno/Bristol Myers Squibb, Novartis, BeiGene, Kite Pharma, and Tessa. The remaining authors declare no competing financial interests.
Correspondence: Daniel J. Landsburg, Perelman Center for Advanced Medicine, University of Pennsylvania, 3400 Civic Center Blvd, 12-182, Philadelphia, PA 19104; e-mail: daniel.landsburg@pennmedicine.upenn.edu.
References
Author notes
Data are available on request from the corresponding author, Daniel J. Landsburg (daniel.landsburg@pennmedicine.upenn.edu).
The full-text version of this article contains a data supplement.