Key Points
MOZ is critical for MOZ/MLL fusion-mediated AML development, Meis1 induction by MOZ fusions, and Hoxa9/Meis1 induction by MLL fusions.
Endogenous MOZ is required to maintain MOZ-target and active histone modifications at the Meis1 gene locus.
Abstract
Monocytic leukemia zinc finger protein (MOZ, MYST3, or KAT6A) is a MYST-type acetyltransferase involved in chromosomal translocation in acute myelogenous leukemia (AML) and myelodysplastic syndrome. MOZ is established as essential for hematopoiesis; however, the role of MOZ in AML has not been addressed. We propose that MOZ is critical for AML development induced by MLL-AF9, MLL-AF10, or MOZ-TIF2 fusions. Moz-deficient hematopoietic stem/progenitor cells (HSPCs) transduced with an MLL-AF10 fusion gene neither formed colonies in methylcellulose nor induced AML in mice. Moz-deficient HSPCs bearing MLL-AF9 also generated significantly reduced colony and cell numbers. Moz-deficient HSPCs expressing MOZ-TIF2 could form colonies in vitro but could not induce AML in mice. By contrast, Moz was dispensable for colony formation by HOXA9-transduced cells and AML development caused by HOXA9 and MEIS1, suggesting a specific requirement for MOZ in AML induced by MOZ/MLL fusions. Expression of the Hoxa9 and Meis1 genes was decreased in Moz-deficient MLL fusion-expressing cells, while expression of Meis1, but not Hoxa9, was reduced in Moz-deficient MOZ-TIF2 AML cells. AML development induced by MOZ-TIF2 was rescued by introducing Meis1 into Moz-deficient cells carrying MOZ-TIF2. Meis1 deletion impaired MOZ-TIF2–mediated AML development. Active histone modifications were also severely reduced at the Meis1 locus in Moz-deficient MOZ-TIF2 and MLL-AF9 AML cells. These results suggest that endogenous MOZ is critical for MOZ/MLL fusion-induced AML development and maintains active chromatin signatures at target gene loci.
Introduction
Acute myelogenous leukemia (AML) is a hematological malignancy derived from hematopoietic stem cells (HSCs) and myeloid progenitors with acquired fusion genes or oncogenic mutations. Various fusion genes involving transcription factors and epigenetic modulators, such as AML1 (RUNX1), CBFβ, MLL1, or PML, have been found in AML. In addition, mutations in genes involved in transcription, such as AML1, CEBPA, DNMT3A, TET2, and EZH2, are also observed in AML. These findings imply that many cases of AML arise because of failure of transcription regulation caused by gene mutations or generation of fusion genes.1-5
Histone modifications, including acetylation, methylation, or ubiquitination of the histone N-terminal tail, are critical epigenetic gene expression regulatory mechanisms.6 Histone modifications are crucial for cancer pathophysiology and molecular targets for therapies, including bromodomain and extraterminal motif or histone deacetylase inhibitors.7,8 In MLL fusion leukemias, DOT1L methyltransferase is critical for leukemia development and induction of aberrant HOXA9/MEIS1 gene expression.9 These reports indicate that various histone modification factors are essential in hematological malignancy through target gene regulation.
Monocytic leukemia zinc finger protein (MOZ; also MYST3 or KAT6A) is one of the MYST-type histone acetyltransferases, which catalyze acetylation of histone H3K9 (H3 lysine 9),10 H3K14 (lysine 14),11 and H3K23 (lysine 23).12MOZ is a component of fusion genes, including MOZ-CREBBP and MOZ-TIF2, found in FAB M4/M5 acute monocytic leukemia and interacts with the AML1 (RUNX1), PU.1, and p53 transcription factors, which are essential in leukemia development.13 In addition, MOZ forms transcriptional complexes with the bromodomain and PHD finger-containing (BRPF) proteins, ING5, and MEAF6,14 and is necessary for HSC self-renewal and appropriate hematopoietic cell lineage development.15-17 Expression levels of the homeobox genes, Hoxa9 and Meis1, and cytokine receptors, c-Kit, c-Mpl, and c-Fms, are reduced in Moz-deficient hematopoietic stem/progenitor cells (HSPCs) and B cells.16,18,19 MOZ localizes to HOXA and MEIS1 loci in normal human CD34+ HSCs and B cells,19,20 while expression of the tumor suppressor gene, p16Ink4a, is elevated in Moz-null HSCs, neural stem cells, and mouse embryonic fibroblasts.21,22 In leukemia, aberrant expression of HOXA9 and MEIS1 is observed in MOZ, MLL, and NUP98 fusion leukemias and NPM1-mutated AML.3,23-25HOXA9 is upregulated in approximately 50% of patients with AML and is essential for MLL fusion-induced transformation and AML development.26,27MEIS1 is also critical for MLL fusion carrying leukemia28 through its roles in cell proliferation,29 homing and engraftment,30 and protection from oxidative stress.31 Although endogenous MOZ is required for self-renewal of HSCs and Hoxa9/Meis1 expression in normal hematopoietic cells, the roles of endogenous MOZ in MOZ/MLL fusion AML development and aberrant expression of Hoxa9/Meis1 in this context have not been addressed. Therefore, we analyzed the role of endogenous MOZ in MOZ/MLL fusion leukemia.
Using Moz-deficient mouse cells transfected with MOZ/MLL fusion genes, we conducted in vitro serial colony replating and in vivo leukemogenesis assays, which demonstrated that endogenous MOZ is essential for MLL fusion-induced immortalization and AML development, as well as MOZ fusion-induced AML development. Further, gene expression analysis revealed notable decreases in Hoxa9/Meis1 expression in Moz-deficient MLL fusion cells and Meis1 expression in MOZ fusion cells. Active histone marks at the Meis1 locus were significantly reduced in Moz-deficient MOZ/MLL fusion AML cells. Moreover, the onset of AML development depended on aberrant Meis1 expression in MOZ fusion AML. These results suggest that endogenous MOZ is a critical factor in MOZ/MLL fusion AML via its induction of aberrant MEIS1 and HOXA9 gene expression.
Materials and methods
Mice
Moz−/− mice16 were backcrossed with C57BL/6 mice more than 10 times. In addition, Meis1 conditionally-deficient (Meis1f/f)32 and ROSA26-Cre-ERT2 knock-in (Cre-ERT2) (TaconicArtemis GmBH; Cologne, Germany) mice were maintained on a C57BL/6 background. C57BL/6J mice were purchased from CLEA Japan (Tokyo, Japan). Animals were kept under specific pathogen-free, temperature-controlled conditions, according to institutional guidelines. Written approval of all animal experiments was obtained from the local Animal Experiments Committee of the National Cancer Center Research Institute. Genotyping was performed using genomic DNA prepared from mouse tail or AML cells boiled in 50 mM NaOH solution at 96°C for 30 minutes. Primers used for genotyping are shown in supplemental Table 1 in the data supplement.
Reagents
Reagents used in this study were: APC-eFluor 780-conjugated streptavidin, anti-CD16/32 (93), anti-CD34-FITC (RAM34), anti-CD115-PE (AFS98), anti-B220-biotin (RA3-6B2), anti-CD11b-PE-Cy7 (M1/70), anti-CD117-APC (2B8), and anti-Gr-1-APC (RB6-8C5) (eBioscience; San Diego, CA); anti-CD3ε-biotin (145-2C11), anti-Gr-1-biotin (RB6-8C5), anti-TER119-biotin (TER-119), anti-CD127-biotin (A7R34), anti-CD16/32-PE-Cy7 (93), and anti-hNGFR-APC (ME20.4) (Biolegend; San Diego, CA); anti-Sca-1-PE (E13-161.7) (BD Biosciences; Franklin Lakes, NJ); polyclonal antibodies against Histone H3 (ab1791), H3K9ac (ab4441), H3K27ac (ab4729), and H3K79me2 (ab3594) (Abcam; Cambridge, United Kingdom); polyclonal antibodies against H3K4me3 (39159), H3K9ac (39137), and H3K23ac (39132) (Active Motif Inc.; Carlsbad, CA); polyclonal antibody against H3K23ac (07-355) (Merck Millipore; Billerica, MA); recombinant murine interleukin-3 (IL-3; 213-03), murine stem cell factor (SCF; 250-03), and murine granulocyte/monocyte-colony stimulating factor (GM-CSF; 315-03) (PeproTech; Rocky Hill, NJ); recombinant murine oncostatin M (OSM; 495-MO-025) (R&D Systems; Minneapolis, MN); and tamoxifen (TAM), 4-OHT (4-hydroxy tamoxifen), corn oil, and rat serum-IgG (Sigma Aldrich; St. Louis, MO).
Purification of HSPCs, c-Kit+, Sca-1+ lineage− cells, and common myeloid progenitors
Fetal liver (FL) samples from E14.5 Moz+/+, Moz+/−, and Moz−/− embryos were dispersed into single-cell suspensions in 2% fetal bovine serum in phosphate-buffered saline. Erythrocytes were lyzed with tris-buffered 0.83% ammonium chloride solution. Whole FL cells were incubated with anti-CD16/32 antibody or rat serum IgG to prevent nonspecific antibody binding. FL cells were then incubated with biotin-labeled antibodies against lineage markers (Ter119, CD3ε, B220, Gr-1, and CD127) (30 min, 4°C), followed by incubation with streptavidin-conjugated magnetic beads (Miltenyl Biotech; Bergisch, Gladbach, Germany) (20 min, 4°C). Cell suspensions containing magnetic bead-bound cells were loaded onto a magnetic column (Miltenyl Biotech) to remove lineage marker-positive magnetic bead-bound cells. The flowthrough of unbound cells was collected (lineage− HSPCs).
For purification of c-Kit+, Sca-1+ lineage− (KSL), and chronic myeloid progenitor (CMP) fractions, lineage− HSPCs were incubated with streptavidin-APC-eFluor 780, c-Kit-APC, Sca-1-PE, PE-Cy7-CD16/32, and CD34-FITC (45 min, 4°C). KSLs and CMPs were distinguished as lineage−, c-Kit+, and Sca-1+, or lineage− c-Kit+, Sca-1−, CD16/32lo, and CD34+, respectively, using a JSAN flow cytometer (Bay Bioscience; Kobe, Japan). Sorted lineage− HSPCs, KSLs, and CMPs were cultured in StemPro34 medium (Thermo Fisher Scientific; Waltham, MA) supplemented with recombinant murine SCF (50 ng/mL), IL-3 (10 ng/mL), OSM (10 ng/mL), tylosin (8 μg/mL) (Sigma Aldrich), penicillin/streptomycin (100 U/mL) (Nacalai Tesque; Kyoto, Japan), and l-glutamine (2 mM; Sigma Aldrich).
In vitro serial colony formation assay
Human MOZ-TIF2, MLL-AF9, MLL-AF10, or HOXA9 genes were introduced into cells using a retrovirus system and cultured for 5 days. Green fluorescent protein (GFP) marker-positive cells were sorted by JSAN flow cytometry. Sorted GFP+ cells (5 × 104) were cultured in Methocult M 3234 methylcellulose medium (Stemcell Technologies; Vancouver, BC, Canada) containing IL-3 (20 ng/mL), SCF (50 ng/mL), GM-CSF (10 ng/mL), and 100 U/mL penicillin/streptomycin. After 3 to 5 days, colonies and total cells were counted, and 3 × 104 or 1 × 105 cells were cultured in methylcellulose medium 3 or 5 times. Cultures were maintained at 37°C in a 5% CO2 humidified atmosphere.
In vivo leukemogenesis assay
Cells transduced with the MOZ/MLL fusion, human MEIS1, or human HOXA9/MEIS1 genes were prepared as described above. Cells (MOZ-TIF2, MLL-AF10, and HOXA9/MEIS1, 2 × 105; MEIS1, 5 × 105) were transplanted into sublethally γ-irradiated (600 rad) recipient mice via the tail vein. MOZ-TIF2 + MEIS1 or mock-transduced cells were generated by the introduction of the MOZ-TIF2 gene into Moz+/− or Moz−/− FL HSPCs. After 3 days of culture, a vector expressing human MEIS1 or empty vector (Mock) was introduced into MOZ-TIF2–infected cells, and these cells were then cultured for 5 days. After infection, MOZ-TIF2 + MEIS1 or MOZ-TIF2 + Mock cells (5 × 105) were transplanted into sublethally γ-irradiated (600 rad) recipient mice. Peripheral blood was collected from recipient mice every 4 weeks and analyzed for populations of GFP+ cells expressing HOXA9, MOZ, or MLL fusion genes and for Mac-1 (myeloid cell marker)-positive cells to monitor AML development, which was defined as a proportion of GFP+ cells >80% in recipient mouse bone marrow (BM), with hepatosplenomegaly and lymphadenopathy.
Statistical analysis
A 2-tailed unequal-variance t test was used to assess the significance of differences among samples. Differences in survival curves among groups were evaluated using the log-rank test in GraphPad Prism 6.0 (GraphPad Software; San Diego, CA).
Supplemental materials and methods
Materials and methods for immunofluorescent staining and flow cytometric analysis, retrovirus production and infection, RNA purification, quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) analysis, chromatin immunoprecipitation (ChIP) assay, ChIP-Seq analysis, and conditional deletion of the Meis1 gene in AML cells in vitro and in vivo are provided as supplemental materials on the Blood Advances website.
Results
Endogenous MOZ is essential for MOZ/MLL fusion-induced immortalization and AML development
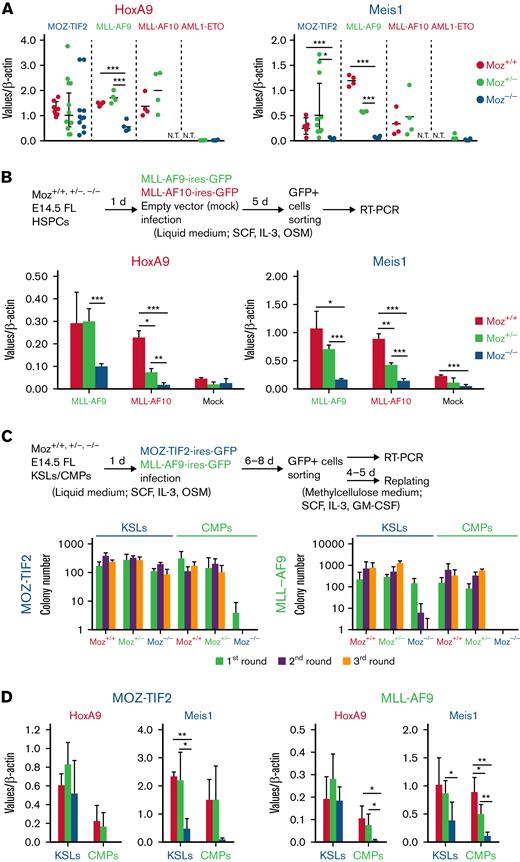
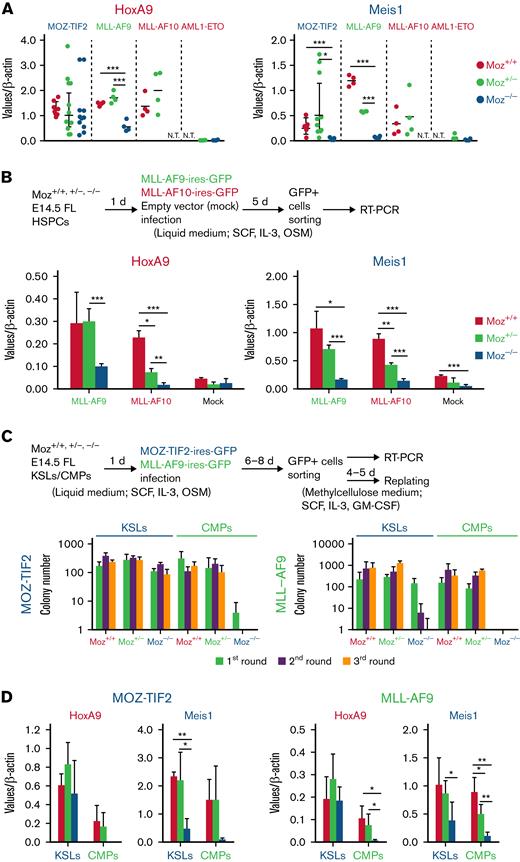
To explore the roles of endogenous MOZ in AML development, we used Moz knockout mice.16 FL HSPCs from E14.5 Moz+/+, Moz+/−, and Moz−/− embryos were transduced with MOZ-TIF2, MLL-AF9, MLL-AF10, or HOXA9 using retrovirus vectors and their abilities to form colonies in vitro tested (Figure 1A). Moz−/− HSPCs transduced with either MOZ-TIF2 or HOXA9 serially formed colonies in methylcellulose medium ≥5 times, whereas Moz−/− HSPCs transduced with MLL-AF10 formed fewer colonies and did not show continuous growth and, although Moz−/− HSPCs transduced with MLL-AF9 transiently formed colonies, their number decreased significantly with successive replating. In addition, Moz−/− HSPCs expressing MLL-AF9 generated fewer cell colonies than Moz+/+ and Moz+/− HSPCs expressing MLL-AF9. To investigate the roles of endogenous MOZ in AML development, Moz+/+, Moz+/−, and Moz−/− FL HSPCs transduced with MOZ-TIF2, MLL-AF10, or HOXA9, together with MEIS1, were transplanted into γ-irradiated recipient mice (Figure 1B). Although Moz−/− HSPCs expressing both HOXA9 and MEIS1 induced AML in recipient mice, Moz−/− HSPCs with either MOZ-TIF2 or MLL-AF10 did not. Further, mice transplanted with Moz+/−MLL-AF10 cells survived longer than those transplanted with Moz+/+MLL-AF10 cells. These results indicate that endogenous MOZ is essential for MOZ/MLL fusion-induced AML development.
Endogenous MOZ is essential for MOZ/MLL fusion-induced leukemia development. (A) Serial colony replating assays were performed in Moz+/+, Moz+/−, and Moz−/− HSPCs bearing MOZ-TIF2, MLL-AF9, MLL-AF10, or HOXA9. A scheme of the experimental workflow is shown at the top. First, Moz+/+, Moz+/−, or Moz−/− FL lineage− HSPCs (2 × 105 cells) were transduced with MOZ-TIF2, MLL-AF10, MLL-AF9, or HOXA9 and cultured in a liquid medium. Subsequently, GFP+ cells (5 × 104) were sorted and cultured in a methylcellulose medium. Every 4 to 5 days, colony and cell numbers (× 105 cells) were counted, and then 1 × 105 cells were serially replated 5 times. Mean values were calculated from 3 to 4 independent experiments. Error bars represent mean ± standard deviation (SD). (B) Overall survival of recipient mice transplanted with Moz+/+, Moz+/−, or Moz−/− HSPCs (2 × 105) bearing MOZ-TIF2, MLL-AF10, or HOXA9 and MEIS1 (MOZ-TIF2, Moz+/+ and Moz−/− [n = 6], Moz+/− [n = 12]; MLL-AF10, Moz+/+ [n = 6], Moz+/− [n = 10], Moz−/− [n = 4]; HOXA9 + MEIS1, [n = 6]). Differences in survival were compared using the log-rank test; ∗P < .05, ∗∗P < .01, and ∗∗∗P < .005.
Endogenous MOZ is essential for MOZ/MLL fusion-induced leukemia development. (A) Serial colony replating assays were performed in Moz+/+, Moz+/−, and Moz−/− HSPCs bearing MOZ-TIF2, MLL-AF9, MLL-AF10, or HOXA9. A scheme of the experimental workflow is shown at the top. First, Moz+/+, Moz+/−, or Moz−/− FL lineage− HSPCs (2 × 105 cells) were transduced with MOZ-TIF2, MLL-AF10, MLL-AF9, or HOXA9 and cultured in a liquid medium. Subsequently, GFP+ cells (5 × 104) were sorted and cultured in a methylcellulose medium. Every 4 to 5 days, colony and cell numbers (× 105 cells) were counted, and then 1 × 105 cells were serially replated 5 times. Mean values were calculated from 3 to 4 independent experiments. Error bars represent mean ± standard deviation (SD). (B) Overall survival of recipient mice transplanted with Moz+/+, Moz+/−, or Moz−/− HSPCs (2 × 105) bearing MOZ-TIF2, MLL-AF10, or HOXA9 and MEIS1 (MOZ-TIF2, Moz+/+ and Moz−/− [n = 6], Moz+/− [n = 12]; MLL-AF10, Moz+/+ [n = 6], Moz+/− [n = 10], Moz−/− [n = 4]; HOXA9 + MEIS1, [n = 6]). Differences in survival were compared using the log-rank test; ∗P < .05, ∗∗P < .01, and ∗∗∗P < .005.
Endogenous MOZ is required for MOZ/MLL fusion-induced Meis1 and Hoxa9 expression, dependent on fusion gene and origin
To investigate the molecular mechanisms underlying the phenotypes observed in Moz−/−MOZ-TIF2, MLL-AF9, and MLL-AF10 AML cells, we analyzed Hoxa9 and Meis1 concentrations, which are aberrantly expressed in MOZ/MLL fusion AML. Hoxa9/Meis1 concentrations were strikingly higher in MOZ/MLL fusion AML cells than in AML1-ETO AML cells that did not exhibit aberrant Hoxa9/Meis1 expression. Hoxa9 and Meis1 concentrations were similar between Moz+/+ and Moz+/− AML cells and between MOZ and MLL fusion AML cells (Figure 2A). Although mean Hoxa9 expression in Moz−/−MOZ-TIF2 AML cells was similar to that in Moz+/+ or Moz+/−MOZ-TIF2 AML cells, expression of the Meis1 gene was dramatically lower in Moz−/−MOZ-TIF2 AML cells. Furthermore, Hoxa9 and Meis1 concentrations were significantly lower in Moz−/−MLL-AF9 AML cells than in Moz+/+ or Moz+/−MLL-AF9 AML cells (Figure 2A).
Endogenous MOZ is critical for MOZ/MLL fusion-mediated induction of target gene expression. (A) Gene expression concentrations of the Hoxa9/Meis1 genes in MOZ-TIF2, MLL-AF9, MLL-AF10, and AML1-ETO AML cells. Colonies of Moz+/+, Moz+/−, or Moz−/− AML cells that were serially replated >3 times were harvested and analyzed for Hoxa9, Meis1, and β-actin expression by quantitative reverse transcriptase-PCR (qRT-PCR). Expression levels of Hoxa9 and Meis1 were normalized to those of β-actin (n = 4 to 8). Error bars represent mean ± SD. Expression levels were compared using the Student t test; ∗P < .05 and ∗∗∗P < .005. N.T., not tested. (B) Expression of the Hoxa9/Meis1 genes in Moz+/+, Moz+/−, or Moz−/− cells bearing MLL-AF9 or MLL-AF10. The experimental scheme is shown at the top. Moz+/+, Moz+/−, or Moz−/− FL lineage− HSPCs were transduced with MLL-AF9, MLL-AF10, or empty vector (Mock) and cultured in a liquid medium. GFP+ cells were then sorted, and the expression levels of Hoxa9, Meis1, and β-actin were analyzed by qRT-PCR (n = 3 to 4). Expression levels of Hoxa9 and Meis1 were normalized to those of β-actin. Error bars represent mean ± SD. Expression levels were compared using the Student t test; ∗P < .05 and ∗∗∗P < .005. (C) Colony formation of Moz+/+, Moz+/−, or Moz−/− KSLs/CMPs bearing MOZ-TIF2 or MLL-AF9. The experimental scheme is shown at the top. First, Moz+/+, Moz+/−, or Moz−/− KSLs/CMPs (1 × 104 cells) were transduced with MOZ-TIF2 or MLL-AF9 fusion genes and cultured in a liquid medium. Subsequently, GFP+ cells (5 × 104) were sorted and cultured in a methylcellulose medium. Colony numbers were counted every 4 to 5 days, and then 3 × 104 cells were subsequently serially replated 3 times. The mean numbers of colonies formed by MOZ-TIF2– or MLL-AF9–expressing Moz+/+, Moz+/−, and Moz−/− cells derived from KSL or CMP fractions were calculated from 3, 4, or 5 independent experiments. (D) Hoxa9 and Meis1 expression in MOZ-TIF2–expressing cells derived from KSLs or CMPs. Moz+/+, Moz+/−, or Moz−/− KSLs and CMPs were transduced with the MOZ-TIF2 or MLL-AF9 fusion genes and cultured in a liquid medium. Then GFP+ cells were sorted, and gene expression levels of Hoxa9, Meis1, and β-actin were analyzed by qRT-PCR (n = 3 to 5). Expression levels of Hoxa9 and Meis1 were normalized to those of β-actin. Error bars represent mean ± SD; ∗P < .05, ∗∗P < .01, and ∗∗∗P < .005.
Endogenous MOZ is critical for MOZ/MLL fusion-mediated induction of target gene expression. (A) Gene expression concentrations of the Hoxa9/Meis1 genes in MOZ-TIF2, MLL-AF9, MLL-AF10, and AML1-ETO AML cells. Colonies of Moz+/+, Moz+/−, or Moz−/− AML cells that were serially replated >3 times were harvested and analyzed for Hoxa9, Meis1, and β-actin expression by quantitative reverse transcriptase-PCR (qRT-PCR). Expression levels of Hoxa9 and Meis1 were normalized to those of β-actin (n = 4 to 8). Error bars represent mean ± SD. Expression levels were compared using the Student t test; ∗P < .05 and ∗∗∗P < .005. N.T., not tested. (B) Expression of the Hoxa9/Meis1 genes in Moz+/+, Moz+/−, or Moz−/− cells bearing MLL-AF9 or MLL-AF10. The experimental scheme is shown at the top. Moz+/+, Moz+/−, or Moz−/− FL lineage− HSPCs were transduced with MLL-AF9, MLL-AF10, or empty vector (Mock) and cultured in a liquid medium. GFP+ cells were then sorted, and the expression levels of Hoxa9, Meis1, and β-actin were analyzed by qRT-PCR (n = 3 to 4). Expression levels of Hoxa9 and Meis1 were normalized to those of β-actin. Error bars represent mean ± SD. Expression levels were compared using the Student t test; ∗P < .05 and ∗∗∗P < .005. (C) Colony formation of Moz+/+, Moz+/−, or Moz−/− KSLs/CMPs bearing MOZ-TIF2 or MLL-AF9. The experimental scheme is shown at the top. First, Moz+/+, Moz+/−, or Moz−/− KSLs/CMPs (1 × 104 cells) were transduced with MOZ-TIF2 or MLL-AF9 fusion genes and cultured in a liquid medium. Subsequently, GFP+ cells (5 × 104) were sorted and cultured in a methylcellulose medium. Colony numbers were counted every 4 to 5 days, and then 3 × 104 cells were subsequently serially replated 3 times. The mean numbers of colonies formed by MOZ-TIF2– or MLL-AF9–expressing Moz+/+, Moz+/−, and Moz−/− cells derived from KSL or CMP fractions were calculated from 3, 4, or 5 independent experiments. (D) Hoxa9 and Meis1 expression in MOZ-TIF2–expressing cells derived from KSLs or CMPs. Moz+/+, Moz+/−, or Moz−/− KSLs and CMPs were transduced with the MOZ-TIF2 or MLL-AF9 fusion genes and cultured in a liquid medium. Then GFP+ cells were sorted, and gene expression levels of Hoxa9, Meis1, and β-actin were analyzed by qRT-PCR (n = 3 to 5). Expression levels of Hoxa9 and Meis1 were normalized to those of β-actin. Error bars represent mean ± SD; ∗P < .05, ∗∗P < .01, and ∗∗∗P < .005.
We also investigated Hoxa9/Meis1 expression in Moz−/− cells bearing MLL-AF10 and found that both Hoxa9 and Meis1 concentrations were significantly lower in MLL-AF10–expressing Moz−/− cells than those in MLL-AF10–expressing Moz+/+ or Moz+/− cells, similar to our findings for MLL-AF9 (Figure 2B). These results demonstrate that endogenous MOZ is essential for aberrant Meis1 induction in MOZ fusion AML cells and both Hoxa9 and Meis1 expression in MLL fusion AML cells.
MOZ or MLL fusions can transform HSCs and myeloid progenitors, CMPs, and granulocyte monocyte progenitors,33 implying that the roles of endogenous MOZ differ between HSCs and myeloid progenitors in MOZ/MLL fusion leukemia. Therefore, we sorted KSL (containing HSCs) and CMP fractions and transduced each with MOZ-TIF2 and MLL-AF9. After transfection, we sorted MOZ or MLL fusion-expressing cells and subjected them to serial colony replating assays and gene expression analysis. Only MOZ-TIF2–expressing Moz−/− cells derived from the KSL fraction could continuously form colonies, which those from CMPs could not. Although Moz−/−MLL-AF9 cells derived from KSLs formed colonies in the first round, colony-forming activity was markedly decreased with each successive generation. Further, Moz−/−MLL-AF9–expressing cells derived from CMPs could not generate colonies (Figure 2C), while Moz−/−MLL-AF9–expressing cells derived from HSPCs only showed colony formation activity in the first round, when they were replated at 3 × 104 cells (supplemental Figure 1). Gene expression analysis showed that induction of both Meis1 and Hoxa9 was decreased in MOZ-TIF2– or MLL-AF9–expressing Moz−/− cells derived from CMPs (Figure 2D), whereas only Meis1 expression was markedly reduced in KSL-derived MOZ-TIF2– or MLL-AF9–expressing cells (Figure 2D). These results indicate that endogenous MOZ is also essential for immortalization and induction of the Hoxa9 gene by the MOZ/MLL fusion in myeloid progenitors.
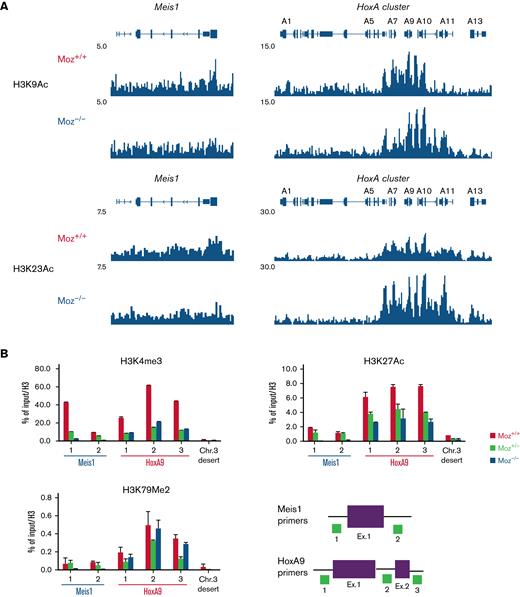
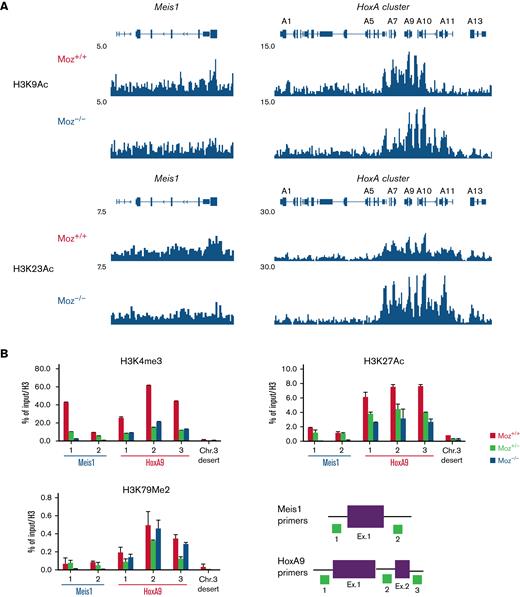
Endogenous MOZ is required for the maintenance of an active chromatin landscape at target gene loci
To reveal the molecular mechanisms underlying the involvement of endogenous MOZ in fusion gene induction of target gene expression, we analyzed the acetylation of H3K9 and H3K23 (target sites for acetylation by MOZ) at the Hoxa cluster and Meis1 loci in MOZ-TIF2 AML cells by ChIP-Seq analysis. In Moz−/−MOZ-TIF2 AML cells, both H3K9 and H3K23 acetylation were reduced at the Meis1 gene locus but not at the Hoxa gene cluster (Figure 3A and supplemental Figure 2A). Other types of active histone marks (H3K4me3, H3K27ac, and H3K79me2) were significantly reduced at the Meis1 gene locus and modestly decreased at the HoxA9 locus in Moz−/−MOZ-TIF2 AML cells (Figure 3B). In Moz−/−MLL-AF9 AML cells, active histone marks were remarkably reduced at the Meis1 locus. In addition, H3K4me3 modification was strikingly decreased at the Hoxa9 locus. Moreover, concentrations of H3K23ac, H3K4me3, and H3K27ac modification were higher in Moz+/+MLL-AF9 AML cells than in Moz+/− and Moz−/−MLL-AF9 AML cells at the Hoxa9/Meis1 loci (supplemental Figure 2B).
Endogenous MOZ is required for active histone modifications at the Meis1 locus.Moz+/+, Moz+/−, or Moz−/−MOZ-TIF2 and MLL-AF9 AML cells were fixed with formalin, and ChIP-Seq or ChIP-qPCR assays were performed. (A) ChIP-Seq analysis of Histone H3 K9 and K23 acetylation at the Hoxa cluster and Meis1 loci in Moz+/+ and Moz−/− MOZ-TIF2 AML cells. (B) Active histone modifications at the Meis1, Hoxa9 loci, and a mouse chromosome 3 desert (Chr. 3 desert) (negative control) loci in MOZ-TIF2 AML cells. Histone modifications at Meis1, Hoxa9 loci, and a Chr. 3 desert loci were measured by quantitative PCR analysis. Representative results of ChIP assays are shown. Seven independent experiments were conducted in MOZ-TIF2 AML cells. Primer sets used to amplify the Meis1 and Hoxa9 loci are indicated at the bottom. Levels of each histone modification were normalized to input DNA and total histone H3 concentrations. Error bars represent mean ± SD.
Endogenous MOZ is required for active histone modifications at the Meis1 locus.Moz+/+, Moz+/−, or Moz−/−MOZ-TIF2 and MLL-AF9 AML cells were fixed with formalin, and ChIP-Seq or ChIP-qPCR assays were performed. (A) ChIP-Seq analysis of Histone H3 K9 and K23 acetylation at the Hoxa cluster and Meis1 loci in Moz+/+ and Moz−/− MOZ-TIF2 AML cells. (B) Active histone modifications at the Meis1, Hoxa9 loci, and a mouse chromosome 3 desert (Chr. 3 desert) (negative control) loci in MOZ-TIF2 AML cells. Histone modifications at Meis1, Hoxa9 loci, and a Chr. 3 desert loci were measured by quantitative PCR analysis. Representative results of ChIP assays are shown. Seven independent experiments were conducted in MOZ-TIF2 AML cells. Primer sets used to amplify the Meis1 and Hoxa9 loci are indicated at the bottom. Levels of each histone modification were normalized to input DNA and total histone H3 concentrations. Error bars represent mean ± SD.
These data suggest that endogenous MOZ is essential for maintaining an active chromatin landscape at the Meis1 gene locus in MOZ-TIF2 and MLL-AF9 AML cells.
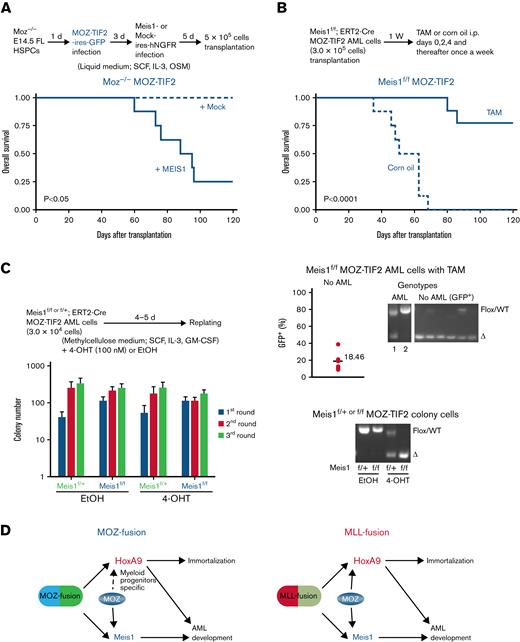
MEIS1 is a critical factor for MOZ fusion-induced AML development but not for immortalization
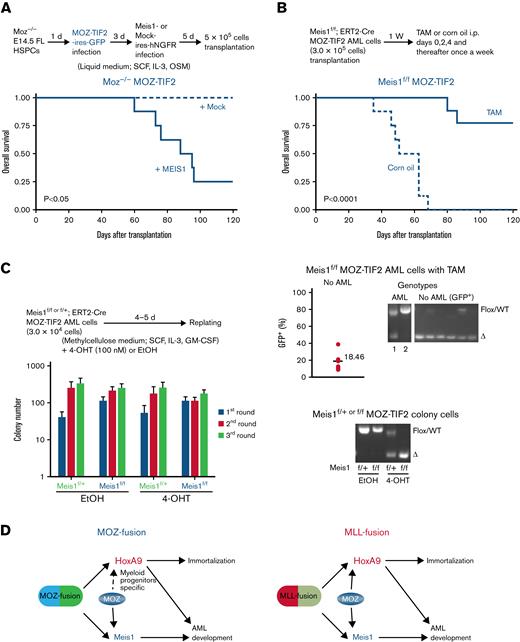
To confirm whether the failure to develop AML in Moz−/−MOZ fusion AML cells was because of an inability to induce aberrant Meis1 expression, we investigated the role of MEIS1 in MOZ fusion-mediated AML development. Both the MOZ-TIF2 fusion and human MEIS1 genes were introduced into Moz−/− HSPCs, and these cells were then transplanted into recipient mice. Expression of the endogenous Meis1 and ectopic MEIS1 genes were significantly higher in HSPCs transduced with MOZ-TIF2 + MEIS1 than those transduced with MOZ-TIF2 + mock vector (supplemental Figure 3A). Most MOZ-TIF2–transduced Moz−/− cells coexpressing the MEIS1 gene caused AML development in recipient mice (Figure 4A). In Moz+/−MOZ-TIF2 AML cells, MEIS1 overexpression did not affect the onset of AML development in recipient mice, similar to that in mice transplanted with Moz+/−MOZ-TIF2 AML cells transduced with the empty vector (Mock control) (supplemental Figure 3B). MEIS1–overexpressing Moz−/−MOZ-TIF2 AML cells expressed myeloid markers, such as Mac-1, Gr-1, and M-CSFR (supplemental Figure 3C). Further, recipient mice transplanted with Moz+/− or Moz−/− HSPCs transduced with MEIS1 did not develop AML, as shown in previous reports34-36 (supplemental Figure 3D).
MEIS1 is essential for MOZ fusion-mediated AML development but not for immortalization. (A) AML development of Moz−/− cells bearing MOZ fusion and MEIS1 genes. The experimental scheme is shown at the top. Moz−/− FL lineage− HSPCs were infected with the MOZ-TIF2 gene, followed by the MEIS1 gene or empty vector (Mock), and cultured in a liquid medium. After 5 days, these cells (5 × 105) were transplanted into recipient mice, and their survival was analyzed (n = 5 to 8 per group). Survival was compared using the log-rank test. (B-C) Conditional deletion of the Meis1 gene in MOZ-TIF2 AML cells. (B) Survival of Meis1-deleted MOZ-TIF2 AML cells. The experimental scheme is shown at the top; Meis1f/fCre-ERT2 MOZ-TIF2 AML cells were transplanted into recipient mice, and then tamoxifen (TAM; 80 mg/Kg) or the same dose of corn oil, administered intraperitoneally (n = 8 to 9 per group). Differences in survival were compared using the log-rank test. The lower right panel shows PCR analysis of Meis1 genotypes in BM samples or BM GFP+ cells from mice that did and did not develop AML following transplantation with Meis1f/fCre-ERT2 MOZ-TIF2 AML cells and treatment with TAM. The lower left graph shows the GFP+ cell population in the BM of recipient mice in which no AML development was observed 120 days after transplantation with Meis1f/fCre-ERT2 MOZ-TIF2 AML cells and treatment with TAM. Values indicate the mean numbers of GFP+ cells in the BM. (C) Colony formation by Meis1-deleted MOZ-TIF2 AML cells. After replating 3 times, Meis1f/+ or Meis1f/fCre-ERT2 MOZ-TIF2 AML cells were treated with 4-hydroxy TAM (4-OHT; 100 nM) or the same dose of ethanol (EtOH) and then serially cultured in methylcellulose medium 3 times. The mean number of colonies was calculated from the results of 3 independent experiments. The lower panel shows genotypes of colonies of Meis1f/+ and Meis1f/fCre-ERT2 MOZ-TIF2 AML cells treated with EtOH or 4-OHT. (D) Summary of the findings of this study.
MEIS1 is essential for MOZ fusion-mediated AML development but not for immortalization. (A) AML development of Moz−/− cells bearing MOZ fusion and MEIS1 genes. The experimental scheme is shown at the top. Moz−/− FL lineage− HSPCs were infected with the MOZ-TIF2 gene, followed by the MEIS1 gene or empty vector (Mock), and cultured in a liquid medium. After 5 days, these cells (5 × 105) were transplanted into recipient mice, and their survival was analyzed (n = 5 to 8 per group). Survival was compared using the log-rank test. (B-C) Conditional deletion of the Meis1 gene in MOZ-TIF2 AML cells. (B) Survival of Meis1-deleted MOZ-TIF2 AML cells. The experimental scheme is shown at the top; Meis1f/fCre-ERT2 MOZ-TIF2 AML cells were transplanted into recipient mice, and then tamoxifen (TAM; 80 mg/Kg) or the same dose of corn oil, administered intraperitoneally (n = 8 to 9 per group). Differences in survival were compared using the log-rank test. The lower right panel shows PCR analysis of Meis1 genotypes in BM samples or BM GFP+ cells from mice that did and did not develop AML following transplantation with Meis1f/fCre-ERT2 MOZ-TIF2 AML cells and treatment with TAM. The lower left graph shows the GFP+ cell population in the BM of recipient mice in which no AML development was observed 120 days after transplantation with Meis1f/fCre-ERT2 MOZ-TIF2 AML cells and treatment with TAM. Values indicate the mean numbers of GFP+ cells in the BM. (C) Colony formation by Meis1-deleted MOZ-TIF2 AML cells. After replating 3 times, Meis1f/+ or Meis1f/fCre-ERT2 MOZ-TIF2 AML cells were treated with 4-hydroxy TAM (4-OHT; 100 nM) or the same dose of ethanol (EtOH) and then serially cultured in methylcellulose medium 3 times. The mean number of colonies was calculated from the results of 3 independent experiments. The lower panel shows genotypes of colonies of Meis1f/+ and Meis1f/fCre-ERT2 MOZ-TIF2 AML cells treated with EtOH or 4-OHT. (D) Summary of the findings of this study.
We also checked the requirement for MEIS1 in MOZ fusion-induced AML development using a Meis1 conditional deficient mouse. Meis1f/+ or Meis1f/fCre-ERT2 MOZ-TIF2 AML cells were transplanted into recipient mice. One week after transplantation, recipient mice were intraperitoneally administered corn oil or TAM to induce Cre recombinase-mediated excision of the Meis1 allele, resulting in alteration of Meis1 genetic status in MOZ-TIF2 AML cells from Meis1 floxed (Meis1f/f) to Meis1 deficient (Meis1Δ/Δ). With transplantation of Meis1f/fCre-ERT2 MOZ-TIF2 AML cells, AML development was significantly delayed or absent in recipient mice injected with TAM compared with those administered with corn oil (Figure 4B). Recipient mice transplanted with Meis1f/fCre-ERT2 MOZ-TIF2 AML cells and treated with TAM showed no sign of AML development and had only 18.46% GFP+ cells in their BM at 120 days after transplantation (Figure 4B).
We also checked the Meis1 genotypes of BM cells from recipient mice transplanted with Meis1f/fCre-ERT2 MOZ-TIF2 AML cells and injected with TAM. GFP+ BM cells from the recipient mice showed no sign of AML development, and only Meis1Δ/ΔMOZ-TIF2 AML cells were detected. In 1 mouse that developed AML, the BM contained Meis1Δ/Δ and Meis1f/fMOZ-TIF2 AML cells. By contrast, only Meis1f/fMOZ-TIF2 AML cells were detected in another mouse, indicating that MOZ-TIF2 AML cells that escaped deletion of the Meis1 gene were mainly responsible for AML development. GFP+ BM cells from the recipient mice showed no sign of AML development, and only Meis1Δ/ΔMOZ-TIF2 AML cells were detected except in 1 sample (Figure 4B). Furthermore, AML development was observed in recipient mice transplanted with Meis1f/+Cre-ERT2 MOZ fusion AML cells treated with either corn oil or TAM (supplemental Figure 3E). These results indicate that MEIS1 is essential for MOZ fusion-mediated AML development.
Next, we analyzed the effects of Meis1 deletion on the maintenance of proliferation or immortalization of MOZ fusion AML cells. Meis1f/fMOZ-TIF2 AML cells were treated with 4-OHT to induce Cre recombinase activation, resulting in the conversion of the Meis1f/+ genotype to Meis1-heterozygous–deleted (Meis1Δ/+) and of Meis1f/f to Meis1-deleted (Meis1Δ/Δ). Meis1 deletion did not affect the proliferation and immortalization activity of MOZ-TIF2 AML cells in vitro (Figure 4C). We confirmed that the Meis1 gene was deleted entirely in Meis1f/fCre-ERT2 MOZ-TIF2 AML cells treated with 4-OHT. This result suggests that MEIS1 is unnecessary for the maintenance of MOZ fusion AML cells to maintain proliferation and immortalization. These findings provide further evidence that Moz−/−MOZ fusion cells failed to promote AML development because aberrant Meis1 expression could not be induced.
Discussion
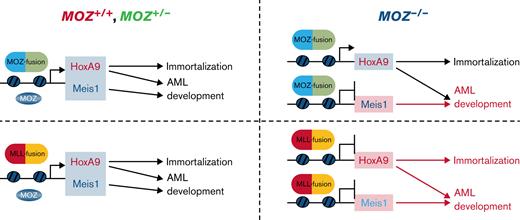
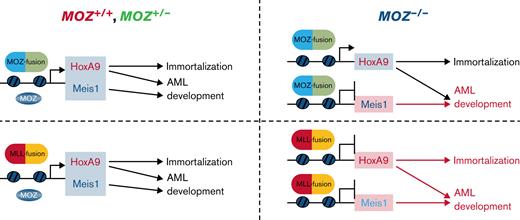
This study revealed that endogenous MOZ was essential for AML development and aberrant Meis1 expression induced by the MOZ/MLL fusion and for immortalization and robust Hoxa9 expression caused by MLL fusion genes (Figure 4D). Furthermore, ChIP analysis showed that endogenous MOZ was critical for maintaining an active chromatin landscape at the Meis1 gene locus. Also, conditional deletion of Meis1 in MOZ fusion AML cells resulted in the absence of AML development or prolonged survival of recipient mice, while Meis1 overexpression rescued AML development from Moz-deficient cells harboring a MOZ fusion.
Previous reports have shown that MOZ and MOZ-related factor (MORF), which share significant sequence similarity, preferentially bind to H3K9ac and H3K14ac sites through their double PHD fingers.37,38 Furthermore, the MOZ transcriptional complex components, BRPF1 and ING5, colocalize at H3K4me3-enriched Hoxa loci via the ING5 PHD finger.39 Consistently, expression of Hoxa9 and Meis1 genes is decreased in Brpf1−/− HSPCs40 and MOZ fusion AML cells with Brpf1 knocked down.41 Moreover, the binding of MOZ fusion proteins at the Hoxa9 locus is reduced in Brpf1 knockdown MOZ fusion AML cells.41 These results suggest that recruitment of a MOZ fusion complex at a target gene locus requires interaction between active histone modifications and the chromatin reader domains of the MOZ transcriptional complex. In addition, H3K4me3 and H3K27ac in Moz+/− and Moz−/− MOZ-TIF2 AML cells were modestly reduced at the HoxA9 locus (Figure 3B). Considered together with our findings that HoxA9 expression is decreased in Moz-deficient MLL fusion-expressing cells but not in Moz-deficient MOZ-TIF2 AML cells. These findings suggest that a certain concentration of active histone modifications may be required to recruit the MOZ complex at a target gene locus and to induce aberrant HoxA9 expression in MOZ-TIF2 AML cells.
Endogenous Moz was also required for transformation and AML development by MLL-AF10, and for proliferation and colony formation activity by MLL-AF9, through aberrant Hoxa9/Meis1 expression. Also, the onset of MLL-AF10–induced leukemia was slower in mice transplanted with Moz+/−MLL-AF10 cells than in those transplanted with Moz+/+MLL-AF10 cells, suggesting that the effect of endogenous Moz in MLL fusion-mediated leukemia is dose-dependent.
Although replating of 1 × 105 cells Moz−/− MLL-AF9 AML cells resulted in successive colony formation activity, there was a significantly decreased cell number of colonies (Figure 1A). By contrast, MLL-AF9 cells derived from Moz−/− KSLs and HSPCs did not show continuous colony formation when replated at 3 × 104 cells (Figure 2D and supplemental Figure 1). These results suggest that a large number of cells may be required for the maintenance of continuous colony formation activity of Moz−/−MLL-AF9 cells in culture media because of a need for intercellular support, for example, via paracrine (cytokine or chemokine) signaling. While HoxA9 expression was similar among MLL-AF9–expressing KSLs with different Moz genotypes (Figure 2D), it was significantly decreased in Moz−/− MLL-AF9 AML cells (Figure 2A). These data suggest that endogenous Moz is required to maintain aberrant HoxA9 expression in MLL fusion AML cells. Taken together, our findings indicate that endogenous Moz is also essential for MLL fusion leukemia development.
Previous studies have demonstrated that H3K9 sites at the HOXA/MEIS1 loci are hyperacetylated in MLL fusion AML cells.42 Indeed, as part of an MLL fusion complex, the Yaf9, ENL, AF9, Taf14 and Sas5 (YEATS) domain of eleven nineteen leukemia (ENL) binds to acetylated H3K9 and is required for AML development.43-45 H3K9ac and other active histone marks were significantly decreased at the Meis1 locus in Moz−/−MLL-AF9 AML cells. Thus, it is conceivable that endogenous MOZ is required for MLL fusion-mediated AML development to facilitate H3K9 acetylation at the MEIS1 locus. By contrast, although Hoxa9 expression was remarkably decreased in Moz−/−MLL fusion AML cells, modification of H3K4me3 was only reduced at the Hoxa9 locus in Moz−/− MLL-AF9 AML cells. H3K4me3 modification by MLL1 is essential for Hoxa9 expression46,47; hence, H3K4me3 appears to be critical for aberrant Hoxa9 expression in MLL-AF9 AML cells.
Previous reports showed that MEIS1 is essential for MLL fusion-induced transformation of HSPCs and Hoxa9 expression.31,36 In this study, we demonstrate that Meis1 is required for AML development (Figure 4A,B) but not for the colony-forming activity or aberrant Hoxa9 expression in MOZ-TIF2 AML cells (Figures 1B, 2A, and 4C). These data suggest that Meis1 is not critical for MOZ fusion-mediated maintenance of colony-forming activity or Hoxa9 expression. A previous report showed that both Meis1- and Moz-deficient cells exhibit increased expression of the tumor suppressor p16Ink4a and reactive oxygen species (ROS).21,22,31,48 Introduction of MOZ-TIF2 into HSPCs represses p16Ink4a expression.49 These data suggest that MOZ-TIF2 can suppress transcription induction of the p16Ink4a gene and ROS production in Meis1-deleted MOZ-TIF2 AML cells, resulting in maintenance of their colony-forming activity.
In the present study, the role of MOZ in the maintenance of AML cells was not addressed. Conditional deletion of Moz in adult hematopoietic cells only engenders defects in the maintenance of quiescent HSCs and lymphocytes,50 in contrast to the severe defects observed in fetal hematopoiesis in Moz−/− mice.13 Further, a MOZ/MORF inhibitor has recently been shown to block Eμ-Myc–driven B-lymphoma development and induction of p16Ink4a expression.51 These results suggest that the role of MOZ in the maintenance of AML cells may differ from its function in AML development. Additional analyses are now required, using a Moz conditional deletion system or an inhibitor, to further explore the role of MOZ in effects on target gene expression in maintaining AML.
Acknowledgments
The authors would like to thank Mika Shino and Noriko Aikawa for their support in conducting animal experiments. The authors are also grateful to all members of our laboratory for their critical discussion and helpful suggestions.
This work was partly supported by Grants-in-Aid for Young Scientists (B), Grant number 23791094 (T.K.). In addition, I.K. was funded by Grants-in-Aid for Scientific Research (B), Grant number 19390268, and Grants-in-Aid for Scientific Research on Innovative Areas, Grant number 22130006.
Authorship
Contribution: T.K., Y.O., K.Y., and I.K. designed the experiments; T.K., Y.O., K.Y., and Y.A. performed experiments; R.G. and T.N. provided materials; and T.K. and I.K. analyzed and interpreted the data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Issay Kitabayashi, Division of Hematological Malignancy, National Cancer Center Research Institute, 5-1-1 Tsukiji, Chuo-ku, Tokyo, 104-0045, Japan; e-mail: ikitabay@ncc.go.jp.
References
Author notes
ChiP-Seq data have been deposited at Gene Expression Omnibus (GSE208741). For original data, please contact iktabay@ncc.go.jp.
The full-text version of this article contains a data supplement.

![Endogenous MOZ is essential for MOZ/MLL fusion-induced leukemia development. (A) Serial colony replating assays were performed in Moz+/+, Moz+/−, and Moz−/− HSPCs bearing MOZ-TIF2, MLL-AF9, MLL-AF10, or HOXA9. A scheme of the experimental workflow is shown at the top. First, Moz+/+, Moz+/−, or Moz−/− FL lineage− HSPCs (2 × 105 cells) were transduced with MOZ-TIF2, MLL-AF10, MLL-AF9, or HOXA9 and cultured in a liquid medium. Subsequently, GFP+ cells (5 × 104) were sorted and cultured in a methylcellulose medium. Every 4 to 5 days, colony and cell numbers (× 105 cells) were counted, and then 1 × 105 cells were serially replated 5 times. Mean values were calculated from 3 to 4 independent experiments. Error bars represent mean ± standard deviation (SD). (B) Overall survival of recipient mice transplanted with Moz+/+, Moz+/−, or Moz−/− HSPCs (2 × 105) bearing MOZ-TIF2, MLL-AF10, or HOXA9 and MEIS1 (MOZ-TIF2, Moz+/+ and Moz−/− [n = 6], Moz+/− [n = 12]; MLL-AF10, Moz+/+ [n = 6], Moz+/− [n = 10], Moz−/− [n = 4]; HOXA9 + MEIS1, [n = 6]). Differences in survival were compared using the log-rank test; ∗P < .05, ∗∗P < .01, and ∗∗∗P < .005.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/6/19/10.1182_bloodadvances.2020003490/11/m_blooda_adv-2020-003490-gr1.jpeg?Expires=1768176576&Signature=M~rCAXZLLypfekJ~StTTyePSqUUCZy7ghDhWHuD2VHG8P4dtPr8PxxiFF3BJoblc5xdCJ~Eh3gykv4IG~oJV2~aYAELPiABdwQ6mU~tDjMIhH78TFEq2hXJk2m2krP8YLQ5eMwXfaiEF8q411rhvw~S9kjXgRbIy0aKzjjBeJCkUKL4V80DvG82ORBpn1-XJlKQX5eqmOiWp0yyucJpRdk5XFZvbI3MV5KZiYOets7VXaYWbA~iSXwB6msHbstoWCKtb2bgR1DzwKDWZhDRCLETWyIkYawlZhzUFxQa3xFx~dbuXuLIxaiwPbb5Sy238J9Z1K4byccrAKq8ivtZ~8Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Endogenous MOZ is essential for MOZ/MLL fusion-induced leukemia development. (A) Serial colony replating assays were performed in Moz+/+, Moz+/−, and Moz−/− HSPCs bearing MOZ-TIF2, MLL-AF9, MLL-AF10, or HOXA9. A scheme of the experimental workflow is shown at the top. First, Moz+/+, Moz+/−, or Moz−/− FL lineage− HSPCs (2 × 105 cells) were transduced with MOZ-TIF2, MLL-AF10, MLL-AF9, or HOXA9 and cultured in a liquid medium. Subsequently, GFP+ cells (5 × 104) were sorted and cultured in a methylcellulose medium. Every 4 to 5 days, colony and cell numbers (× 105 cells) were counted, and then 1 × 105 cells were serially replated 5 times. Mean values were calculated from 3 to 4 independent experiments. Error bars represent mean ± standard deviation (SD). (B) Overall survival of recipient mice transplanted with Moz+/+, Moz+/−, or Moz−/− HSPCs (2 × 105) bearing MOZ-TIF2, MLL-AF10, or HOXA9 and MEIS1 (MOZ-TIF2, Moz+/+ and Moz−/− [n = 6], Moz+/− [n = 12]; MLL-AF10, Moz+/+ [n = 6], Moz+/− [n = 10], Moz−/− [n = 4]; HOXA9 + MEIS1, [n = 6]). Differences in survival were compared using the log-rank test; ∗P < .05, ∗∗P < .01, and ∗∗∗P < .005.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/6/19/10.1182_bloodadvances.2020003490/11/m_blooda_adv-2020-003490-gr1.jpeg?Expires=1768196195&Signature=UYLFkB2khvknbL7CirejjGxfE2Aik3T3rvAxX4Sd3KrVTuwv3YTvMaFNtCuBHlh1c9zXnVYvmbrdOMegdvYTpwxpg4ICcPZIkAqe~LM6WGKXFVEd4u9I6bajJP~~RQuYoJQDLsDSrXe975pDcpSXIbaIXtD0wT3YKYFCy7e9zGcIwQLRejlWbpgBN7LGNOgVvyHKCZ06OQMY8G9EjTzSGr63DMSrVXxjNDzUe8CjMvayDCYil3vu2SCId3AU6fVOX~OFKd~IP4ASZPet~72OJ6r1SnJNwkCAaKiZmg8jlbUNsHjYW4HbuHO2mOqw4wyoSoenOkrRPXYhyLrUu4d2aQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)