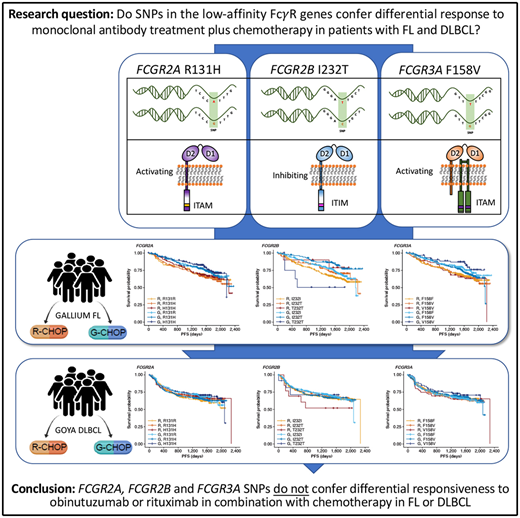
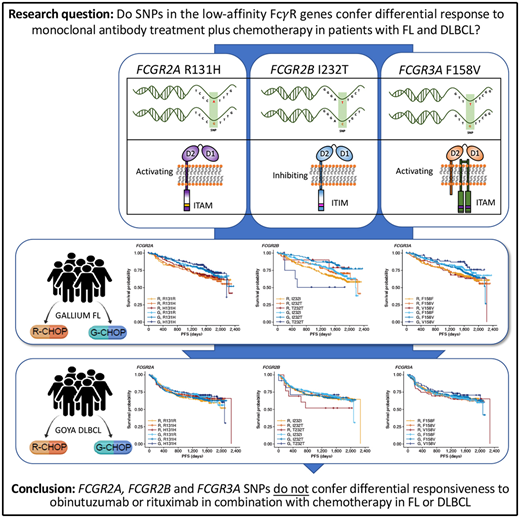
Key Points
FcγR genotype is not associated with response to obinutuzumab/rituximab combined with chemotherapy in untreated patients with FL or DLBCL.
Abstract
Single-nucleotide polymorphisms (SNPs) have been shown to influence Fcγ receptor (FcγR) affinity and activity, but their effect on treatment response is unclear. We assessed their importance in the efficacy of obinutuzumab or rituximab combined with chemotherapy in untreated advanced follicular lymphoma (FL) and diffuse large B-cell lymphoma (DLBCL) in the GALLIUM (www.clinicaltrials.gov #NCT01332968) and GOYA (#NCT01287741) trials, respectively. Genomic DNA was extracted from patients enrolled in GALLIUM (n = 1202) and GOYA (n = 1418). Key germline SNPs, FCGR2A R131H (rs1801274), FCGR3A F158V (rs396991), and FCGR2B I232T (rs1050501), were genotyped and assessed for their impact on investigator-assessed progression-free survival (PFS). In both cohorts there was no prognostic effect of FCGR2A or FCGR3A. In FL, FCGR2B was associated with favorable PFS in univariate and multivariate analyses comparing I232T with I232I, with a more modest association for rituximab-treated (univariate: hazard ratio [HR], 0.78; 95% confidence interval [CI], 0.54-1.14; P = .21) vs obinutuzumab-treated patients (HR, 0.56; 95% CI, 0.34-0.91; P = .02). Comparing T232T with I232I, an association was found for obinutuzumab (univariate: HR, 2.76; 95% CI, 1.02-7.5; P = .0459). Neither observation retained significance after multiple-test adjustment. FCGR2B was associated with poorer PFS in multivariate analyses comparing T232T with I232I in rituximab- but not obinutuzumab-treated patients with DLBCL (HR, 4.40; 95% CI, 1.71-11.32; P = .002; multiple-test–adjusted P = .03); however, this genotype was rare (n = 13). This study shows that FcγR genotype is not associated with response to rituximab/obinutuzumab plus chemotherapy in treatment-naive patients with advanced FL or DLBCL.
Introduction
Obinutuzumab is a humanized, glycoengineered, type II anti-CD20 antibody with enhanced direct cell killing and antibody-dependent cellular phagocytosis and cytotoxicity (ADCC) compared with type I anti-CD20 antibodies, such as rituximab.1-5 Obinutuzumab has been approved for the first-line treatment of chronic lymphocytic leukemia, in combination with chlorambucil; follicular lymphoma (FL), in combination with chemotherapy followed by obinutuzumab maintenance, and in combination with bendamustine followed by obinutuzumab maintenance for relapsed/rituximab-refractory FL.6-8
Fcγ receptors (FcγRs), of which there are 6 in humans, encoded by the FCGR1A, FCGR2A, FCGR2B, FCGR2C, FCGR3A, and FCGR3B genes, are critical mediators of anti-CD20 monoclonal antibody (mAb)–mediated cell killing.9 Activating FcγR (FcγRI, FcγRIIa, FcγRIIc, and FcγRIIIa) deliver immunoreceptor tyrosine-based activation motif–mediated active signaling, capable of driving cytotoxic granule release (leading to ADCC) and/or antibody-dependent cellular phagocytosis from various cellular effectors, including natural killer cells, macrophages, monocytes, and neutrophils. In contrast, FcγRIIb, the sole inhibitory FcγR, impairs this signaling through Src homology 2 domain–containing inositol phosphatase- and Src homology 2–containing tyrosine phosphatase-1–mediated phosphatase activation.10
Importantly, the FcγR genes are highly polymorphic, exhibiting multiple single-nucleotide polymorphism (SNP) and copy number variation events (reviewed in Hargreaves et al11). The R131H and F158V SNPs in FCGR2A and FCGR3A, respectively, have been shown to alter affinity for different subclasses of immunoglobulin G,12,13 which, in the case of the high-affinity FCGR3A 158V allele, leads to clear enhancement of natural killer cell–mediated ADCC at lower mAb concentrations.14 Other SNPs, such as FCGR2B I232T appear to affect receptor activity by impairing inhibitory signaling,15 but have yet to be definitively assessed in the context of anticancer mAb activity, due to their low prevalence.11
Although the impact of these SNPs on FcγR function has been demonstrated in vitro,16 their influence on overall patient response is less clear. Initial retrospective studies, based on low numbers of patients, suggested that the FCGR3A and FCGR2A genotypes affect the efficacy of rituximab. Cartron et al showed inferior response rates for the FCGR3A 158F genotype compared with 158V carriers in 49 patients with FL treated with rituximab.17 In a retrospective cohort, Weng and Levy demonstrated improved response rates and more durable remissions in 87 rituximab-treated patients with FL with the FCGR3A V158V and FCGR2A H131H genotypes, compared with F and R carriers, respectively.18 Finally, in a small cohort of rituximab-treated patients with FL or mantle cell lymphoma, Ghielmini et al reported that patients with FCGR3A V158V exhibited superior event-free survival among patients with FL.19 More recent analyses have failed to identify an impact of FCGR2A and FCGR3A genotypes in rituximab-treated patients with FL. The RESORT (www.clinicaltrials.gov; #NCT00075946) and PRIMA (#NCT00140582) trials and a retrospective cohort of newly diagnosed patients all failed to show an influence of the FCGR2A R131H and FCGR3A F158V genotypes on patient outcome after rituximab and chemotherapy.20-22 Similarly, data from previously published studies demonstrated that FCGR2A and FCGR3A genotype status did not correlate with treatment response in patients with diffuse large B-cell lymphoma (DLBCL).23-25 The inconsistent effects reported by these multiple cohort studies are likely due to their limited size, the heterogeneity of the cohorts, and the variation in the use of rituximab (monotherapy vs combination therapy, first vs second/subsequent line, among others).17,19-22,26-30
To overcome these limitations and to provide definitive results of the impact of these SNPs on anti-CD20 mAb therapy, large, prospective, uniform studies using carefully designed SNP assays are needed. Accordingly, we retrospectively analyzed data from 2 recently completed, large (>1000-patient) phase 3 studies, to assess the potential impact of these FcγR genotypes on the efficacy of obinutuzumab or rituximab in combination with chemotherapy in patients with previously untreated advanced FL (GALLIUM; www.clinicaltrials.gov/ #NCT01332968)6 or DLBCL (GOYA; #NCT01287741).31
Methods
Study design, patients, and treatments
GALLIUM and GOYA were phase 3, multicenter, open-label, randomized studies. Full details of their design and primary end points have been published.6,31 In GALLIUM, patients with previously untreated advanced-stage FL were randomized to receive either obinutuzumab- or rituximab-based immunochemotherapy, followed by maintenance with the same antibody in the responders.6 The chemotherapy arm (cyclophosphamide, doxorubicin, vincristine, and prednisone [CHOP]; cyclophosphamide, vincristine, and prednisone [CVP], or bendamustine) was selected by each study center (33.2%, 9.7%, and 57.1%, respectively). In GOYA, previously untreated patients with DLBCL were randomly assigned to receive eight 21-day cycles of obinutuzumab or rituximab plus 6 or 8 cycles of CHOP.31 The number of CHOP cycles for both arms was agreed on in advance with each study site. If only 6 CHOP cycles were administered, the antibody was administered as monotherapy during cycles 7 and 8. The primary end point for both studies was investigator-assessed progression-free survival (PFS).6,31 Both studies were conducted in accordance with the Declaration of Helsinki, Good Clinical Practice guidelines, and all applicable local laws and regulations. The study protocols and amendments and other study-related materials were approved by institutional review boards or ethics committees at the participating centers. Written informed consent was provided by all patients.
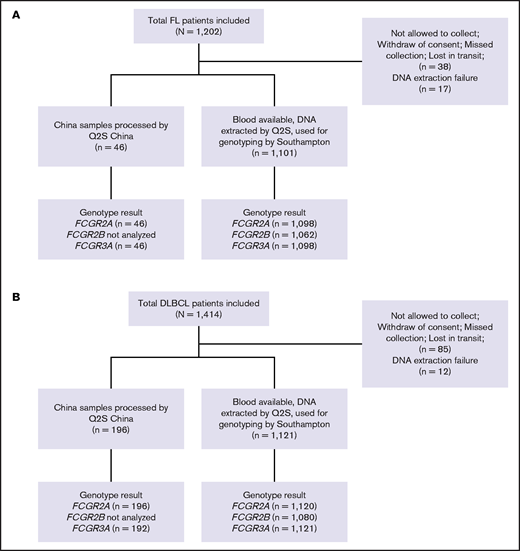
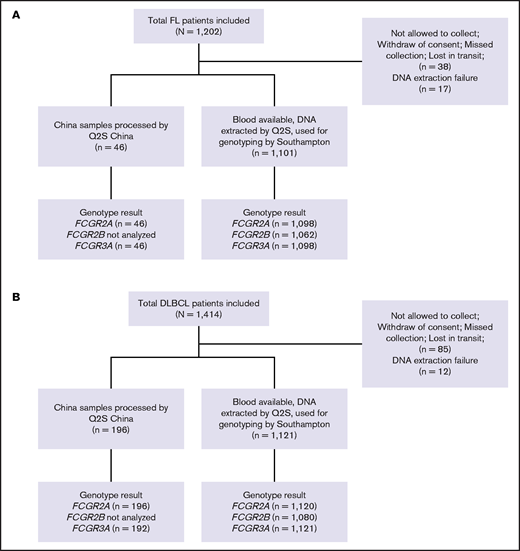
A total of 1202 and 1418 patients enrolled in the GALLIUM and GOYA studies, respectively, were included in this study, with updated clinical data available for 1202 and 1414 patients, respectively.32,33 Of these, 55 and 97 patients could not be genotyped because of lack of consent, lack of blood samples, or DNA extraction failure (Figure 1). Median duration of follow-up was 57.4 months (range, 0.0-77.4 months) in GALLIUM and 47.7 months (range, 0.1-78.2 months) in GOYA.
CONSORT diagrams. (A) Patients with FL in the GALLIUM trial and (B) patients with DLBCL in the GOYA trial. Q2S, Q2Solutions.
CONSORT diagrams. (A) Patients with FL in the GALLIUM trial and (B) patients with DLBCL in the GOYA trial. Q2S, Q2Solutions.
Sample preparation, FcγR genotyping, and molecular confirmation
Genomic DNA was extracted from whole blood from 2222 trial patients by UK Q2Solutions with Autopure, using Gentra Purgene Kits (Qiagen). Nucleic acid extraction and genotyping were performed as previously described.34 DNA was quantified with UV absorbance and quality was controlled with 260/280 optical density ratio measurements prior to dilution to the appropriate concentration in distilled water. For genotyping, sample triplicates were prepared using the CAS-1200 polymerase chain reaction (PCR) setup robot (Corbett Life Science, Qiagen); amplification and allelic discrimination were performed with a Corbett Rotor-Gene 6000 (Corbett Life Science) and Rotor-Gene Q series Software 2.0.2 (Build 4), respectively. The SNPs, FCGR2A R131H (rs1801274), FCGR3A F158V (rs396991), and FCGR2B I232T (rs1050501), were genotyped with commercially available (C_9077561_20 and C_25815666_10) and custom-designed TaqMan discrimination assays (Life Technologies, Paisley, United Kingdom), according to the manufacturer’s instructions. Quality control samples were included in each batch as follows: known FCGR2A, FCGR3A and FCGR2B wild-type homozygotes, heterozygotes, homozygote variants (Coriell Cell Repository, Camden, NJ), and nontemplate negative control. For a small number of Chinese patients (n = 242), DNA extraction and subsequent FCG2A and FCGR3A genotyping were performed by Q2Solutions in China (ViiA 7 Real-Time PCR System; Applied Biosystems-Life Technologies, Carlsbad, CA).
Sanger sequencing using the FcγR-gene–specific PCR primers (supplemental Table 1) was performed for samples that failed TaqMan assays or as a confirmatory step. PCR products were purified using the MinElute 96 UF PCR purification kit (Qiagen) or the ExoSAP method (ExoSAP-IT protocol 2000; USB Corporation, Cleveland, OH), and SNPs were confirmed by Sanger sequencing (Source BioScience, Cambridge, United Kingdom), as previously described.34 Approximately 10% of the samples were randomly selected for confirmatory sequencing. As they can be technically challenging to identify, we also confirmed all FCGR2B non-II genotypes using the same Sanger approach. Genotyping by Taqman assay for FCGR2A, FCGR2B, and FCGR3A failed in 1% to 2%, 4% to 5%, and 13% to 16% of samples, respectively. FCGR2A and FCGR3A genotypes by TaqMan assay were confirmed in 100% and 99% of cases by Sanger sequencing, respectively. For FCGR2B, the II genotype was confirmed in 100% of cases, and the non-II genotypes were confirmed in 85% to 89% of cases. The final FCGR2A and FCGR3A genotypes were reported as per the TaqMan assay result, except for those cases that failed genotyping, where the Sanger result was utilized. The final FCGR2B genotype was reported as the TaqMan assay result where cases were discordant with Sanger sequencing.34
Statistical analysis
For both trials, the effect of SNP genotype on investigator-assessed PFS was evaluated per treatment arm and for the pooled treatment arms. The SNP effect, expressed as a hazard ratio (HR) with 95% confidence interval (CI), was estimated in a univariate Cox proportional hazards regression analysis adjusted for treatment arm and a multivariate Cox regression analysis adjusted for stratification factors (GALLIUM: follicular lymphoma (FL) International Prognostic Index [FLIPI], chemotherapy regimen; GOYA: geographic region, International Prognostic Index [IPI], number of CHOP cycles). In GOYA, adjustments were also made for cell of origin (COO) and BCL2 protein expression, which have been shown to be prognostically significant in DLBCL.31,35 The analysis of pooled data additionally accounted for the treatment arm. SNP effect in the prognostic analysis was assessed by using the Wald test at a 5% level. Multiple test correction was performed with the Benjamini and Hochberg methodology. The Kaplan-Meier method was used to estimate PFS distribution for genotypes in each treatment arm.
Results
Patients and genotyping results
Patient demographic and disease characteristics for the FL and DLBCL patient populations in the GALLIUM and GOYA trials, respectively, are summarized in Table 1. These characteristics, including the prevalence of each FcγR genotype (Table 2), were comparable across trials and across treatment arms. The prevalence of FcγR genotypes by ethnicity in GALLIUM and GOYA are presented in supplemental Table 2. A higher frequency of patients with the FCGR2B 232T allele were observed in the Asian populations of both GALLIUM and GOYA, compared with the White population (supplemental Table 2). Of note, because of missing data for COO and BCL2, the number of samples included in the multivariate Cox regression analyses were ∼50% and 97% of those included in the univariate analyses in GOYA and GALLIUM, respectively.
Prognostic effect of FcγR genotype in patients with FL from the GALLIUM trial
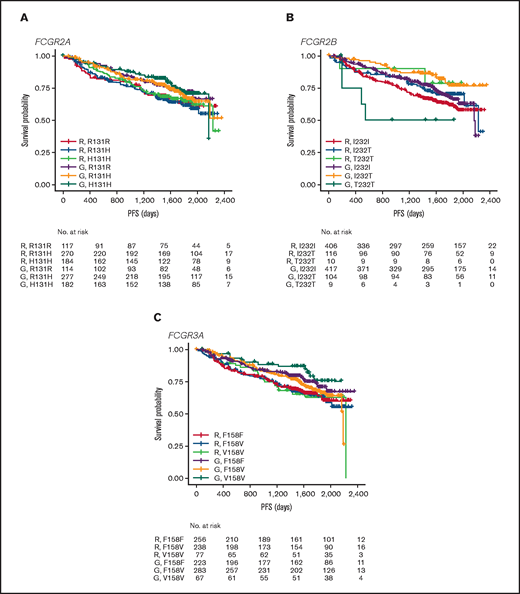
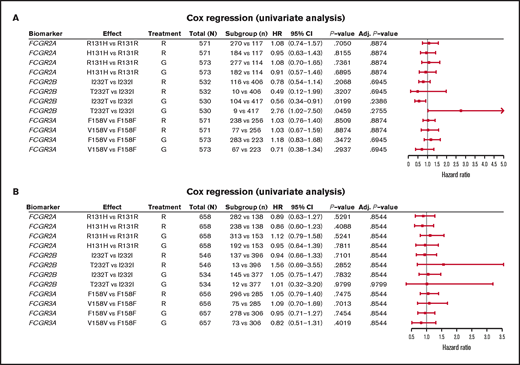
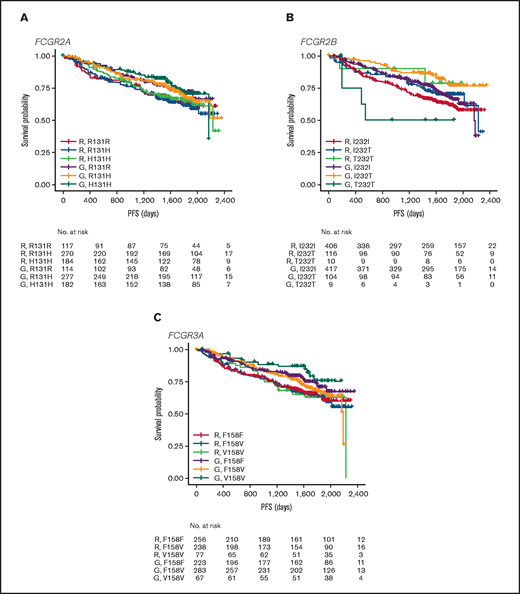
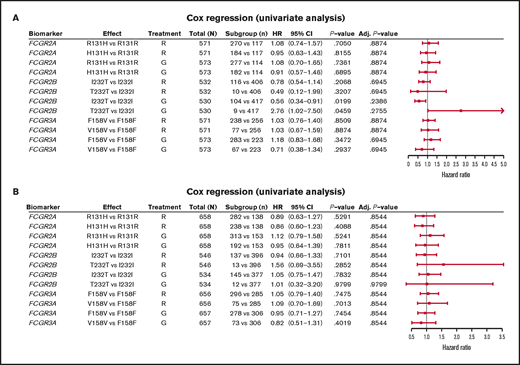
In FL, no significant association was found between PFS and genotypes of either FCGR2A or FCGR3A (Figures 2 and 3; supplemental Figure 1). For obinutuzumab, the PFS curve for patients with FCGR3A V158V was above that for those with F158V and F158F, but this finding is attributable to the statistical variation caused by the lower number of patients with V158V. Only FCGR2B was associated with superior PFS in univariate analyses of pooled treatment arms comparing the I232T with the I232I genotype (HR, 0.69; 95% CI, 0.51-0.93; P = .01), with a more modest association for rituximab-treated patients (HR, 0.78; 95% CI, 0.54-1.14; P = .21) compared with obinutuzumab-treated patients (HR, 0.56; 95% CI, 0.34-0.91; P = .02; Figure 3; supplemental Figure 1). A similar effect was observed in the multivariate analyses (supplemental Figure 1). When comparing the T232T and I232I genotypes, obinutuzumab was associated with a worse prognosis in the univariate analysis (HR, 2.76; 95% CI, 1.02-7.5; P = .0459). However, the sample size and number of events for the T232T genotype were very low at 9 and 4, respectively. Neither observation retained statistical significance after P-value adjustment for multiple comparison.
PFS. Data show PFS in patients with the FCGR2A 131 (A), FCGR2B 232 (B), or FCGR3A 158 (C) SNP genotype with FL treated with rituximab (R) or obinutuzumab (G) plus chemotherapy in the GALLIUM trial.
PFS. Data show PFS in patients with the FCGR2A 131 (A), FCGR2B 232 (B), or FCGR3A 158 (C) SNP genotype with FL treated with rituximab (R) or obinutuzumab (G) plus chemotherapy in the GALLIUM trial.
Forest plot representing survival analysis and the impact of the Fcγ receptor genotypes on PFS per treatment arm. (A) Patients with FL in the GALLIUM trial; (B) patients with DLBCL in the GOYA trial. Time to event was defined by PFS (in days). All analyses were unadjusted and unstratified. Variables for the univariate analysis included biomarker and treatment arm. P-value (for hypothesis testing of whether the biomarker effect was equal to 0) was calculated with the Wald test. Adjusted P-values were corrected for multiple comparisons by the Benjamini and Hochberg method. Adj, adjusted; G, obinutuzumab; R, rituximab.
Forest plot representing survival analysis and the impact of the Fcγ receptor genotypes on PFS per treatment arm. (A) Patients with FL in the GALLIUM trial; (B) patients with DLBCL in the GOYA trial. Time to event was defined by PFS (in days). All analyses were unadjusted and unstratified. Variables for the univariate analysis included biomarker and treatment arm. P-value (for hypothesis testing of whether the biomarker effect was equal to 0) was calculated with the Wald test. Adjusted P-values were corrected for multiple comparisons by the Benjamini and Hochberg method. Adj, adjusted; G, obinutuzumab; R, rituximab.
Prognostic effect of FcγR genotype in patients with DLBCL enrolled in the GOYA trial
In the patients with DLBCL enrolled in GOYA, PFS results similar to those of patients enrolled in GALLIUM were observed (Figure 4). There was no evidence of a univariate prognostic effect for any FcγR genotype (Figures 3 and 4; supplemental Figure 2). However, in the multivariate analysis, there was an association in the rituximab treatment arm with the FCGR2B T232T SNP (HR, 4.40; 95% CI, 1.71-11.32; P = .002; multiple-test adjusted P = .03) compared with the FCGR2B I232I SNP. However, this observation should be interpreted with caution, given the low prevalence of this genotype (n = 6; supplemental Figure 2). The multivariate analysis also showed an initial association in the rituximab treatment arm with the FCGR2A R131H SNP (HR, 0.57; 95% CI, 0.33-0.99; P = .047) compared with the FCGR2A R131R SNP; however, this was not significant after P-value adjustment for multiple tests (adjusted P = .282). There was no association between any of the FCGR genotypes and COO in the GOYA trial (supplemental Table 3), and no significant association of the FcγR genotypes with PFS was observed when stratified by COO (supplemental Table 4).
PFS. Data show PFS according to FCGR2A 131 (A), FCGR2B 232 (B), and FCGR3A 158 (C) SNP genotype for patients with DLBCL treated with rituximab (R) or obinutuzumab (G) plus chemotherapy in the GOYA trial.
PFS. Data show PFS according to FCGR2A 131 (A), FCGR2B 232 (B), and FCGR3A 158 (C) SNP genotype for patients with DLBCL treated with rituximab (R) or obinutuzumab (G) plus chemotherapy in the GOYA trial.
Discussion
We explored the clinical significance of the 3 well-described FcγR SNPs, FCGR2A R131H (rs1801274), FCGR3A F158V (rs396991), and FCGR2B I232T (rs1050501), in 2 large, international trials (GALLIUM6 and GOYA31) comparing the efficacy and safety of obinutuzumab and rituximab in combination with chemotherapy in treatment-naive patients with FL and DLBCL. As our cohort included many more patients than previously published studies, our analyses provided greater statistical power to establish the clinical importance of these FcγR genotypes. After analyzing the FcγR genotype status in 2464 patients with indolent and aggressive lymphoma and available DNA, we identified no clear prognostic impact for any of these FcγR SNPs in patients treated with immunochemotherapy.
As considerable inconsistency remains in the literature regarding the prognostic importance of FcγR SNPs, we performed our analysis on 2464 clinical trial patients. The analysis of GALLIUM and GOYA provided the ideal opportunity to (1) evaluate the importance of the 3 best-understood functional FcγR SNPs in a homogeneous cohort of rituximab-treated patients, and (2) to perform a novel assessment of the impact of FcγR SNPs on obinutuzumab-treated patients with FL or DLBCL. Our findings are definitive in this context and further demonstrate the importance of prospective clinical trials for the accurate evaluation of putative biomarkers. The main strengths of the present study were (1) the cost-effective genotyping approach, which permitted the analysis of a number of candidate SNPs at high sensitivity; (2) the size and homogeneous nature of both cohorts, enabling us to overcome many of the limitations befalling previous studies (eg, inadequate statistical power and heterogeneous study cohorts); and (3) the prospective nature of the patient cohorts, permitting assessment of the impact of FcγR SNPs on disease pathophysiology at multiple survival end points. The main limitation of our study was the targeted nature of our analysis, focusing on only the 3 most well-characterized FcγR SNPs. As such, we did not evaluate the importance of other functionally relevant FcγR SNPs (eg, those in promoters regulating gene expression), nor did we assess the importance of germline CNV, which is known to target key FcγR genes.11 Furthermore, our analysis of nontumoral DNA precluded the detection of somatic FcγR gene alterations. For example, it has been demonstrated that the FCGR2B locus is targeted by focal somatic duplications in DLBCL, which are associated with increased gene transcription and poor patient survival.36 We also failed to assess the integrated nature of these various FcγR changes (eg, assessing the combined impact of the 3 FcγR SNPs), largely due to insufficient sample size, even in these large trials (the largest of their kind to date designed to assess the efficacy of obinutuzumab and rituximab treatment in combination with chemotherapy). A similar limitation concerns the definitive interpretation of the FCGR2B I232T SNP results, given that the T-genotype is so rare, with only ∼1% to 2% of patients carrying the TT genotype, it is difficult to make firm conclusions with regard to their impact. This small sample size may explain the apparent reduction in PFS for obinutuzumab- and rituximab-treated T232T patients in GALLIUM and GOYA, respectively. In previous studies, the FCGR2B 232T allele was found at a 1% frequency in European populations and varied between a 5% and 11% frequency in African and South East Asian populations.37,38 The impact of these differences on patient outcomes in ethnically diverse lymphoma cohorts is worthy of further research, particularly in centers across Asia and Africa, where the prevalence of the 232T genotype is higher than in European populations.37,38 Indeed, our results should be interpreted in the context of the ethnic composition of our cohorts, as our study was not designed to investigate the clinical impact of FcγR SNPs in all ethnicities.
In conclusion, our study, the largest of its type performed to date, provides clear evidence that the FcγR SNPs (FCGR2A R131H, FCGR3A F158V, and FCGR2B I232T) do not confer differential responsiveness to obinutuzumab or rituximab in combination with chemotherapy in treatment-naive patients with advanced FL or aggressive DLBCL.
Acknowledgments
The authors thank all patients who participated in and clinicians who contributed to the study; and the Experimental Cancer Medicine Centre (ECMC)–funded University of Southampton Faculty of Medicine Human Tissue Bank (Human Tissue Authority license 12009) for sample storage. Editorial support under the direction of Malgorzata Nowicka was provided by Zoe Toland and Louise Profit of Ashfield MedComms, an Ashfield Health company, and funded by F. Hoffmann-La Roche Ltd.
This work was supported by funding from Bloodwise (grant 12050 and 12036) and Cancer Research United Kingdom (grants A18087, A24721, A25139, and A15581) (M.S.C.), NC3R CRACKIT (grant 15402-106217) (J.C.S. and M.S.C.), and F. Hoffmann-La Roche Ltd. GALLIUM was sponsored by F. Hoffmann-La Roche Ltd and GOYA was sponsored by F. Hoffmann-La Roche Ltd with the scientific support of the Fondazione Italiana Linfomi.
Authorship
Contribution: J.C.S., A.D., W.H., W.K., M.M., L.H.S., M.T., U.V., C.R.B., C.K., A.K., M.Z.O., and M.S.C. designed the study; J.C.S., C.E.H., C.B., R.G., C.I., K.V.L., H.P., K.N.P., and M.J.J.R.-Z. conducted the study; C.B., A.D., W.H., M.M., L.H.S., M.T., U.V., C.R.B., C.K., A.K., and M.Z.O. contributed to the recruitment and follow-up of patients; J.C.S., M.N., C.E.H., C.B., A.D., R.G., W.H., C.I., W.K., M.M., H.P., M.J.J.R.-Z., L.H.S., M.T., U.V., C.R.B., C.K., A.K., M.Z.O., and M.S.C. collected the data; J.C.S., M.N., C.E.H., C.I., F.M., H.P., M.J.J.R.-Z., M.Z.O., and M.S.C. analyzed the data; and J.C.S., M.N., F.M., M.Z.O., and M.S.C. interpreted the data.
Conflict-of-interest disclosure: J.C.S. has received research funding from F. Hoffmann-La Roche Ltd. M.N. is an employee of F. Hoffmann-La Roche Ltd. A.D. has received grants and personal fees from F. Hoffmann-La Roche Ltd for serving on advisory boards, honoraria, and travel expenses. W.H. reports consultancy fees, research funding, honoraria, speaker bureau fees, and membership on boards of directors or advisory committees of F. Hoffmann-La Roche Ltd, Janssen, and Gilead. W.K. has received grants from F. Hoffmann-La Roche Ltd, Amgen, Takeda, and Regeneron. M.M. has received personal fees from Gilead Sciences, Novartis, Janssen, F. Hoffmann-La Roche Ltd, Sandoz, and Servier and travel expenses from F. Hoffmann-La Roche Ltd and Servier. F.M. was previously employed by F. Hoffmann-La Roche Ltd. L.H.S. has received consultancy fees from F. Hoffmann-La Roche Ltd, Genentech, Inc., AbbVie, Amgen, Apobiologix, AstraZeneca, Acerta, Celgene, Gilead Sciences, Janssen, Kite, Karyopharm, Lundbeck, Merck, MorphoSys, Seattle Genetics, Teva, Takeda, TG Therapeutics, and Verastem and research funding from F. Hoffmann-La Roche Ltd and Genentech, Inc. M.T. has received honoraria from Janssen, Gilead Sciences, Takeda, Bristol-Myers Squibb, Amgen, AbbVie, F. Hoffmann-La Roche Ltd, MorphoSys, and Incyte and has served in an advisory role to Celgene. U.V. has received personal fees from Celgene, F. Hoffmann-La Roche Ltd, AbbVie, Gilead Sciences, Janssen, and Regeneron. C.R.B. is employee of Genentech, Inc. and has equity ownership in F. Hoffmann-La Roche Ltd. C.K. is an employee of and has patents with and stock ownership in F. Hoffmann-La Roche Ltd. A.K. is an employee of F. Hoffmann-La Roche Ltd. M.Z.O. is a former employee at Roche with patent royalties in F. Hoffmann-La Roche Ltd and is an employee of Novo Nordisk. M.S.C. reports grants from Cancer Research United Kingdom and Bloodwise, grants and personal fees from Bioinvent, and patents with Bioinvent. The remaining authors declare no competing financial interests.
The current affiliation for M.Z.O. is Novo Nordisk, Zurich, Switzerland.
Correspondence: Jonathan C. Strefford, Cancer Genomics, Cancer Sciences Academic Unit, Faculty of Medicine, University of Southampton, Southampton, United Kingdom; e-mail: jcs@soton.ac.uk.
References
Author notes
J.C.S., M.N. and C.E.H. share first authorship.
Presented in part at the 60th annual meeting of the American Society of Hematology, San Diego CA, 1-5 December 2018 (Abstract P-1409).
Qualified researchers may request access to individual patient-level data through the clinical study data request platform (https://vivli.org/). Further details on Roche’s criteria for eligible studies are available here (https://vivli.org/members/ourmembers/). For further details on Roche’s Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see here (https://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing.htm).
The full-text version of this article contains a data supplement.