Key Points
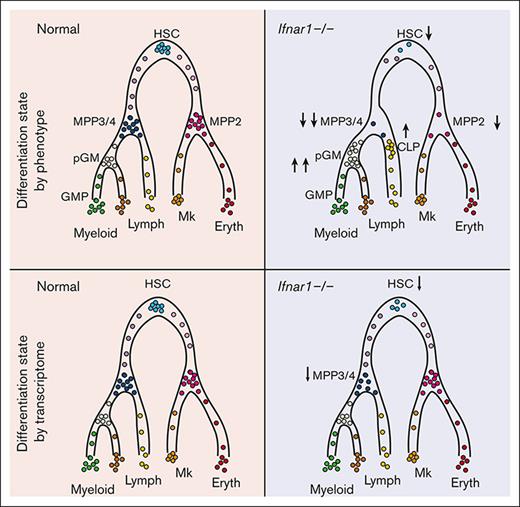
Loss of IFN-1 signaling in neonatal mice depletes phenotypic blood progenitors without compromising postnatal hematopoiesis.
Progenitor populations remain intact when measured based on single-cell transcriptomes rather than surface marker phenotypes.
Abstract
Type I interferon (IFN-1) regulates gene expression and hematopoiesis both during development and in response to inflammatory stress. We previously showed that during development in mice, hematopoietic stem cells (HSCs) and multipotent progenitors (MPPs) induce IFN-1 target genes shortly before birth. This coincides with the onset of a transition to adult hematopoiesis, and it drives the expression of genes associated with antigen presentation. However, it is not clear whether perinatal IFN-1 modulates hematopoietic output, as has been observed in contexts of inflammation. We have characterized hematopoiesis at several different stages of blood formation, from HSCs to mature blood cells, and found that loss of the IFN-1 receptor (IFNAR1) leads to depletion of several phenotypic HSC and MPP subpopulations in neonatal and juvenile mice. Committed lymphoid and myeloid progenitor populations expand simultaneously. These changes had a surprisingly little effect on the production of more differentiated blood cells. Cellular indexing of transcriptomes and epitopes by sequencing resolved the discrepancy between the extensive changes in progenitor numbers and modest changes in hematopoiesis, revealing stability in most MPP populations in Ifnar1-deficient neonates when the populations were identified based on gene expression rather than surface marker phenotype. Thus, basal IFN-1 signaling has only modest effects on hematopoiesis. Discordance between transcriptionally and phenotypically defined MPP populations may affect interpretations of how IFN-1 shapes hematopoiesis in other contexts, such as aging or inflammation.
Introduction
Inflammatory cytokines modulate hematopoiesis throughout life, particularly in response to infections or other inflammatory stressors.1,2 Several cytokines, including interferon alfa (IFN-α), IFN-γ, and interleukin-1 beta (IL-1β) have been shown to drive quiescent hematopoietic stem cells (HSCs) into cycle and alter hematopoietic output.3-6 Short-term exposure to type I IFNs (IFN-1), which include IFN-α and IFN-β, promotes HSC proliferation and emergency megakaryopoiesis.3,7 Likewise, IL-1β exposure induces transient HSC proliferation and myelopoiesis.5,8 These changes enhance blood production and immune function under conditions of stress. Inflammatory cytokines also help maintain basal hematopoietic output. For example, gut microflora stimulate IFN-α and IL-1β production, which in turn promotes myeloid progenitor expansion and myelopoiesis.9,10 Over time, chronic cytokine stimulation promotes myeloid lineage bias that typifies aged HSCs.9 These observations show that infection and inflammation can shape hematopoietic output in adults by reprogramming immature progenitors.
Inflammatory cytokines also regulate fetal and neonatal hematopoiesis, though not necessarily in response to infectious stimuli. During midgestation in mice, IFN-α, IFN-γ, and IL-1β promote the emergence of definitive HSCs from hemogenic endothelium in the dorsal aorta.11-13 During late gestation, IFN-1 levels spike and trigger the expression of antiviral transcripts (eg, Ifit1, Oas2, and Ifit3), several transcription factors associated with IFN-1 signal transduction (Stat1, Irf7 and Irf9), and major histocompatibility complex–I genes within HSCs and non–self-renewing multipotent progenitors (MPPs).14 The perinatal IFN-1 spike does not reflect a response to infection or a change in the fetal/maternal microbiome because it occurs even in germ-free mice.14 It coincides with the onset of a transition from fetal to adult transcriptional states in both HSCs and MPPs. Deleting the IFN-1 receptor gene, Ifnar1, slows the transition, indicating a causal interaction between sterile IFN-1 signaling and adult gene expression programs.14
It is less clear whether sterile IFN-1 signaling is necessary to support hematopoietic output during late gestation and in neonates. We previously showed that Ifnar1 deletion prevents the expansion of phenotypic MPP populations that takes place between embryonic day (E)16 and postnatal day (P)0.14 This finding aligns with recent observations in adult mice, showing that Stat1 deletion causes a reduction in MPP numbers.10,15 However, Ifnar1-deficient mice develop normally, despite the aforementioned transcriptional changes, suggesting that the loss of phenotypic MPPs does not markedly impair hematopoiesis. Assessments of HSC and MPP frequencies via flow cytometry are confounded by the fact that Sca1, an HSC/MPP surface marker, is itself positively regulated by IFN-1.3 This raises the question of whether changes in phenotypic HSC, MPP, and myeloid progenitor numbers in Ifnar1- or Stat1-deficient mice underlie functional changes that shape hematopoietic output, or whether they simply reflect changes in population surface marker phenotypes.
We have used Ifnar1-deficient mice to further characterize the role of IFN-1 signaling in neonatal and juvenile hematopoiesis. We found that Ifnar1 deletion led to a pronounced decrease in phenotypic HSC and MPP numbers in newborn mice, with a concomitant increase in more mature myeloid and lymphoid progenitors. These changes were also evident in 14-day-old juvenile mice and in adult mice after conditional or germ line Ifnar1 deletion. Neonatal myelopoiesis, lymphopoiesis, and blood production were unaffected by Ifnar1 deletion despite the extensive changes within immature progenitor populations. Cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq)16 resolved this discrepancy. Ifnar1 deletion had only minor effects on neonatal hematopoiesis, as defined by single-cell transcriptomes, despite extensive changes in surface marker identity. We observed a modest decrease in HSC numbers and a T-lineage–primed MPP population in newborn mice, but functional changes were minor compared with changes in phenotypic populations. The data indicate that noninfectious IFN-1 signaling induces transcriptional changes that promote antigen presentation and antiviral activity within HSCs/MPPs beginning in late gestation, but it has only modest effects on hematopoiesis.
Methods
Mouse lines and treatments
Ifnar1-null,17Ubc-CreER,18and Ifnar1-flox19 mice were obtained from The Jackson Laboratory and housed in a standard pathogen-free barrier facility. For tamoxifen treatments, mice were fed 100 μg/g body weight of tamoxifen in corn oil daily for 5 days by oral gavage. HSCs were cultured in MethoCult and genotyped to confirm >90% Ifnar1 deletion efficiency. IL1-β and IFN-γ stress responses were assayed by administering recombinant IL-1β (20 ng on day 1 and 50 ng on day 3) or IFN-γ (0.6 μg on day 1 and 1.25 μg on day 3) subcutaneously before spleen and thymus analysis on day 5. All procedures were performed in accordance with an institutional animal care and use committee–approved protocol at the Washington University School of Medicine.
Flow cytometry
Cell cycle, BrdU incorporation, and annexin V assays
Cell cycle, 5′-bromo-2′-deoxyuridine (BrdU), and annexin V assays were performed as previously described.14,21 For BrdU incorporation assays, pregnant female mice were injected with BrdU (150 mg/kg body weight) on P19, 24 hours before cell collection and then administered BrdU dissolved in drinking water (1 mg/mL). They gave birth overnight, and livers were isolated from newborn pups for analysis.
Western blot
Lineage-Sca1+c-Kit+ (LSK) cells were isolated from P0 and 8-week-old adult mice, stimulated with IFN-α (12.5 IU/μL, Miltenyi) ex vivo for 30 minutes, washed, and transferred to 10% trichloracetic acid. Western blotting was performed as previously described, using antibodies from Cell Signaling Technologies (supplemental Table 1).20
CITE-seq analysis
P0 liver cells from 3 pooled mice per genotype or P14 bone marrow cells from 2 to 4 mice per genotype were c-Kit selected and stained with TotalSeq B antibodies (supplemental Table 1; BioLegend). We isolated lineage-c-Kit+ (LK) and LSK cells via flow cytometry and generated libraries with the Chromium Next GEM Single Cell 3′ kit version 3.1 (PN-1000268), Chromium Single Cell 3′ Feature Barcode Library kit (PN-1000079), Dual Index kit NT Set A (PN-1000242), and Dual Index kit TT Set A (PN-1000215). Libraries were sequenced on an Illumina Novaseq S4.
The Cell Ranger version 6.0.1 pipeline was used to process data generated from the 10× Chromium platform. Digital gene expression files were filtered and normalized as previously described.14 We performed iterative clustering and guide-gene selection (ICGS, version 2) analysis with the AltAnalyze toolkit.22 Thresholds for surface barcode expression were set by visual inspection of histograms to identify levels that distinguished the positive and negative populations (supplemental Figure 8). Populations were defined based on phenotypes described in supplemental Table 2. Comparisons of population sizes were evaluated with a χ2 test. Comparisons of gene expression between Ifnar1+/− and Ifnar1–/– progenitors were performed using Seurat with the Wilcoxon rank sum test.23 Analysis of cell cycle and apoptosis gene sets were performed using gene set variation analysis.24
Colony formation and B- and T-cell potential assays
Colony-forming unit (CFU) frequencies were assayed by sorting single cells from the indicated populations into MethoCult (Stem Cell Technologies, M3434). Colonies were scored 14 days later. B- and T-cell potential were assayed as previously described by Pietras et al.25 To measure B- and T-cell potential, OP9 or OP9-Δ cells, respectively, were seeded in 96-well plates at 10 000 cells per well in minimum essential medium–α, 20% fetal bovine serum, and penicillin-streptomycin. Limiting doses of LK or MPP cells were sorted into at least 48 independent wells per genotype and cell dose. When we measured T-cell output from MPPs at different ages (Figure 1), we plated single MPPs rather than a range of cell doses because most E16 MPPs had T-cell potential. Cells were cultured with FLT3L and IL-7 (B cells, 10 ng/mL for 2 weeks; T cells, 5 ng/mL for 2 weeks and then 2.5 ng/mL for 1 week). The media was changed every 4 days. After 14 days (B cells) or 21 days (T cells), each well was analyzed via flow cytometry for the presence of CD19+ or B220+ B cells or CD4 or CD8 T cells. Extreme limiting dilution analysis 26 was used to compare limiting dilution curves. Fisher exact test was used to compare the numbers of single MPPs that gave rise to T cells at E16, P0, and adulthood.
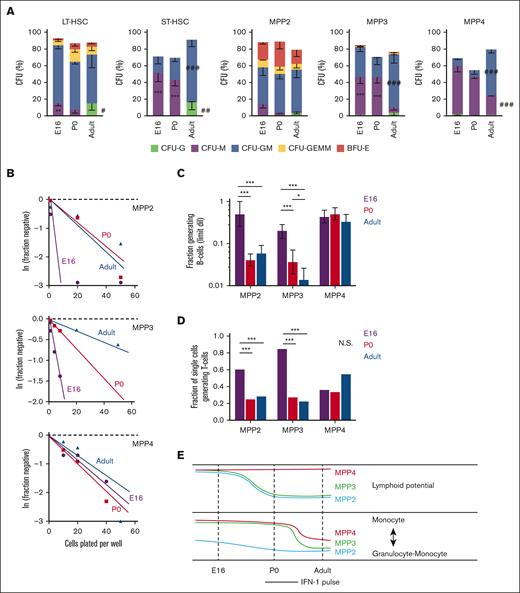
MPP potential changes between the fetal, neonatal, and adult stages of development. (A) CFU assays for single LT-HSC, ST-HSC, MPP2, MPP3, and MPP4. Across several populations, CFU-M frequency decreased between birth and adulthood, and CFU-G and CFU-GM frequency increased. Error bars show standard deviations from 3 independent experiments performed with at least 60 cells per population per experiment. ∗∗∗P < .001; ∗∗P < .01 in either E16 or P0 mice relative to adult. ∗∗∗P < .001; ∗∗P < .01 in adult relative to both E16 and P0. P values were calculated by one-way ANOVA with Holm-Sidak post hoc test. (B) Limiting dilution curves for B-cell production from MPP2, MPP3, and MPP4. (C) Fractions of each MPP subtype that gave rise to B cells based on limiting dilution curves in panel B. Frequencies were calculated and compared using ELDA. (D) Fraction of individually plated MPPs that gave rise to T cells. The data reflect 3 independent experiments with at least 20 individually plated MPP per experiment. ∗∗∗P < .001 using Fisher exact test. (E) Schematic overview of dynamic changes in MPP potential between E16 and adulthood. Lymphoid potential is shown in the top panel, and the balance between monocyte and mixed granulocyte-monocyte output is shown in the bottom panel. ANOVA, analysis of variance; ELDA, extreme limiting dilution analysis.
MPP potential changes between the fetal, neonatal, and adult stages of development. (A) CFU assays for single LT-HSC, ST-HSC, MPP2, MPP3, and MPP4. Across several populations, CFU-M frequency decreased between birth and adulthood, and CFU-G and CFU-GM frequency increased. Error bars show standard deviations from 3 independent experiments performed with at least 60 cells per population per experiment. ∗∗∗P < .001; ∗∗P < .01 in either E16 or P0 mice relative to adult. ∗∗∗P < .001; ∗∗P < .01 in adult relative to both E16 and P0. P values were calculated by one-way ANOVA with Holm-Sidak post hoc test. (B) Limiting dilution curves for B-cell production from MPP2, MPP3, and MPP4. (C) Fractions of each MPP subtype that gave rise to B cells based on limiting dilution curves in panel B. Frequencies were calculated and compared using ELDA. (D) Fraction of individually plated MPPs that gave rise to T cells. The data reflect 3 independent experiments with at least 20 individually plated MPP per experiment. ∗∗∗P < .001 using Fisher exact test. (E) Schematic overview of dynamic changes in MPP potential between E16 and adulthood. Lymphoid potential is shown in the top panel, and the balance between monocyte and mixed granulocyte-monocyte output is shown in the bottom panel. ANOVA, analysis of variance; ELDA, extreme limiting dilution analysis.
Results
Phenotypic MPPs have distinct lineage potentials at E16, P0, and 8 weeks old
Adult MPP subpopulations can be distinguished based on surface marker expression and have different lymphoid, myeloid, erythroid, and megakaryocytic biases.25,27-29 For the purposes of this work, we have adopted the nomenclature and surface marker phenotypes previously assigned by Pietras et al (supplemental Figure 1A; supplemental Table 2).25 Population names and associated biases include long-term HSCs (LT-HSC; multipotent with long-term self-renewal capacity), short-term HSC (ST-HSC; multipotent with limited self-renewal capacity), MPP2 (erythroid/megakaryocytic bias), MPP3 (myeloid bias), and MPP4 (lymphoid bias).25 It is not clear whether these phenotypically defined populations have similar biases in the fetal or neonatal stages. Given that IFN-1 may regulate perinatal MPP fate, we sought to clarify the lineage biases of each fetal/neonatal MPP subpopulation.
To assess myeloid potential, we sorted individual LT-HSC, ST-HSC, MPP2, MPP3, and MPP4 onto methylcellulose and scored CFUs as previously described.25 There were several notable changes in CFU production with age. At E16 and P0, both LT-HSC and ST-HSC had greater monocyte bias (CFU-monocyte) and reduced granulocyte bias (CFU-granulocyte [CFU-G] and CFU–granulocyte-macrophage [CFU-GM]) relative to adult HSCs (Figure 1A). MPP3 and MPP4 also showed greater CFU-M, and fewer CFU-GM, relative to adult MPP3/4. The balance between monocyte and granulocyte priming therefore appears to shift after birth (Figure 1E).
To assess B- and T-cell potential, we plated MPPs onto OP9 (B-cell) or OP9-Δ(T-cell) stroma at single cell or limiting dilution doses and cultured as previously described.25 MPP2 and MPP3 had higher B- and T-cell potential at E16 relative to equivalent populations at P0 or 8 weeks old (Figure 1B-D). Indeed, a majority of E16 MPPs had lymphoid potential, irrespective of whether they exhibited MPP2, MPP3, or MPP4 surface phenotypes. These data show that MPP lymphoid potential declined in the MPP2 and MPP3 populations between E16 and birth (Figure 1E). These changes coincide with the perinatal spike in IFN-1 signaling,14 raising the question of whether IFN-1 regulates neonatal hematopoiesis, particularly lymphoid output.
Ifnar1 deletion alters phenotypic HSC, MPP, and committed progenitor populations in neonatal and juvenile mice without disrupting hematopoiesis
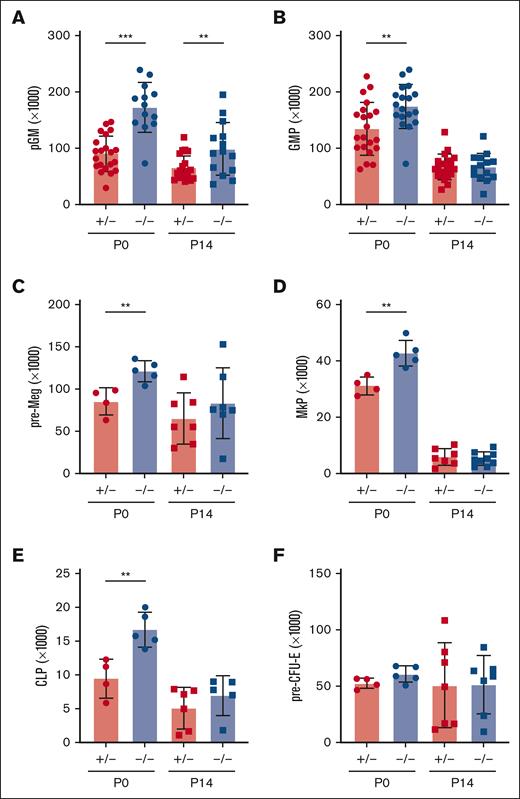
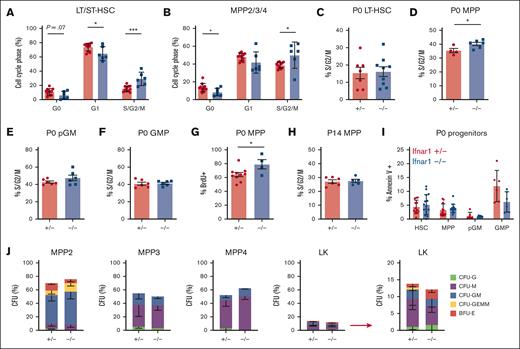
We used Ifnar1-deficient mice to thoroughly characterize the effects of IFN-1 signaling on neonatal and juvenile hematopoiesis. Neonatal and adult HSCs/MPPs were similarly responsive to IFN-1, as measured by STAT1 and STAT3 phosphorylation (supplemental Figure 2), indicating that IFN-1 signal transduction does not change with age. To simplify breeding strategies, Ifnar1+/− were used as controls rather than Ifnar1+/+ mice. We confirmed that Ifnar1+/+ and Ifnar1+/− mice had equivalent numbers of HSCs, MPPs, and myeloid progenitors (supplemental Figure 3). We previously showed that Ifnar1–/– mice have normal HSC and MPP numbers at E16, and that MPP numbers are reduced in Ifnar1–/– mice by P0.14 We reaffirmed the previous findings, observing depletion of several phenotypic HSC and MPP subpopulations in livers of P0 Ifnar1–/– mice (Figure 2A-E). Significant, but less severe, reductions in HSC/MPP numbers were observed in P14 juvenile mice (Figure 2F-J). We also conditionally deleted Ifnar1 in adulthood by administering tamoxifen to Ubc-CreER; Ifnar1f/f mice at 6 weeks old and then evaluating HSC and MPP numbers 4 weeks later. All MPP subpopulations (MPP2, MPP3, and MPP4) were significantly reduced in Ifnar1-deleted mice, whereas HSC numbers were normal (Figure 2K-O). Similar reductions in MPP3 and MPP4 numbers were observed in adult mice with germ line Ifnar1 deletions (supplemental Figure 4). Altogether, the data show that Ifnar1 deletion reduces phenotypic MPP numbers across a range of developmental stages.
Ifnar1 deletion leads to reductions in phenotypic HSCs and MPPs during the neonatal, juvenile, and adult stages of life. (A-E) Numbers of indicated HSC and MPP subpopulations in livers of P0 Ifnar1+/− and Ifnar1–/– mice, n = 21-35. (F-J) Numbers of indicated HSC and MPP subpopulations in bone marrow (2 hindlimbs) of P14 Ifnar1+/− and Ifnar1–/–-mice, n = 12-27. (K-O) Numbers of indicated HSC and MPP subpopulations in bone marrow of 10-week-old control (Cre–) and Ubc-CreER; Ifnar1f/f mice after tamoxifen treatment at 6 weeks old, n = 5. For all panels, error bars reflect standard deviation. Surface marker phenotypes are specified in supplemental Table 2, and gating strategies are shown in supplemental Figure 2. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001 using two-tailed t test.
Ifnar1 deletion leads to reductions in phenotypic HSCs and MPPs during the neonatal, juvenile, and adult stages of life. (A-E) Numbers of indicated HSC and MPP subpopulations in livers of P0 Ifnar1+/− and Ifnar1–/– mice, n = 21-35. (F-J) Numbers of indicated HSC and MPP subpopulations in bone marrow (2 hindlimbs) of P14 Ifnar1+/− and Ifnar1–/–-mice, n = 12-27. (K-O) Numbers of indicated HSC and MPP subpopulations in bone marrow of 10-week-old control (Cre–) and Ubc-CreER; Ifnar1f/f mice after tamoxifen treatment at 6 weeks old, n = 5. For all panels, error bars reflect standard deviation. Surface marker phenotypes are specified in supplemental Table 2, and gating strategies are shown in supplemental Figure 2. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001 using two-tailed t test.
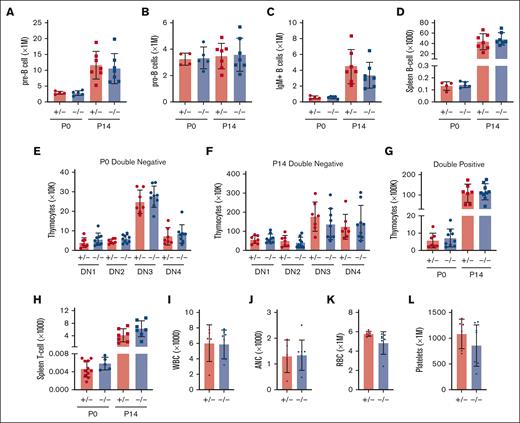
In light of the marked depletion of HSCs and MPPs in Ifnar1-deficient mice, we next evaluated whether more mature stages of blood production are similarly disrupted. We measured pre-granulocyte-monocyte progenitor (pGM), granulocyte-monocyte progenitor (GMP), premegakaryocyte progenitor (pre-Meg), megakaryocyte progenitor (MkP), common lymphoid progenitor (CLP), and preerythroid CFU (pre-CFU-E) numbers at neonatal (P0) and juvenile (P14) stages using surface marker phenotypes described by Pronk et al (supplemental Figure 1B,C; supplemental Table 2).30 We also measured more mature blood cells, including B- and T-cell progenitors and fully differentiated peripheral blood cells (supplemental Figure 5). We observed significant expansion, rather than depletion, of pGM, GMP, pre-Meg, MkP, and CLP populations (Figure 3), particularly at P0, when IFN-1 target gene expression is highest.14 These increases in population size were paradoxical, given the depletion of HSC and MPP precursors. Interestingly, Ifnar1 deletion neither impaired nor enhanced production of mature blood progenitors or fully differentiated blood cells, as B-cell progenitors (Figure 4A-D), T-cell progenitors (Figure 4E-H), and peripheral blood counts were all normal in Ifnar1–/– mice at P0 and P14 (Figure 4I-L). Thus, profound shifts in the composition of immature HSC and MPP populations in Ifnar1–/–- neonates does not result in altered hematopoietic output.
Ifnar1 deletion leads to expansion of phenotypic committed myeloid and lymphoid progenitor populations in neonates and juveniles. (A-F) Absolute numbers of committed myeloid (A,B), megakaryocyte (C,D), lymphoid (E), and erythroid (F) progenitors in P0 liver or P14 bone marrow from mice of Ifnar1+/− and Ifnar1–/–-mice. For panels A and B, n = 13 to 21. For panels C-F, n = 4 to 7. For all panels, error bars reflect standard deviation. Surface marker phenotypes are specified in supplemental Table 2, and gating strategies are shown in supplemental Figure 2. ∗∗P < .01; ∗∗∗P < .001 using two-tailed t test.
Ifnar1 deletion leads to expansion of phenotypic committed myeloid and lymphoid progenitor populations in neonates and juveniles. (A-F) Absolute numbers of committed myeloid (A,B), megakaryocyte (C,D), lymphoid (E), and erythroid (F) progenitors in P0 liver or P14 bone marrow from mice of Ifnar1+/− and Ifnar1–/–-mice. For panels A and B, n = 13 to 21. For panels C-F, n = 4 to 7. For all panels, error bars reflect standard deviation. Surface marker phenotypes are specified in supplemental Table 2, and gating strategies are shown in supplemental Figure 2. ∗∗P < .01; ∗∗∗P < .001 using two-tailed t test.
Ifnar1 deletion does not impair lymphopoiesis or mature blood production. (A-D) Absolute numbers of indicated B-cell progenitor populations in P0 liver or P14 bone marrow from mice of Ifnar1+/− and Ifnar1–/–-mice, or spleen B-cell populations from the same mice. (E-H) Absolute numbers of thymocyte subpopulations Ifnar1+/− and Ifnar1–/–-mice, or spleen T-cell populations from the same mice. (I-L) White blood cell (H), absolute neutrophil (I), red blood cell (J), and platelet (K) counts presented for 1 μL of blood from P14 mice. For all panels, n = 4 to 11 and error bars reflect standard deviation. Surface marker phenotypes are specified in supplemental Table 2, and gating strategies are shown in supplemental Figure 5. Comparisons performed using two-tailed t test did not show significant differences between Ifnar1+/− and Ifnar1–/–-mice for any measurement.
Ifnar1 deletion does not impair lymphopoiesis or mature blood production. (A-D) Absolute numbers of indicated B-cell progenitor populations in P0 liver or P14 bone marrow from mice of Ifnar1+/− and Ifnar1–/–-mice, or spleen B-cell populations from the same mice. (E-H) Absolute numbers of thymocyte subpopulations Ifnar1+/− and Ifnar1–/–-mice, or spleen T-cell populations from the same mice. (I-L) White blood cell (H), absolute neutrophil (I), red blood cell (J), and platelet (K) counts presented for 1 μL of blood from P14 mice. For all panels, n = 4 to 11 and error bars reflect standard deviation. Surface marker phenotypes are specified in supplemental Table 2, and gating strategies are shown in supplemental Figure 5. Comparisons performed using two-tailed t test did not show significant differences between Ifnar1+/− and Ifnar1–/–-mice for any measurement.
Basal IFN-γ and IL-1β signaling do not modulate neonatal progenitor population sizes
In addition to IFN-1, IL-1β and IFN-γ have both been shown to regulate fetal HSC emergence and adult HSC/MPP stress responses.11-13 We, therefore, tested whether either cytokine could modulate HSC/MPP numbers alone or in cooperation with IFN-1. We crossed mice deficient for either the IL-1β receptor (Il1r1) or the IFN-γ receptor (Ifngr1) with Ifnar1-deficient mice to test for genetic interactions between the inflammatory cytokine receptors. Il1r1 deletion did not lead to HSC or MPP depletion in P0 mice, nor did it amplify the effects of Ifnar1 deletion (supplemental Figure 6A-E). Indeed, Il1r1 deletion rescued HSC depletion caused by Ifnar1 deletion. Ifngr1 deletion did not lead to HSC or MPP depletion in P0 mice by itself, nor did it amplify the effects of Ifnar1 deletion (supplemental Figure 6F-J). These findings show that IFN-1 is selectively responsible for the expansion of phenotypic HSC and MPP populations in neonates.
IFN-1–dependent changes in neonatal MPP population sizes do not reflect changes in proliferation, cell death, or differentiation
We sought to understand why HSC and MPP populations decline in Ifnar1–/–- neonates, even as committed progenitor populations expand. We tested whether these changes reflect differences in cell proliferation rates or cell death. To measure proliferation, we first analyzed cell cycle phases using Ki67/4′,6-diamidino-2-phenylindole (DAPI) staining (supplemental Figure 7A). Ifnar1 deletion caused a significant increase in the percentages of P0 HSCs and MPPs in the S/G2/M phases of the cell cycle relative to heterozygous controls (Figure 5A,B). We observed a small but significant reduction in quiescent (G0) MPPs and a trend toward the reduction in quiescent HSCs (Figure 5A,B). These findings suggest that Ifnar1 deletion increases rather than decreases HSC/MPP proliferation. As an orthogonal strategy, we isolated HSCs, MPPs, pGMs, and GMPs from P0 livers and performed propidium iodide staining to assess the fraction of each population in S/G2/M phase of the cell cycle (supplemental Figure 7B). Ifnar1–/–- MPPs again showed a modest but significant increase in proliferation relative to controls (Figure 5D). The remaining populations showed no differences in proliferation between Ifnar1+/− and Ifnar1–/– mice (Figure 5C,E,F). We confirmed that P0 Ifnar1–/–- MPPs proliferate at a slightly higher rate than Ifnar1+/− controls based on BrdU incorporation (Figure 5G; supplemental Figure 7C). Interestingly, we did not observe a similar increase in MPP proliferation at P14 (Figure 5H). There were no Ifnar1-dependent differences in programmed cell death rates in P0 HSCs, MPPs, pGMs, or GMPs, as measured with annexin V staining (Figure 5I; supplemental Figure 7D,E). Altogether, the data indicate that changes in proliferation and cell death do not adequately account for the decrease in HSCs/MPPs and the increase in pGMs/GMPs observed in newborn Ifnar1–/– mice.
Ifnar1 deletion does not impair proliferation or promote death or neonatal HSCs or MPPs. (A,B) Cell cycle distributions of HSCs (CD150+CD48-LSK) and MPPs (CD48+LSK) as measured using Ki67/DAPI staining, n = 5-6. (C-F) Percentages of P0 HSCs, MPPs, pGMs, or GMPs in S/G2/M phase of the cell cycle in Ifnar1+/- and Ifnar1–/–-mice, as measured by propidium iodine staining, n = 4-9. (G) Percentage of BrdU-positive MPPs (CD48+LSK) after a 24 hour pulse between E19 and P0, n = 4-10. (H) Percentage of P14 MPPs (CD48+LSK), in S/G2/M phase of the cell cycle as measured by propidium iodine staining n = 5-6. (I) Percentage of Annexin V+, DAPI-negative apoptotic HSCs, MPPs, pGM, and GMP in the P0 livers of Ifnar1+/– and Ifnar1–/–mice, n = 6-15. For all panels, error bars reflect standard deviation. Representative gating strategies are shown in supplemental Figure 5. ∗∗∗P < .001; ∗P < .05 using two-tailed t test. (J) CFU assays for individually plated MPP2, MPP3, MPP4, or LK cells. Error bars reflect standard deviations from 3 independent experiments with at least 60 cells plated per population per experiment.
Ifnar1 deletion does not impair proliferation or promote death or neonatal HSCs or MPPs. (A,B) Cell cycle distributions of HSCs (CD150+CD48-LSK) and MPPs (CD48+LSK) as measured using Ki67/DAPI staining, n = 5-6. (C-F) Percentages of P0 HSCs, MPPs, pGMs, or GMPs in S/G2/M phase of the cell cycle in Ifnar1+/- and Ifnar1–/–-mice, as measured by propidium iodine staining, n = 4-9. (G) Percentage of BrdU-positive MPPs (CD48+LSK) after a 24 hour pulse between E19 and P0, n = 4-10. (H) Percentage of P14 MPPs (CD48+LSK), in S/G2/M phase of the cell cycle as measured by propidium iodine staining n = 5-6. (I) Percentage of Annexin V+, DAPI-negative apoptotic HSCs, MPPs, pGM, and GMP in the P0 livers of Ifnar1+/– and Ifnar1–/–mice, n = 6-15. For all panels, error bars reflect standard deviation. Representative gating strategies are shown in supplemental Figure 5. ∗∗∗P < .001; ∗P < .05 using two-tailed t test. (J) CFU assays for individually plated MPP2, MPP3, MPP4, or LK cells. Error bars reflect standard deviations from 3 independent experiments with at least 60 cells plated per population per experiment.
Next, we performed CFU assays to test whether Ifnar1 deletion promotes precocious MPP differentiation and concomitant loss of colony-forming capacity. We sorted single P0 MPP2, MPP3, and MPP4 into methylcellulose and scored colonies. We observed no difference in CFU frequencies between Ifnar1+/− and Ifnar1–/–- MPPs (Figure 5J), and colonies were similar in size (data not shown). There was also no difference in CFU frequencies in less pure LK cells, which include pGMs and GMPs (Figure 5J). Overall, the data show that Ifnar1–/–- neonatal MPPs can self-renew and differentiate normally. Changes in proliferation, apoptosis, and differentiation do not offer suitable explanations for the MPP depletion and concomitant myeloid progenitor expansion observed in Ifnar1-deficient neonates.
Ifnar1 deletion has only modest effects on progenitor populations as defined by single-cell transcriptomes, in contrast to extensive changes observed by surface phenotype
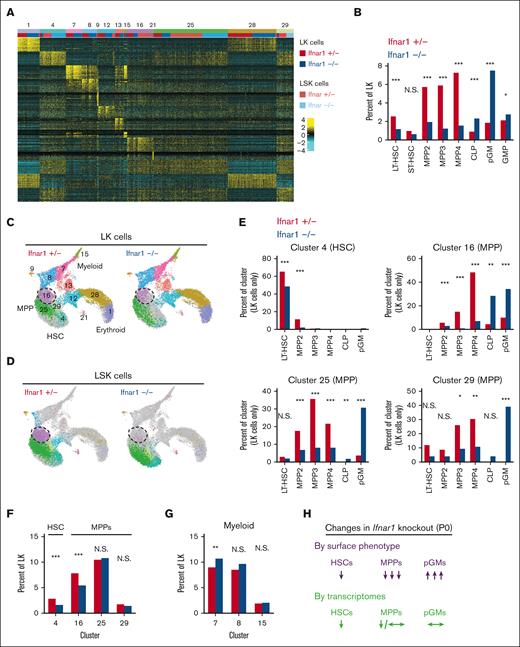
We used CITE-seq to better characterize the effects of IFN-1 signaling on neonatal and juvenile hematopoiesis, independent from surface marker phenotypes. The goal was to understand why committed progenitor populations paradoxically expand in Ifnar1–/– mice while HSC and MPP populations decline, and why changes in immature progenitor populations do not alter hematopoietic output. The CITE-seq assays incorporated feature barcodes for 9 different surface markers, thus enabling us to resolve HSC, MPP, pGM, GMP, and CLP subpopulations while simultaneously assessing single-cell gene expression (supplemental Figure 8). We obtained transcriptomes for LK and LSK cells and generated libraries independently. We then aggregated the data after sequencing. We took this approach so that we could compare changes in progenitor population frequencies when Sca1 was both included and not included as a selection marker, given that Sca1 is an IFN-1 target gene.3 We used ICGS222,31 to cluster the data and overlay surface marker phenotypes (Figure 6A,B; supplemental Tables 2 and 3). The CITE-seq surface barcodes redemonstrated the depletion of HSC and MPP populations and the concomitant expansion of CLP, pGM, and GMP populations in Ifnar1–/– mice (Figure 6B).
Ifnar1 deletion has only modest effects on immature progenitor populations when the populations are defined by single-cell transcriptomes rather than surface marker phenotypes. (A) Heatmap identifying 13 distinct clusters after iterative clustering via ICGS2. The clusters are numbered (ranging, 1-29), as defined by the ICGS2 algorithm. The clustering reflects aggregates of separately sorted LK and LSK cells from Ifnar1+/− and Ifnar1–/–-mice. Genotypes and cell types are indicated by color coding in the second horizontal bar above the heatmap. (B) HSC, MPP, and committed progenitor populations were defined based on surface markers as per supplemental Table 2. The frequencies of each phenotypically defined population within the LK samples are shown for Ifnar1+/− and Ifnar1–/–-mice. (C,D) UMAP plots showing clustering results for Ifnar1+/− and Ifnar1–/–-LK and LSK cells. Differentiation states are indicated based on marker gene expression for each cluster. Clusters are color coded to match panel A. Cluster 16, a T-cell–biased MPP population, is indicated by the hashed circle. (E) Percentages of LK cells in clusters 4, 16, 25, and 29 with the indicated surface marker phenotypes. (F) Percentages of LK cells in HSC cluster 4 or MPP clusters 16, 25, and 29 in Ifnar1+/− and Ifnar1–/–-mice. (G) Percentages of LK cells in myeloid clusters 7, 8, and 15 in Ifnar1+/− and Ifnar1–/–-mice. (H) Summary of changes in HSC, MPP, and pGM frequencies when the populations are defined by surface marker phenotype as compared with single-cell transcriptomes. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001 by the χ2 test.
Ifnar1 deletion has only modest effects on immature progenitor populations when the populations are defined by single-cell transcriptomes rather than surface marker phenotypes. (A) Heatmap identifying 13 distinct clusters after iterative clustering via ICGS2. The clusters are numbered (ranging, 1-29), as defined by the ICGS2 algorithm. The clustering reflects aggregates of separately sorted LK and LSK cells from Ifnar1+/− and Ifnar1–/–-mice. Genotypes and cell types are indicated by color coding in the second horizontal bar above the heatmap. (B) HSC, MPP, and committed progenitor populations were defined based on surface markers as per supplemental Table 2. The frequencies of each phenotypically defined population within the LK samples are shown for Ifnar1+/− and Ifnar1–/–-mice. (C,D) UMAP plots showing clustering results for Ifnar1+/− and Ifnar1–/–-LK and LSK cells. Differentiation states are indicated based on marker gene expression for each cluster. Clusters are color coded to match panel A. Cluster 16, a T-cell–biased MPP population, is indicated by the hashed circle. (E) Percentages of LK cells in clusters 4, 16, 25, and 29 with the indicated surface marker phenotypes. (F) Percentages of LK cells in HSC cluster 4 or MPP clusters 16, 25, and 29 in Ifnar1+/− and Ifnar1–/–-mice. (G) Percentages of LK cells in myeloid clusters 7, 8, and 15 in Ifnar1+/− and Ifnar1–/–-mice. (H) Summary of changes in HSC, MPP, and pGM frequencies when the populations are defined by surface marker phenotype as compared with single-cell transcriptomes. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001 by the χ2 test.
We next tested whether frequencies of transcriptionally defined progenitor populations shifted in Ifnar1–/–- neonates. ICGS2 identified 13 unique clusters (Figure 6A,C,D). Cluster 4 expressed guide genes consistent with LT-HSC identity (supplemental Table 4; Figure 7A) and a majority of cells within the cluster bore LT-HSC surface markers (CD201+CD150+CD48–LSK) in wild-type mice (Figure 6E). The phenotypic LT-HSC population declined within cluster 4 in Ifnar1–/– mice. Note that for this analysis, we focused exclusively on the LK cell data so that cells that downregulate Sca1 in Ifnar1-deficient mice would still be included in the analysis. We also identified 3 clusters, numbers 16, 25, and 29, with guide-gene profiles and surface marker phenotypes indicative of MPPs (Figure 6C-E). These clusters, along with cluster 4, accounted for almost all LSK cells (Figure 6D), consistent with MPP identities. Cells within clusters 16, 25, and 29 shifted from MPP to CLP and pGM surface barcode identities in Ifnar1–/– mice, which is consistent with flow cytometry findings (Figure 6E). There were no expression changes in genes associated with cell proliferation and apoptosis within these populations (supplemental Figure 9). IFN-1 target gene expression was reduced in clusters 16, 25, and 29, consistent with our prior studies (supplemental Table 5), but only cluster 16 was modestly reduced in size (Figure 6F), despite the much larger reduction in MPPs by surface phenotype. Likewise, myeloid progenitor clusters 7, 8, and 15 showed little to no expansion despite increases in phenotypic pGMs (Figure 6G). A similar analysis at P14 revealed only modest changes in MPP population frequencies, as defined by gene expression (supplemental Figure 10A-E). Thus, IFN-1 is not necessary to sustain neonatal or juvenile MPPs when one uses gene expression rather than surface markers to identify the populations (Figure 6H). The simplest explanation for this phenomenon is that Sca1 expression declines in Ifnar1-deficient MPPs and causes them to appear as CLPs and pGMs.
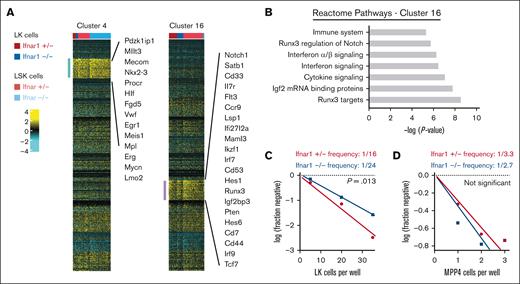
Ifnar1 deletion causes a modest reduction in T-cell potential in neonatal mice. (A) Clusters 4 and 16 were modestly depleted in Ifnar1–/–- neonates as compared with Ifnar1+/− neonates. Cluster 4 cells expressed genes consistent with HSC identity. Cluster 16 cells expressed some genes associated with myeloid bias (eg, Cd33) and many genes associated with T-lymphoid bias (eg, Notch, Hes1, Runx3, and so on). (B) Reactome pathway analysis of marker genes for cluster 16 showed interferon-related genes as well as Notch/Runx3-associated genes that would align with T-cell bias. (C,D) Results from limiting dilution assays of LK cells (C) or MPP4 (D) plated on OP9-delta stroma to assess T-cell potential at P0. The frequencies of LK and MPP4 cells with T-cell potential in Ifnar1+/- and Ifnar1–/– mice were calculated and compared with the findings from ELDA, n = 48-96 wells per cell dose and genotype across ∼2 to 3 independent experiments.
Ifnar1 deletion causes a modest reduction in T-cell potential in neonatal mice. (A) Clusters 4 and 16 were modestly depleted in Ifnar1–/–- neonates as compared with Ifnar1+/− neonates. Cluster 4 cells expressed genes consistent with HSC identity. Cluster 16 cells expressed some genes associated with myeloid bias (eg, Cd33) and many genes associated with T-lymphoid bias (eg, Notch, Hes1, Runx3, and so on). (B) Reactome pathway analysis of marker genes for cluster 16 showed interferon-related genes as well as Notch/Runx3-associated genes that would align with T-cell bias. (C,D) Results from limiting dilution assays of LK cells (C) or MPP4 (D) plated on OP9-delta stroma to assess T-cell potential at P0. The frequencies of LK and MPP4 cells with T-cell potential in Ifnar1+/- and Ifnar1–/– mice were calculated and compared with the findings from ELDA, n = 48-96 wells per cell dose and genotype across ∼2 to 3 independent experiments.
IFN-1 sustains a T-cell–biased MPP population in neonates
As noted earlier, we observed modest decreases in transcriptionally defined HSCs (cluster 4) and an MPP population (cluster 16) in Ifnar1–/– neonates as a fraction of the LK population (Figures 6F and 7A). We have previously shown that Ifnar1-deficient HSCs function normally in repopulating assays and therefore did not characterize them further. Instead, we sought to characterize the function of the MPPs that populated cluster 16. Guide genes for cluster 16 included Notch1, Il7r, Runx3, Pten, and several Notch signal transduction intermediates (Maml3, Hes1, and Hes6), all suggesting a T-lineage bias (Figure 7A,B). To test whether T-cell potential was modestly reduced in Ifnar1–/–- neonates, as predicted by the single-cell transcriptomes, we plated limiting numbers of LK cells on OP9-delta stromal cells and counted wells that gave rise to T cells. We observed a ∼50% reduction in T-cell potential in P0 Ifnar1-/- LK cells (Figure 7C), but not in P14 Ifnar1-/- LK cells (supplemental Figure 10F), consistent with the changes observed by CITE-seq. Because cluster 16 was heavily enriched for phenotypic MPP4 cells in wild-type neonates (Figure 6E), we also plated limiting doses of MPP4 on OP9-Δ stroma. Ifnar1+/− and Ifnar1–/–- MPP4 had similar T-cell potential (Figure 7D). Thus, Ifnar1 deficiency reduces the frequency of T-cell progenitors within the larger LK fraction rather than impairing the function of individual T-cell progenitors. The remaining T-cell progenitors were sufficient to maintain normal T-cell output in Ifnar1–/–- neonates, even when they were stressed by other inflammatory cytokines, such as IL-1β and IFN-γ (supplemental Figure 11). The depletion of transcriptionally and functionally defined T-cell progenitors in Ifnar1–/–- neonates is far less severe than the depletion of phenotypic MPPs, particularly MPP4. This likely accounts for normal hematopoietic output and normal lymphopoiesis in Ifnar1–/–- neonates.
Discussion
Inflammatory cytokines have well-established roles in promoting myelopoiesis and megakaryopoiesis under stress conditions,1,2 but their roles in baseline hematopoiesis are just starting to emerge. Here, we have carefully evaluated the effects of basal IFN-1 signaling on neonatal and juvenile hematopoiesis. Consistent with prior work,14 deleting the IFN-1 receptor (Ifnar1) had profound effects on many immature progenitor populations, at least based on surface marker phenotypes. However, despite extensive efforts, we found that Ifnar1 deletion does not impair mature blood cell production in neonates or adults. Furthermore, the depletion of MPPs in Ifnar1-deficient neonates appears to reflect shifts in surface marker expression rather than the loss of the populations altogether, given that transcriptionally defined MPP populations remained largely intact in our CITE-seq assays (Figure 6). We did observe a modest decline in a T-cell–biased MPP population at P0 that correlated with a reduction in T-cell potential (Figure 7), though the decline was far more modest than would be anticipated based on MPP4 numbers. Transcriptionally defined HSCs also declined in Ifnar1-deficient neonates, but our prior studies showed that HSC function remains intact.14 Altogether, the data do not support a prominent role for physiologic IFN-1 signaling in sustaining neonatal or juvenile hematopoiesis, and other cytokines (eg, IFN-γ and IL-1β) also appeared to have negligible effects in the absence of infection (supplemental Figure 6).
Our findings pertain specifically to IFN-1 signaling during normal ontogeny, and they do not address conditions of stress or aging. Maternal infections can alter postnatal blood production,32 and our data do not directly address that context. Likewise, hematopoietic changes in response to microbial colonization and infection in adulthood may be different from responses to sterile IFN-1 signaling during fetal/neonatal stages.10 Nevertheless, our CITE-seq data indicate that even under basal conditions, IFN-1 signaling can have profound effects on HSC/MPP surface marker phenotypes. Changes in Sca1 expression that have previously been shown to accompany hyperinflammation3 can also affect HSC/MPP phenotypes in noninflamed states. Thus, efforts to use surface markers to characterize hematopoiesis under germ-free or cytokine-deficient contexts should be made with some caution. CITE-seq and other single-cell techniques can provide valuable complementary insights.
Our findings raise an important question: What is the role of the perinatal IFN-1 spike if it does not modulate hematopoietic output? One key function is to promote the expression of genes associated with antigen presentation and antiviral responses, particularly major histocompatibility complex–I genes, within the postnatal HSC and MPP populations. We described these changes previously and redemonstrate them here.14 IFN target gene expression has also been shown to increase between middle and late gestation in human HSCs/MPPs, suggesting that the late-gestation IFN-1 response may be conserved.33,34 Our data suggest that perinatal IFN-1 may prime postnatal progenitors to present antigens and activate antiviral programs without substantially influencing hematopoietic output. It is also possible that flux through the HSC/MPP/pGM populations changes in response to IFN-1 signaling, even if the populations themselves remain stable in size. Future lineage-tracing studies can resolve this question.
In addition to clarifying the role of IFN-1 signaling in perinatal hematopoiesis, our data illuminate previously unappreciated ontological shifts in the lineage biases of phenotypically defined MPP subpopulations (Figure 1). Fetal and perinatal MPP3 and MPP4 demonstrate more monocyte bias in fetal and perinatal stages than in adult stages of development. Likewise, MPP2 and MPP3 demonstrate greater lymphoid bias during midgestation than at birth and in adulthood. The transcriptional programs that underlie these shifts remain an active area of investigation because they may contribute to age-related changes in immune output and leukemia initiation.
Acknowledgments
This work was financially supported by grants to J.A.M. from the National Heart, Lung, and Blood Institute (R01 HL152180 and R01 HL136504), Alex's Lemonade Stand Foundation “A” Award, Hyundai Hope on Wheels, the Alvin J. Siteman Cancer Center Investment Program (supported by the Foundation for Barnes Jewish Hospital and National Cancer Institute Cancer Center Support grant P30 CA091842), and the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital. J.A.M. is a Leukemia and Lymphoma Society Scholar.
Authorship
Contribution: J.A.M. designed and oversaw all experiments, conducted experiments, interpreted data, wrote the manuscript, and secured funding; Y.L. conducted experiments, interpreted data, and wrote the manuscript; W.Y. performed all bioinformatic analyses; H.C.W., R.M.P., E.B.C., and E.D. performed experiments and interpreted data; and all authors reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing interests.
Correspondence: Jeffrey A. Magee, Washington University School of Medicine, 660 S. Euclid Ave, Box 8208, St. Louis, MO 63110; e-mail: mageej@wustl.edu.
References
Author notes
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE205544).
Data are available on request from the corresponding author, Jeffrey A. Magee (mageej@wustl.edu).
The full-text version of this article contains a data supplement.