Key Points
We identify USP9X as a female-specific B-cell ALL susceptibility gene associated with multiple congenital anomalies.
In sporadic pediatric B-cell ALL, USP9X acts a tumor suppressor gene in both males and females.
Abstract
We recently reported that children with multiple birth defects have a significantly higher risk of childhood cancer. We performed whole-genome sequencing on a cohort of probands from this study with birth defects and cancer and their parents. Structural variant analysis identified a novel 5 kb de novo heterozygous inframe deletion overlapping the catalytic domain of USP9X in a female proband with multiple birth defects, developmental delay, and B-cell acute lymphoblastic leukemia (B-ALL). Her phenotype was consistent with female-restricted X-linked syndromic intellectual developmental disorder-99 (MRXS99F). Genotype-phenotype analysis including previously reported female probands (n = 42) demonstrated that MRXS99F probands with B-ALL (n = 3) clustered with subjects with loss-of-function (LoF) USP9X variants and multiple anomalies. The cumulative incidence of B-ALL among these female probands (7.1%) was significantly higher than an age- and sex-matched cohort (0.003%) from the Surveillance, Epidemiology, and End Results database (P < .0001, log-rank test). There are no reports of LoF variants in males. Males with hypomorphic missense variants have neurodevelopmental disorders without birth defects or leukemia risk. In contrast, in sporadic B-ALL, somatic LoF USP9X mutations occur in both males and females, and expression levels are comparable in leukemia samples from both sexes (P = .54), with the highest expressors being female patients with extra copies of the X-chromosome. Overall, we describe USP9X as a novel female-specific leukemia predisposition gene associated with multiple congenital, neurodevelopmental anomalies, and B-ALL risk. In contrast, USP9X serves as a tumor suppressor in sporadic pediatric B-ALL in both sexes, with low expression associated with poorer survival in patients with high-risk B-ALL.
Introduction
Genetic syndromes are one of the strongest known risk factors for developing acute leukemia. For example, children with chromosomal aneuploidies, in particular trisomy 21, have 20-fold increased risk of leukemia (both acute myeloid leukemia and acute lymphoblastic leukemia [ALL]).1 In addition, autosomal recessive (eg, ataxia telangiectasia) and X-linked disorders (eg, Wiskott-Aldrich syndrome) are also associated with leukemia risk.2 Many other leukemia predisposition syndromes are autosomal dominant disorders associated with variants in genes related to hematopoietic development, for example, PAX5, and present without syndromic or neurodevelopmental features.3
In our recent Genetic Association Between Anomalies and Cancer in Kids (GOBACK) study, we evaluated the associations between birth defects and childhood cancer. We found that children with nonchromosomal birth defects are 1.3 times (95% confidence interval [CI], 1.1-1.5) more likely to develop ALL compared with children without birth defects.3 When evaluating specific birth defects, we observed several novel associations. For example, children with choanal atresia were 9.2 times more likely (95% CI, 3.8-22.1) to develop acute leukemia compared with children without birth defects. In addition, the risk of any cancer was greater with increasing number of birth defects. For hematologic malignancies, the hazard ratio (HR) was 3.7 for ≥4 birth defects vs none (95% CI, 3.1-4.5). Based on these findings, we hypothesized that genomic sequencing of children with birth defects and cancer would reveal novel cancer predisposition syndromes.
Here, we describe the results of whole-genome sequencing (WGS) of proband/parent trios of a small heterogeneous cohort of participants from the GOBACK study leading to the identification of a de novo inframe single-exon deletion in USP9X in a female proband with multiple anomalies and cancer consistent with the recently described female-restricted X-linked syndromic intellectual developmental disorder-99 (MRXS99F; MIM# 300968).4,5 We provide evidence that the MRXS99F syndrome resulting from loss-of-function (LoF) USP9X germline variants is a female-limited leukemia predisposition disorder, and in contrast, USP9X acts as a tumor suppressor gene with an impact on survival in sporadic (non-germline) pediatric B-ALL in both males and females.
Methods
Enrollment of probands with birth defects and childhood cancer
The GOBACK research protocol was described previously6 and approved by the Institutional Review Board for Human Subjects Research at Baylor College of Medicine. Parents of children with birth defects and cancer were identified through state-based cancer registries and then contacted for study participation. Consented families provided medical records, completed questionnaires, and biological samples from the child, parents, and siblings (if available) using Oragene DNA saliva self-collection kits (Genotek, Ottawa, Ontario, Canada). Medical records, questionnaires, and saliva samples were returned to the Population Sciences Biorepository at Baylor College of Medicine to be processed, extracted, and cataloged.
Germline WGS and variant analysis
WGS was performed at the Human Genome Sequencing Center at Baylor College of Medicine on an Illumina HiSeqX platform to generate paired-end sequence reads with an average 40× coverage. Alignment was performed using Burrows-Wheeler Alignment-Maximal Exact Match (BWA-MEM)7 against the human hs37d5 assembly, followed by base score recalibration and indel realignment using Genome Analysis Toolkit (GATK).8 Single nucleotide variants (SNVs) were called using the Platypus variant caller.9 SNVs were annotated using ANNOVAR10 and filtered for rarity using a minor allele frequency cutoff of <0.01 in gnomAD.11 Variants were visualized in the integrative genomics viewer (IGV).12
An ensemble approach was used to detect structural variants (SVs) including deletions, duplications, and inversions based on a consensus rule from 5 SV callers, namely Delly,13 Lumpy,14 CNVnator,15 Manta,16 and BreakDancer,17 from the WGS data of the trios (Figure 1). A consensus SV was defined by greater than 50% reciprocal overlap of the location of the predicted event. For interchromosomal break-ends, consensus SVs were called if 2 SVs were within 500 bp for both break-ends (determined using pgltools18). For each proband, SVs that were detected by more than 1 caller were carried forward to limit false variant calls. De novo SVs were detected by comparing SVs in the proband to SVs identified by any caller in either parent and further filtered using an internal data set of SVs identified in 74 individuals without cancer or congenital anomalies and the SV data set from the 1000 Genomes Project.19 Each putative de novo SV was also visually evaluated using IGV.
Identification of a novel de novo germline deletion in USP9X in a child with congenital anomalies and B-ALL. (A) Schematic of the SV analysis pipeline used to analyze WGS of 78 DNA samples. Five SV callers (orange boxes) were used based on their ability to detect distinct patterns in aligned reads. SV calls were merged, filtered, prioritized, and visualized as described (blue boxes). (B) IGV image showing break points of the de novo heterozygous deletion in USP9X in the GOBACK proband. (C) PCR validation of the deletion in DNA from the GOBACK proband, family members, and lymphoblastoid cell line (HG148) control.
Identification of a novel de novo germline deletion in USP9X in a child with congenital anomalies and B-ALL. (A) Schematic of the SV analysis pipeline used to analyze WGS of 78 DNA samples. Five SV callers (orange boxes) were used based on their ability to detect distinct patterns in aligned reads. SV calls were merged, filtered, prioritized, and visualized as described (blue boxes). (B) IGV image showing break points of the de novo heterozygous deletion in USP9X in the GOBACK proband. (C) PCR validation of the deletion in DNA from the GOBACK proband, family members, and lymphoblastoid cell line (HG148) control.
Polymerase chain reaction (PCR) and Sanger sequencing
PCR was performed on the DNA from saliva samples of the GOBACK proband, parents, and siblings using the TaKaRa LA Taq Kit (cat #: KK2502). Genomic DNA was extracted from the HG00148 LCL line using QIAGEN’s DNeasy Blood and Tissue Kit (cat #: 69506). Primers were designed using Primer3Plus software.20 The genomic region around the 5 kb deletion detected on chromosome X was amplified using the following primers: forward primer, GCCTCCTGAGTAGCTAGGGT and reverse primer, AACACCCCCACAAACCACAT. The expected amplicon size was 589 bp (with the deletion) and 5589 bp (without the deletion). Deletion break points were sequenced using the following primers: forward primer, TAACAGGGGTTACAGGCGTG and reverse primer, GTGTTTCTTGCAGAGGGGGA.
Description of the B-ALL cohorts of the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative
The TARGET ALL pilot project included 190 children and adolescents with high-risk precursor B-ALL who had been enrolled on the Children’s Oncology Group (COG) Trial P9906. These children had a disease onset age of >9 years, had a predicted 4-year event-free survival of 44% based on previous Pediatric Oncology Group studies and did not carry known favorable (ETV6-RUNX1 rearrangements or trisomy 4 and 10) or unfavorable genetic alterations (BCR:ABL1 translocation or hypodiploidy). These probands underwent WGS and gene expression analysis using the Affymetrix U133 Plus 2.0 Array.21
The expansion effort or phase 2 of TARGET included tissues and clinical data from fully characterized patients enrolled in the following COG protocols: AALL03B1, AALL0232, P9906, AALL0331, and ALL0434, including a broad spectrum of ALL subtypes. These patients were included if they experienced early bone marrow relapse (defined as ≤4 years from the time of diagnosis) and adequate DNA/RNA for genomic profiling extracted from peripheral blood or bone marrow. In this phase, gene expression was profiled using Affymetrix U133 Plus 2.0 Array, chromosome copy number and loss of heterozygosity analysis were performed using the Affymetrix SNP6.0 Array, DNA methylation was assessed using the HpaII tiny fragment enrichment by ligation-mediated PCR (HELP) assay, WGS was performed through Complete Genomics Incorporated, and whole-exome sequencing (WES), messenger RNA (mRNA) sequencing, and microRNA sequencing were performed using Illumina Hi-Seq 2000.
Leukemia incidence and outcome analyses
Leukemia prevalence estimates in the general population were derived from age- and sex-adjusted population estimates of B-ALL prevalence from the Surveillance, Epidemiology, and End Results (SEER) Program software, SEER∗Stat 8.3.9.2, using the “SEER Research Data, 13 Registries” database, which included data on cancer prevalence from a representative sample of the United States population from 2000 to 2018.
Survival data of patients with B-ALL in phase 2 of TARGET was obtained from the “TARGET_ALL_ClinicalData_PhaseII_Discovery_20170525.xlsx” file in the TARGET Data Matrix using the column labeled “Overall Survival Time in Days”, and vital status was obtained from the “Vital Status” column. Survival differences were assessed between patients with low (lowest quartile) vs intermediate (middle 2 quartiles) vs high (highest quartile) expression of USP9X. Survival curves were generated using the Kaplan-Meier method.
The online oncogenomics database (OncoGenomics DB) maintained by the Khan laboratory at NCI (http://pob.abcc.ncifcrf.gov/cgi-bin/JK) was used to investigate the association between USP9X mRNA expression and event-free survival in patients with B-ALL in the TARGET ALL pilot project with the Gene Expression Omnibus accession number: GSE11877.21 Results using probe set 201100_s_at are described. The cutoff for analysis was determined using a P value minimization check in OncoGenomics DB.
Statistical analyses
The Kaplan-Meier survival probabilities were compared across different groups using the log-rank test. Cox proportional hazards models were used for estimating HRs and 95% CIs after adjusting for sex and the COG protocol in which they were initially enrolled. All statistical analyses were conducted in R version 4.0.3.
Results
Identification of novel germline deletion in USP9X in female proband with multiple congenital anomalies and B-cell precursor ALL
WGS was performed on DNA isolated from saliva from 12 probands with varying combinations of birth defects and childhood cancer diagnoses, as well as their parents. Using our SV analysis pipeline (Figure 1A), we identified a novel heterozygous 5 kb deletion (hg19 chrX:41066285-41071603) in a female GOBACK proband in the USP9X gene, which belongs to the ubiquitin-specific proteases subfamily of deubiquitinating enzymes. The germline deletion in USP9X was called by 3 of the 5 callers, each of which takes different features of short read sequencing data into account: Delly (read pairs and split read), CNVNator (read depth), and BreakDancer (read pairs). The deletion was de novo in the proband and absent in her unaffected sister. It passed the visual inspection criteria in IGV (Figure 1B). In addition, we validated the size of the deletion and the inheritance pattern using PCR (Figure 1C). It is an inframe deletion encompassing exon 33 (58 amino acids), which codes for part of the catalytic domain of USP9X. Neither this deletion nor any exonic intragenic deletions of the USP9X gene were present in the gnomAD database (gnomad.broadinstitute.org).
No potential LoF variants in cancer susceptibility genes were identified by SNV analysis. As previously reported by another team, this proband also has compound heterozygous missense variants in NUP214, NM_005085:c.3367 G>A, and c.4585G>A22 with the c.3367 G>A, also found in her unaffected sister. However, NUP214 is not constrained for missense variants in gnomAD with an observed/expected ratio of 0.95 and has only been associated with acute myeloid leukemia as a translocation partner. There was no other evidence to suggest pathogenicity, and we concluded they were unlikely to contribute to the phenotype observed in the GOBACK proband.
A review of the state birth defect registry data for this proband revealed 5 different anomalies: cataract, heart defects, choanal atresia, growth retardation, genital anomalies, and ear anomalies. In addition, the proband was diagnosed with B-cell precursor ALL (BCP-ALL) at 3 years of age. The set of birth defects seen in this proband overlaps with anomalies described in CHARGE syndrome.23,24 A review of the WGS sequence variant and SV calls revealed no rare coding variants in CHD7. Independently, her medical records also contained a negative CHD7 test report.
Access to the GOBACK proband’s electronic health record revealed additional and more specific details on the proband’s phenotypes, including intellectual disability (detailed in Table 1). This constellation of birth defects resembled the relatively recently described female-specific syndromic intellectual disability disorder (MRXS99F, MIM: 300968)4 associated with de novo heterozygous LoF variants and whole gene deletions in USP9X. Subsequently, Jolly et al, in 2020 described pathogenic missense variants and a single amino acid deletion of USP9X in an expanded cohort of females who shared the core phenotypic features, including multiple birth defects, with the previous LoF cohort but with slight phenotypic variation.5
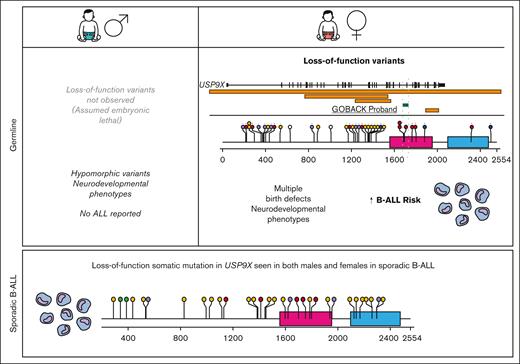
Males with pathogenic or likely pathogenic missense variants in USP9X present with a neurodevelopmental disorder called XLID99 (MIM: 300919). Although their (n = 16) neurological phenotypes overlap with those found in females, they do not present with major birth defects, and strikingly, there are no reports to date of any malignancies among male probands.25,26 In addition, no LoF variants have been described in males, suggesting that USP9X is essential for embryonic viability. Therefore, subsequent analysis was restricted to females.
Genotype-phenotype correlations in females
To date, including the 2 published cohorts and the GOBACK proband, 42 female patients with MRXS99F syndrome have been described with detailed clinical information.4,5 The vital status or early mortality of these patients were not described in these papers. Their ages at the last examination ranged from 9 days to 23 years, in addition to 2 of them being described as female fetuses. The GOBACK proband, diagnosed with B-ALL at age 3, was last examined at 8 years of age in the available medical records. Molecularly, this cohort was reported with nonsense, frameshift, missense variants, and intragenic and inframe germline deletions in USP9X (Figure 2A).4,5,27-33 The clinical importance of exon 33 (deleted in the GOBACK proband) is implicated by the overlap of this exon with 2 pathogenic missense variants p.D1685N and p.L1693W, previously reported in 3 female patients: Jolly et al females 26, 27, and Jolly et al. female 8 with this disorder4,5 (Figure 2A). The variant p.D1685N was de novo in Jolly et al female 27 and inherited from a somatic mosaic mother in Jolly et al female 26.5
Similar patterns of germline variants in USP9X in females with congenital anomalies and developmental delay and somatic mutations in leukemia samples from males and females. (A) Germline SVs in USP9X. The intragenic deletion reported here is highlighted. (B) Sequence variants in USP9X previously reported in female patients. Nonsense variants are shown in purple, frameshift in yellow, missense in red, inframe deletions in blue and splice site alterations in green. (C) Somatic mutations in USP9X previously reported in patients with B-ALL. Mutation type denoted by the same colors.
Similar patterns of germline variants in USP9X in females with congenital anomalies and developmental delay and somatic mutations in leukemia samples from males and females. (A) Germline SVs in USP9X. The intragenic deletion reported here is highlighted. (B) Sequence variants in USP9X previously reported in female patients. Nonsense variants are shown in purple, frameshift in yellow, missense in red, inframe deletions in blue and splice site alterations in green. (C) Somatic mutations in USP9X previously reported in patients with B-ALL. Mutation type denoted by the same colors.
We investigated the genotype-phenotype correlations in all 42 female patients described to date with germline variants in USP9X (including the GOBACK proband) by developing an oncoprint-type34,35 analysis based on the phenotypes (including malignancy, if noted) and genotypes reported (Figure 3A). Speech delay (present in 37 of the 38 patients in which it was described) and intellectual disability (present in 34 of the 36 patients in which it was described) were the most common phenotypic features across all the patients. Those subjects with typical LoF variants, for example, nonsense and frameshift alleles, clustered around the top with a higher number of clinical features. The most common features in this group included speech delay, intellectual disability, eye anomalies, hearing defects, ventriculomegaly, and dental anomalies. The GOBACK proband was the only proband with an intragenic inframe deletion in this cohort whose features clustered with those of the group of patients with pathogenic LoF variants.
Germline variants in USP9X are associated with multiple congenital anomalies, intellectual disability, and an increased risk of B-ALL. (A) Oncoprint analysis showing major categories of phenotypes (columns) found in individual probands (rows) in decreasing order of frequency (right to left and top to bottom, respectively) and also summarized with the percentages in the bottom right of the figure. The left 3 columns depict malignancy (presence of B-ALL is shown in black and osteosarcoma in dark grey), mutation types (deletion of 5' UTR shown in dark green, deletion >5kb in brown, frameshift in yellow, inframe in blue, missense in red, nonsense in purple and splice site in green), and whether the variant was de novo or not (shown in black vs blue, respectively). (B) Cumulative incidence plot of female patients with germline LoF variants in USP9X diagnosed with B-ALL compared with age- and sex-adjusted patients with B-ALL in the SEER database.
Germline variants in USP9X are associated with multiple congenital anomalies, intellectual disability, and an increased risk of B-ALL. (A) Oncoprint analysis showing major categories of phenotypes (columns) found in individual probands (rows) in decreasing order of frequency (right to left and top to bottom, respectively) and also summarized with the percentages in the bottom right of the figure. The left 3 columns depict malignancy (presence of B-ALL is shown in black and osteosarcoma in dark grey), mutation types (deletion of 5' UTR shown in dark green, deletion >5kb in brown, frameshift in yellow, inframe in blue, missense in red, nonsense in purple and splice site in green), and whether the variant was de novo or not (shown in black vs blue, respectively). (B) Cumulative incidence plot of female patients with germline LoF variants in USP9X diagnosed with B-ALL compared with age- and sex-adjusted patients with B-ALL in the SEER database.
B-ALL risk in female patients with germline variants in USP9X
Of note, 3 of the 42 female patients have been diagnosed with pediatric B-ALL, including the GOBACK proband, Jolly et al female 27 (diagnosed at age 4), and Reijnders et al female 1 (diagnosed at age 7). No additional description of the B-ALL subtypes was provided in the original studies describing Jolly et al female 27 and Reijnders et al female 1. The GOBACK proband has standard risk pre–B-cell ALL with no additional molecular features described in the medical record. In addition, 1 patient (Reijnders et al female 14) had an osteosarcoma diagnosis at age 22.4,5 Among the remaining female patients, additional diagnoses of ALL might develop later in childhood given the young age of the subjects. For example, Jolly et al. female 26 whose age at last exam was 17 months has the same missense variant as Jolly et al female 27 who was diagnosed with ALL at 4 years of age. The 3 patients with B-ALL clustered with the individuals with the largest number of syndromic features and had almost all the core phenotypic features of the syndrome, suggesting that B-ALL does not represent a rare subtype of the disorder.
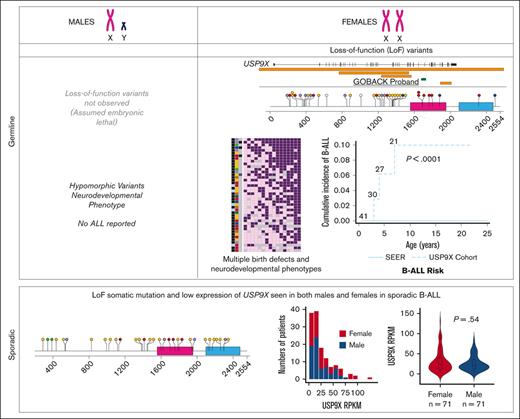
We compared the prevalence of B-ALL in this cohort of female patients with pathogenic germline variants in USP9X with that of females in the general population. The cumulative incidence of B-ALL in this cohort of female patients was 7.1% (3/42), whereas the age- and sex-adjusted cumulative incidence of B-ALL among females in the SEER database from 2000 to 2018 was 0.003% (P < .0001, log-rank test), as shown in Figure 3B. Taken together, this analysis suggests that B-ALL risk is a phenotypic feature of the female-specific MRXS99F syndrome.
Somatic mutational landscape of USP9X in sporadic B-ALL
We evaluated the medical literature and databases for somatic USP9X mutations in sporadic B-ALL. Downregulation of USP9X through somatic mutation has been reported in Down syndrome–associated B-ALL (DS-ALL), in which it was shown to help buffer JAK-STAT hypersignaling and enhance B-ALL cell survival in 25% of patients with CRLF2-positive DS-ALL.36 Somatic LoF mutations have also been reported in USP9X in non-DS sporadic B-ALL.37-44 The pattern of somatic mutation accumulated from published reports (Figure 2C) is similar to germline variants (Figure 2B) with a preponderance of LoF variants, thus consistent with a tumor suppressor mechanism in leukemia. In a previous assessment of somatic mutations in patients with B-ALL across different studies, 1.1% of patients (10/906) with B-ALL carried somatic mutations in USP9X.45 A distinct difference between germline and somatic findings is that somatic LoF USP9X mutations occur in both male and female pediatric patients with leukemia. Thus, although germline LoF variants in males appear to be inviable, somatic USP9X mutations occur in leukemia precursor cells in both males and females.
mRNA expression analysis of USP9X in a childhood B-ALL cohort
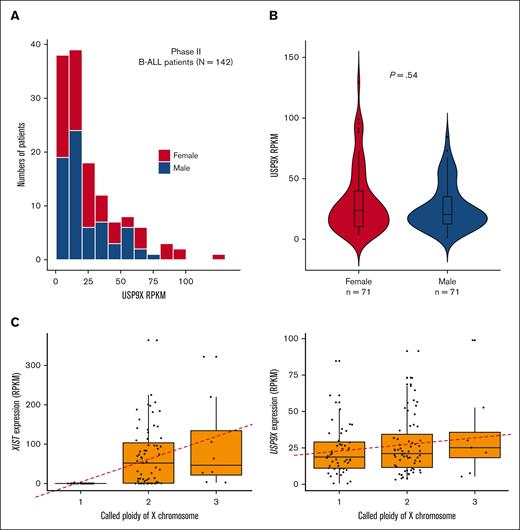
We analyzed open-access RNA sequencing (RNA-seq) data from 142 patients with high-risk B-ALL from the TARGET Data Matrix enrolled as part of phase 2 of the TARGET Study. This unique set of patients did not overlap with patients analyzed in other phases of the TARGET ALL project. The overall expression profile of USP9X in the cohort is shown in Figure 4A. Similar to the data on somatic mutations, we observed no significant difference in the expression of USP9X between males and females (Figure 4A-B), although the samples with the highest USP9X expression were from females.
USP9X expression in sporadic B-ALL samples does not differ between males and females with maintenance of normal patterns of X inactivation. (A) Expression of USP9X mRNA across 142 male (blue) and female (red) patients with B-ALL enrolled in phase 2 of TARGET. (B) No significant difference in expression levels of USP9X between males (n = 71) and females (n = 71) graphed as a violin plot. (C) Reads Per Kilobase of transcript per Million reads mapped (RPKM) of control gene XIST plotted against X chromosome copy number across patients with B-ALL (n = 127) on the left and RPKM of USP9X across patients with B-ALL (n = 123) on the right, with the regression line shown as a red dashed line.
USP9X expression in sporadic B-ALL samples does not differ between males and females with maintenance of normal patterns of X inactivation. (A) Expression of USP9X mRNA across 142 male (blue) and female (red) patients with B-ALL enrolled in phase 2 of TARGET. (B) No significant difference in expression levels of USP9X between males (n = 71) and females (n = 71) graphed as a violin plot. (C) Reads Per Kilobase of transcript per Million reads mapped (RPKM) of control gene XIST plotted against X chromosome copy number across patients with B-ALL (n = 127) on the left and RPKM of USP9X across patients with B-ALL (n = 123) on the right, with the regression line shown as a red dashed line.
Overall, the available genomic sequencing data from the TARGET leukemia samples from female patients presents no evidence for second somatic events or “hits,” with leukemia samples retaining both copies of USP9X or gaining additional copies of the X chromosome with wild-type USP9X sequence. Second hits are not necessarily expected for genes on the X chromosome, as most X-linked genes are only expressed by the one X in male cells or the active X in female cells. Recent analysis performed on gene expression from X chromosomes in noncancer cells revealed that USP9X was expressed, albeit at a reduced level, from the inactive X.46 We therefore aimed to determine whether leukemic bone marrow maintains a similar expression pattern for USP9X and other genes reported to be expressed by the inactive X. Using RNA-seq and copy number data from the TARGET Data Matrix of the leukemia cohort of n = 123 samples, we found that each inactive X in a sporadic leukemia sample was contributing on average 24.6% of the level of expression of the active X to the overall expression of USP9X in B-ALL (Figure 4C), which is only slightly increased compared with 14% to 17% reported in lymphoblastoid cells and fibroblasts of patients with sex chromosomal aneuploidies.46 The level of contribution from the inactive X for gene XIST in leukemia samples (n = 127) was 100% consistent with the previously reported germline analysis (Figure 4D). This analysis suggests that leukemia cells retain the same state of X inactivation as found in the germline and may explain why the leukemia samples with the highest USP9X expression derive from leukemia samples with extra copies of the X chromosome in female patients.
Survival analysis of patients with B-ALL based on USP9X expression
We examined the association between USP9X expression and overall survival in a set of patients with high-risk B-ALL who were enrolled in COG trials and analyzed during phase 1 of TARGET,47 in which expression data was captured using microarray. Patients with lower expression of USP9X had significantly poorer survival (OncoGenomics DB: P = .023) (Figure 5B). Next, we assessed survival outcomes among 142 patients with high-risk B-ALL who experienced early bone marrow relapse (≤4 years from time of diagnosis) from phase 2 of TARGET (Figure 5A), in which additional clinical information was included. After adjusting for sex and treatment protocol, we observed an elevated hazard of death among those with low expression of USP9X compared with patients with higher expression of USP9X (HR, 2.0; 95% CI, 1.09-3.71) (Table 2). These low expressors included a comparable number of males (n = 16) and females (n = 20). Of these 142 patients with RNA-seq data on their primary sample, 26 underwent WES, and 133 underwent WGS with copy number data on 123 samples. A review of the WES data demonstrates that none of the 26 patients had a somatic mutation in USP9X. Two patients among the 133 with WGS data had a single LoF mutation in USP9X. Copy number loss involving USP9X was found in 2 additional patients. Two of these 4 patients with somatic variation in USP9X died at 31 and 38 months.
Low USP9X expression is associated with poor survival in patients with high-risk B-ALL. (A) Kaplan-Meier curves are shown for overall survival in patients with high-risk B-ALL (n = 142; TARGET phase 2) expressing low (lowest quartile (in purple [n = 36]), intermediate (two middle quartiles in green [n = 70]), or high (highest quartile in yellow [n = 36]) levels of USP9X. (B) Kaplan-Meier curves are shown for event-free survival in patients with high-risk B-ALL (n = 207; TARGET ALL pilot project) from OncoGenomics DB comparing the lower quartile (in purple) versus the top three quartiles (in blue). For statistical analysis refer to the “Methods.”
Low USP9X expression is associated with poor survival in patients with high-risk B-ALL. (A) Kaplan-Meier curves are shown for overall survival in patients with high-risk B-ALL (n = 142; TARGET phase 2) expressing low (lowest quartile (in purple [n = 36]), intermediate (two middle quartiles in green [n = 70]), or high (highest quartile in yellow [n = 36]) levels of USP9X. (B) Kaplan-Meier curves are shown for event-free survival in patients with high-risk B-ALL (n = 207; TARGET ALL pilot project) from OncoGenomics DB comparing the lower quartile (in purple) versus the top three quartiles (in blue). For statistical analysis refer to the “Methods.”
Discussion
Here, we describe a novel germline inframe de novo deletion in USP9X in a female proband with MRXS99F-associated (MIM: 300968) birth defects, intellectual disability, and B-cell precursor ALL. The presence of multiple birth defects and B-ALL is consistent with our prior study demonstrating that cancer risk was highest in children with 4 or more birth defects, as well as a specific association of leukemia in children with choanal atresia (HR, 9.2; 95% CI, 3.8-22.1). The intragenic 5 kb deletion break points in the introns spanning exon 33 of USP9X were detected using WGS and multiple SV callers. This proband was previously described as part of an independent WES cohort and was noted to remain undiagnosed by the authors, although compound heterozygous missense variants in NUP214 were noted.22 The inability to detect small heterozygous deletions by exome sequencing highlights the importance of genome sequencing for gene discovery efforts.48
Several lines of evidence suggest this inframe deletion acts as a LoF variant. The genotype-phenotype analysis shows that the GOBACK proband with the deletion had 12 of the 16 phenotypes, including speech delay, intellectual disability, eye anomalies, hearing defects, ventriculomegaly, and dental anomalies. The deletion overlaps part of the catalytic domain of the protein, and 3 other MRXS99F probands have been described with pathogenic missense variants in exon 33.
USP9X codes for a deubiquitinating enzyme that removes both mono- and polyubiquitin chains from at least 35 substrates in humans, thus protecting them from proteasomal-mediated degradation and/or altering subcellular localization.49,50 Depending on the cancer context, USP9X has been described as both an oncogene or a tumor suppressor. Overexpression of USP9X has been reported in malignancies such as lung cancers and multiple myeloma.51,52 In contrast, in KRAS mutant pancreatic cancer, USP9X has been shown to act as a tumor suppressor by stabilizing ITCH.53 However, in the 4 leukemic samples with somatic USP9X mutations in the TARGET data set, there were no somatic mutations in KRAS or NRAS.
Previous studies of female probands with germline variants in USP9X had described 2 subjects with B-ALL; however, a formal analysis of leukemia risk was not performed. The cumulative incidence of B-ALL in this cohort of female patients was 7.1%, with pathogenic USP9X germline variants, and significantly increased compared with age- and sex-matched controls from the SEER database. Germline LoF variants in males do not appear to be tolerated, presumably because of embryonic lethality, and males with hypomorphic missense variants have neurodevelopmental phenotypes without birth defects or reports of B-ALL or other pediatric cancers. Thus, USP9X is the first female-specific pediatric B-ALL predisposition gene, even though somatic mutations in USP9X occur in sporadic leukemias in both sexes, suggesting that hematopoietic cells can tolerate absent USP9X function (Figure 6).
Model of the role of germline vs somatic mutation of USP9X in leukemia development. In the germline, males do not tolerate LoF variants, presumably because of embryonic cells without any USP9X function. Males with hypomorphic missense variants have neurodevelopmental phenotypes with no reported B-ALL risk. Female embryos with germline LoF variants will contain a mixture of near normal USP9X levels from cells with expression from the nonmutant X and cells with small amount of USP9X from the nonmutant inactive X. They present with severe congenital anomalies, intellectual disability, and an increased risk of B-ALL. In sporadic B-ALL, somatic LoF mutations are seen in samples from both sexes.
Model of the role of germline vs somatic mutation of USP9X in leukemia development. In the germline, males do not tolerate LoF variants, presumably because of embryonic cells without any USP9X function. Males with hypomorphic missense variants have neurodevelopmental phenotypes with no reported B-ALL risk. Female embryos with germline LoF variants will contain a mixture of near normal USP9X levels from cells with expression from the nonmutant X and cells with small amount of USP9X from the nonmutant inactive X. They present with severe congenital anomalies, intellectual disability, and an increased risk of B-ALL. In sporadic B-ALL, somatic LoF mutations are seen in samples from both sexes.
Female patients with MRXS99F should be considered at increased risk for B-ALL in childhood and referred to a hematology-oncology specialist for assistance with cancer surveillance, as indicated for other leukemia predisposition syndromes.3 To date, the latest age of B-ALL diagnosis is 8 years. Overall, the risk of developing B-ALL is highest in children below 5 years of age.54 It would be of interest to determine the risk of leukemia in patients with more longitudinal data as well as whether those with germline USP9X LoF variants and a lack of cancer diagnoses demonstrate skewed X inactivation in bone marrow cells with the preponderance of the active X containing the wild-type USP9X gene,55-57 which could potentially diminish cancer development. One of the previously described female patients was diagnosed with osteosarcoma at 22 years of age. Therefore, as this cohort of female patients gets older, there is the possibility of an increased risk of other cancer types developing. However, in a recent study describing the frequency of germline variants in susceptibility genes in 1244 osteosarcoma patients, no USP9X variants were found.58
In 2017, Schwartzman et al. were the first to describe the tumor-suppressive role of somatic mutations in USP9X in B-ALL.36 However, their assessment was limited to DS-ALL and to a subset of Ph-like ALLs, where CRLF2 rearrangements drive pathogenesis through activation of the JAK-STAT pathway. In their analysis of paired samples from diagnosis and relapse patients with DS-ALL, they found that USP9X was inhibited in 25% of CRLF2-positive DS-ALLs through focal deletions or frameshift mutations. This and other studies have reported LoF mutations in USP9X in major clones in diagnostic and relapse B-ALL samples, such that mutations occur early in tumorigenesis and persist after treatment, consistent with a tumor-suppressive mechanism.39-41,45 However, USP9X expression data in high-risk B-ALL and somatic variation in non-DS ALL had not been extensively studied previously.
The lack of difference in USP9X expression from sporadic ALL samples between male and female patients was initially surprising. Prior work in a limited number of lymphoblastoid cell lines (n = 4 each for males and female-derived lines) by Reijnders et al demonstrated almost twice the amount of USP9X in females compared with males, suggesting a complete lack of X inactivation.4 However, a recent study used gene expression in normal lymphoblasts or fibroblasts from patients with sex chromosomal aneuploidy and demonstrated that for USP9X, each inactive X contributes only 14% to 17% of expression compared with the active X.46 Using data from the TARGET phase 2 cohort of patients with sporadic ALL (n = 142 samples), which frequently demonstrate sex chromosome aneuploidy, we were able to demonstrate that leukemic cells retain the same pattern of expression from the inactive X as seen in nonmalignant samples.
Although this study was not specifically designed to evaluate the prognostic utility of USP9X expression in ALL survival, we did observe inferior overall survival among those with lower expression of USP9X. This was true both in both TARGET phase 1 and phase 2 data, where we also adjusted for sex and treatment protocol. Based on this, we recommend that a future assessment following REMARK guidelines be developed to comprehensively evaluate this association. It is important to highlight that these low expressors consisted of a comparable number of males and females, and thus this observation is independent of sex for an X-linked gene in sporadic B-ALL.
In summary, we demonstrate that USP9X is a sex-limited leukemia predisposition gene with leukemia risk associated with LoF variants found in girls with a complex neurodevelopmental and multibirth defect disorder. In contrast, this gene demonstrates features of a tumor suppressor among both males and females with sporadic pediatric B-ALL, with somatic mutations and low expression observed.
Acknowledgments
The authors acknowledge the families that participated in the GOBACK study. The authors would also like to thank David C. Page for helpful discussions regarding X-inactivation.
This work was supported by the Cancer Prevention and Research Institute of Texas Grant (RP140258, and RP160283), Department of Defense Grant (W81XWH-20-1-0567), and the National Cancer Institute (R03CA272955).
Authorship
Contribution: S.D.S. designed the study, performed research, collected data, analyzed and interpreted data, performed statistical analysis, and wrote the manuscript; P.M. contributed vital analytical tools, collected data, analyzed and interpreted data, and revised the manuscript; H.L. performed research, collected data, analyzed and interpreted data, and revised the manuscript; J.M.S. designed research, collected data, analyzed and interpreted data, performed statistical analysis, and revised the manuscript; M.E.S. designed research, performed statistical analysis, and revised the manuscript; S.S., H.D., D. Muzny, D. Mitchell, and O.T. collected data and revised the manuscript; A.S. analyzed and interpreted data, secured funding, and revised the manuscript; P.J.L. designed the study, secured funding, and provided overall guidance of project and manuscript; and S.E.P. designed the study, secured funding, directed the research, and provided overall guidance of project and manuscript.
Conflict-of-interest disclosure: S.E.P. is a member of the scientific advisory panel of the Baylor Genetics laboratory. The remaining authors declare no competing financial interests.
Correspondence: Sharon E. Plon, Feigin Tower, Suite 1200, 1102 Bates St, Houston, TX 77030; e-mail: splon@bcm.edu.
References
Author notes
The sequence data of this cohort will be available in the ANViL database (https://anvilproject.org/overview). However, permission to access the data through the dbGAP permissions process under NHGRI GREGoR Consortium: Genomics Research to Elucidate the Genetics of Rare Disease dbGaP Study (Accession: phs003047.v1.p1).
Other forms of data are available on request from the corresponding author, Sharon E. Plon (splon@bcm.edu).




![Low USP9X expression is associated with poor survival in patients with high-risk B-ALL. (A) Kaplan-Meier curves are shown for overall survival in patients with high-risk B-ALL (n = 142; TARGET phase 2) expressing low (lowest quartile (in purple [n = 36]), intermediate (two middle quartiles in green [n = 70]), or high (highest quartile in yellow [n = 36]) levels of USP9X. (B) Kaplan-Meier curves are shown for event-free survival in patients with high-risk B-ALL (n = 207; TARGET ALL pilot project) from OncoGenomics DB comparing the lower quartile (in purple) versus the top three quartiles (in blue). For statistical analysis refer to the “Methods.”](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/16/10.1182_bloodadvances.2023009814/2/m_blooda_adv-2023-009814-gr5.jpeg?Expires=1763493259&Signature=SpV4A2vRPIQeUAs6M98AB33yl-h-oDTAagc24EHPyJb7z4RCtqWqcJRkjAOm~IZVQF-yg5EWCydZg9tuIo3y85AijlhPiOGKNX38ga--pe-pIQGEebArZA16gAsKAeVQWMA3-hGznP1yja-tLli8pT-9stw5-JgPph-maIPX7NVWX--gPUxWQ4fpFtuZEfBTRLwrylwdxgxYXNANbRmA3oZJq6n85t81n0XAi5xkzVF4jdyiOmd6WpPrh5wwLg65kGRMG0LCfv8XwcHzKdPIQwsr1MNqb6fKLDgHnPUPbBpN4dtXb6RBQoJRf6hVsHvyV2ZhiyK7dIf~nSsxv~rTlg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)