Key Points
MAPPs allows more accurate T-cell epitope identification as the assay needs MHC protein proteolytic processing and antigen presentation.
In a MAPPs assay, antigen-presenting cells presented more T-cell epitopes when matured with rFVIII compared with pdFVIII.
Abstract
The immunogenicity of protein therapeutics is an important safety and efficacy concern during drug development and regulation. Strategies to identify individuals and subpopulations at risk for an undesirable immune response represent an important unmet need. The major histocompatibility complex (MHC)–associated peptide proteomics (MAPPs) assay directly identifies the presence of peptides derived from a specific protein therapeutic on a donor’s MHC class II (MHC-II) proteins. We applied this technique to address several questions related to the use of factor VIII (FVIII) replacement therapy in the treatment of hemophilia A (HA). Although >12 FVIII therapeutics are marketed, most fall into 3 categories: (i) human plasma-derived FVIII (pdFVIII), (ii) full-length (FL)–recombinant FVIII (rFVIII; FL-rFVIII), and (iii) B-domain–deleted rFVIII. Here, we investigated whether there are differences between the FVIII peptides found on the MHC-II proteins of the same individual when incubated with these 3 classes. Based on several observational studies and a prospective, randomized, clinical trial showing that the originally approved rFVIII products may be more immunogenic than the pdFVIII products containing von Willebrand factor (VWF) in molar excess, it has been hypothesized that the pdFVIII molecules yield/present fewer peptides (ie, potential T-cell epitopes). We have experimentally tested this hypothesis and found that dendritic cells from HA patients and healthy donors present fewer FVIII peptides when administered pdFVIII vs FL-rFVIII, despite both containing the same molar VWF excess. Our results support the hypothesis that synthesis of pdFVIII under physiological conditions could result in reduced heterogeneity and/or subtle differences in structure/conformation which, in turn, may result in reduced FVIII proteolytic processing relative to FL-rFVIII.
Introduction
The most severe complication of factor VIII (FVIII) replacement therapy, used to treat hemophilia A (HA), is the development of FVIII-neutralizing antibodies or “inhibitors.”1 More broadly, immunogenicity is a safety-and-efficacy concern during the development and licensure of therapeutic proteins.2 Numerous FVIII products, either purified from human plasma (plasma-derived FVIII [pdFVIII]) or generated using recombinant DNA technology (recombinant FVIII [rFVIII]), are in clinical use.3,4 Recent epidemiological studies5-8 and a prospective randomized clinical control trial9 suggest that the rFVIII products may be more immunogenic than the pdFVIII products. Although hypotheses have been advanced to explain this difference,10 testing these experimentally has been challenging. The few experimental studies that have been conducted suggest that von Willebrand factor (VWF) inhibits FVIII endocytosis into monocyte-derived dendritic cells (MoDCs) and, therefore, limits their presentation of FVIII-derived peptides.11-13
The major histocompatibility complex (MHC)–associated peptide proteomics (MAPPs) assay is a powerful tool that identifies the therapeutic protein-derived peptides presented on the MHC class II (MHC-II) molecules expressed by a subject’s antigen-presenting cells.14-16 Studies have shown that peptide–MHC-II affinity is a good predictor of immunogenicity.17-19 However, evaluation of peptide–MHC-II affinity alone incorrectly presupposes that all potential peptides will be generated. Protein processing and presentation are both necessary to elicit antigen-specific T-cell responses.20,21 Using peptide pools to identify T-cell epitopes does not address the question of whether the peptide(s) identified as candidate epitopes can be generated by the MoDC proteolytic machinery. Conversely, T-cell proliferation mediated by the intact protein does not allow identification of specific T-cell epitope(s).
The mass spectrometry (MS)–based MAPPs assay is an analytical tool that provides information about both protein processing and peptide presentation.22 In studying immunogenicity, we used this approach to characterize a neosequence in an engineered variant of FVIIa that was more immunogenic than the wild-type molecule.23 The study used a range of in silico assessments and in vitro and ex vivo assays for the immunological characterization of the neosequences; the MAPPs assay was the only analytical tool that could demonstrate that the foreign antigen was both processed and presented by the immune system. Several studies have also used the MAPPs assay to identify the FVIII-derived peptides presented by MHC-II proteins.24-26
The MAPPs technology offers an experimental platform for testing hypotheses related to product-specific immunogenicity of different FVIII concentrates. For instance, the protection of T-cell epitopes by VWF10,27 and differences in the cellular processing of pdFVIII and rFVIII have been proposed to explain differences in clinical immunogenicity.11,12 These hypotheses can be tested using MAPPs assays, which permit the comparison of peptide–MHC-II repertoires when cells are treated with the various therapeutic FVIII products.
Here, using MAPPs, we provide experimental evidence that: (i) the number of unique FVIII-derived peptides isolated, average length of peptides, and range of peptide lengths were comparable for MHC-II proteins immunoprecipitated from MoDCs from HA patients or healthy blood donors; (ii) for each subject, FVIII-derived peptides identified by the MAPPs assay were enriched for peptides with high affinities for the MHC-II variants from which they were eluted compared with a million peptides of comparable lengths randomly obtained from the human proteome; (iii) when MoDCs from the same donor were exposed to full-length (FL)-rFVIII or B-domain–deleted (BDD)-rFVIII, similar peptides were identified on their MHC-II molecules (as expected, cells incubated with BDD-rFVIII did not present peptides originating from the B domain); and (iv) when MoDCs from the same donor were incubated with FL-rFVIII or pdFVIII (both in the presence of pdVWF), fewer FVIII peptides were recovered from the pdFVIII compared with FL-rFVIII. These results indicate that the cellular sources (ie, healthy human pooled plasma-derived vs Chinese hamster ovary–expressed) of the FVIII and/or manufacturing processes may differentially affect their processing and presentation onto MHC-II molecules and thus their immunogenic potential.
Materials and methods
Materials
Cells.
Peripheral blood mononuclear cells were isolated from 6 HA patients by leukopheresis at the University of North Carolina Apheresis Unit. The procedure was performed following informed consent under institutional review board study number 14-0582. The cells were cryopreserved in 10% dimethyl sulfoxide. Each 1.8-mL cryotube contained ∼20 × 106 cells. Peripheral blood mononuclear cells were also obtained from an additional 15 unrelated healthy blood donors at the Addenbrooke’s Hospital (Cambridge, United Kingdom) after informed consent.
FVIII and VWF sample preparation.
Beriate, a marketed pdFVIII product provided by CSL Behring GmbH, contains pdVWF (supplemental Figure 1). For this study, pdVWF was removed from reconstituted Beriate using a VIIISelect affinity resin (GE Healthcare) using an VIIISelect affinity column. FVIII activity in the final samples was determined by chromogenic FVIII activity assay (Chromogenix Coamatic Factor VIII; Diapharma). pdVWF (supplemental Figure 2) was provided as an intermediate precipitate of the plasma-fractioning process by CSL Behring GmbH. pdFVIII bound to pdVWF was separated from the precipitate through a size-exclusion chromatography step using HiPrep Sephacryl S500 (GE Healthcare) (supplemental Figure 3) using a N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) buffer containing 400 nM calcium chloride, as described previously.28 Fractions 1.A.2 to 2.A.2. from the size-exclusion chromatography were used. Residual FVIII in final preparations was negligible (0.004% mass). Using human MoDC-based assays, the uptake of tFVIIIs in the presence of pdVWF was evaluated in a concentration-dependent manner, as described previously.12 We identified a minimum molar ratio of FVIII to VWF, which is required to effectively inhibit FVIII endocytosis by MoDCs in vitro (data not shown).29 Both pdFVIII and rFVIII products (145 nM final concentration) were individually combined with pdVWF at a molar ratio of 1:13.4 prior to use in the MAPPs assays.
Methods
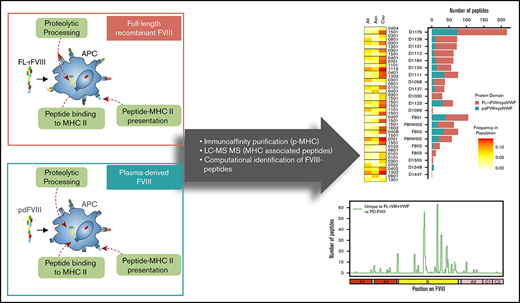
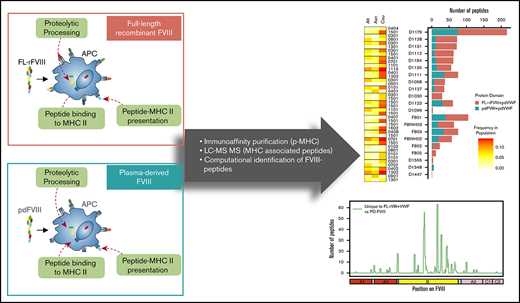
In vitro profiling of peptides displayed by HLAcII-DR, -DP, and -DQ after uptake by MoDCs.
The study was performed by ProImmune using the ProPresent Antigen Presentation Assay (see Rombach-Riegraf et al,30 Xue et al,31 Ventura et al,32 and Gouw et al33 for details). Monocytes were isolated from the mononuclear cell fraction of peripheral blood samples by positive selection and differentiated in vitro into immature MoDCs. The immature MoDCs were: (i) cultured in the presence of 1 of the 3 FVIIIs (pdFVIII, FL-rFVIII, BDD-rFVIII), each containing 13.4 molar excess pdVWF, and (ii) matured in the presence of the same preincubated FVIII-plus-pdVWF mix. The mature MoDCs were harvested and lysed. HLA class II (HLAcII)-DR, -DQ, and -DP molecules were recovered from the cell lysate using immune-affinity columns. The HLAcII-bound peptides were then eluted and processed for further analysis by high-resolution-sequencing MS (liquid chromatography–tandem MS). The resulting data were compiled and analyzed using sequence analysis software referencing the Swiss-Prot Human Proteome Database with the incorporated test item sequence(s). Liquid chromatography–tandem MS–based identification of peptide sequences was based on scoring algorithms and statistical significance determination. The likelihood of peptides to be real identities is described by their expect value (EV) and by the false discovery rate (FDR) obtained by searching an unrelated database. The EV considers specific features of the experimental MS/MS spectrum, for example, the number of peaks that match predicted fragments from a peptide sequence in the database. The EV provides a statistical significance for the peptide identification and it is defined as being reflective of the number of assignments with equal (or better) scores that are expected to occur by chance alone. The lower this value, the higher the probability the assignment is correct. EV ≤ 0.05. was used to report significant hits as recommended by current guidelines for documentation of peptide and protein identification by MS.34 An FDR is determined by repeating the search using identical searching parameters against a “decoy” database, where the sequences have been reversed or randomized. The number of matches that are found with the decoy database is an estimate of the number of false positives that are present in the results from the real or “target” database. The FDR can be expressed as the total false positives divided by the sum of true positives and false positives. Identifications reported with EV ≤ 0.05 typically have FDRs <1%.
Reads with sequences that matched both the FL protein sequence and a variant were counted once if the polymorphism was not included in the matched region. Peptides derived from BDD-rFVIII are numbered based on their position in the FL-FVIII. When cells from the same subject were treated with 2 products, peptides were identified as unique to 1 product if no peptides at that location were identified for the other product. In some cases, peptides with identical cores but different lengths were identified following treatment with different products; fragments <9 amino acids (aa) in length were disregarded.
HLA typing.
For HLA typing, we used next-generation sequencing for typing the DRB1, DPB1, and DQB1 loci. Next-generation sequencing typing allows precise resolution of HLA types to precise 4-digit allelic level.
Computational model of FVIII structure.
PyMOL (Schrödinger) was used for mapping of the MAPPs assay–derived peptides onto the FVIII crystal structure. Structure of FVIII was obtained from https://www.rcsb.org/ using Protein Data Bank identifier (PDB ID) 2R7E. The information about the interaction between FVIII and VWF was obtained from published sources.35-39
Data analysis
The probability density plot is the comparison of 4 independent sets of peptides: (i) a random set of 1 000 000 15-mer sequences computationally generated; (ii) all human proteins in the Swiss-Prot database were computationally cleaved into 15-mer peptides, and 1 000 000 random peptides from this list resulted in a data set of 912 997 nonredundant peptides; (iii) all overlapping 15-mer peptides derived from the FVIII and VWF sequences (5117 peptides); and (iv) the set of peptides from the MAPPs assay consists of all peptides identified in the assay with an EV score <0.05. For each subject, the minimum percent-rank score of each peptide for that subject’s HLA alleles was recorded, and the 4 sets of scores were plotted as histograms and smoothed using Gaussian kernel density estimation. For each peptide, the predicted affinity to each HLA variant identified in that subject was computed using netMHCIIpan version 3.2.40
Data analyses were performed using Python (https://www.python.org) and R (https://www.R-project.org/). Graphics were generated using ggplot241 matplotlib.42
Results
FVIII peptides presented by MHC-II molecules on MoDCs from HA patients and healthy blood donors
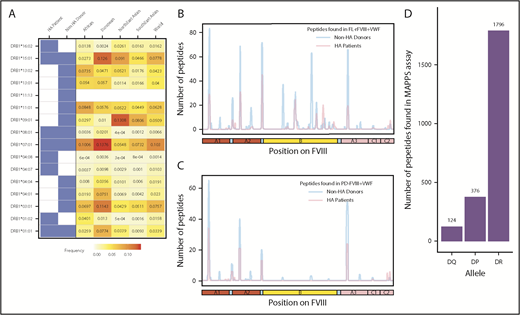
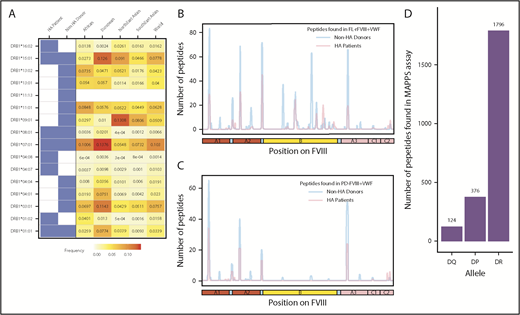
FVIII peptides were identified on MHC-II proteins isolated from MoDCs incubated with FL-rFVIII plus VWF in a MAPPs assay. The cells were obtained from 6 HA patients (supplemental Table 1 describes patient characteristics) and 15 unrelated healthy blood donors. Figure 1A summarizes the distribution of HLAcII-DRB1 alleles in the 2 groups and frequencies of these alleles in different populations. The DR, DP, and DQ isomers of the MHC-II proteins were captured separately using specific immunoaffinity columns. Peptides associated with the MHC-II proteins were identified by mass-spectrometric analyses. Peptides from endogenous proteins and the FVIII products coincubated with the MoDCs are presented by the MHC-II. The computational identification of FVIII-derived peptides with a FDR of <1% are described in “Methods.” The number of unique FVIII peptides isolated, total number of peptides identified, the average length of peptides, and range of peptide lengths in the 2 groups are shown in Table 1. There are no significant differences in these parameters between the HA patients and healthy donors. Similarly, when MoDCs obtained from HA patients or healthy donors were incubated with FVIII (either FL-rFVIII plus pdVWF or pdFVIII plus pdVWF), all unique FVIII peptides identified on MHC-II proteins showed comparable locations on the FVIII primary sequence (Figure 1B-C). Unique peptides are defined as nonoverlapping regions of the proteins found in only 1 of the 2 proteins compared. We also compared the number of peptides identified on DR, DP, and DQ isomers and determined that almost 80% of the peptides identified were presented by the DR molecules (Figure 1D).
Identification of FVIII peptides on MoDCs obtained from HA patients and healthy blood donors using the MAPPs assay. (A) HLAcII-DRB1 alleles of subjects used in this study. The blue shading shows whether a particular allele was found in HA patients or healthy donors. Frequencies of each allele in various demographic groups are listed and identified on a scale from white (0) to red (15%). (B-C) Peptides identified in the MAPPs assay on DRB1-based DR molecules when MoDCs were incubated with FL-rFVIII plus pdVWF (B) or pdFVIII plus pdVWF (C). The peptides identified in cells from HA patients are shown in red; peptides identified in healthy donors are shown in blue. (D) Number of FVIII peptides identified on DR, DP, and DQ molecules in the MAPPs assay when APCs were incubated with FL-rFVIII plus pdVWF.
Identification of FVIII peptides on MoDCs obtained from HA patients and healthy blood donors using the MAPPs assay. (A) HLAcII-DRB1 alleles of subjects used in this study. The blue shading shows whether a particular allele was found in HA patients or healthy donors. Frequencies of each allele in various demographic groups are listed and identified on a scale from white (0) to red (15%). (B-C) Peptides identified in the MAPPs assay on DRB1-based DR molecules when MoDCs were incubated with FL-rFVIII plus pdVWF (B) or pdFVIII plus pdVWF (C). The peptides identified in cells from HA patients are shown in red; peptides identified in healthy donors are shown in blue. (D) Number of FVIII peptides identified on DR, DP, and DQ molecules in the MAPPs assay when APCs were incubated with FL-rFVIII plus pdVWF.
Peptide–MHC-II affinity is a necessary but not sufficient condition for peptide presentation on MHC-II molecules
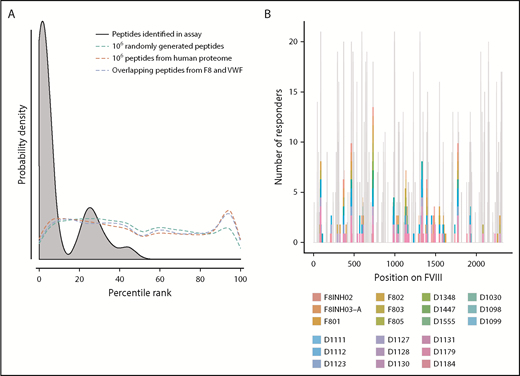
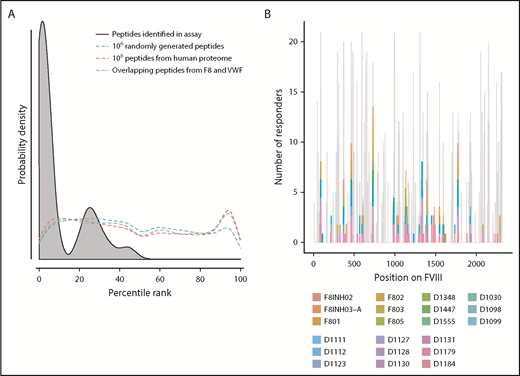
Peptide–MHC-II–binding affinity is often used as a surrogate marker for immunogenicity.17,43,44 We tested the hypothesis that peptides with higher affinity to MHC-II proteins are more likely to be presented by the MoDCs. We generated 3 virtual data sets of peptides (see “Methods” for details): (i) overlapping 15-mer peptides derived from FVIII and VWF, (ii) million randomly generated peptides, and (iii) million peptides from the human proteome. We estimated the peptide–MHC-II affinities (expressed as a percentile score) for each of these data sets using the MHC-II variants identified in each subject (Figure 2A). The percentile score expresses the rank of a particular peptide–MHC-II affinity in an ordered list of all peptide–MHC-II affinities for that particular MHC-II variant. In large data sets, peptides of all percentile scores (affinities) are equally likely to be found. Thus, as expected, the probability distributions of peptides in all 3 virtual data sets are a flat line. In sharp contrast, the FVIII peptides identified on the MHC-II proteins isolated from MoDCs are skewed to the left, which indicates a greater probability of finding peptides with higher affinity. This observation was true for FVIII peptides identified on MHC-II proteins isolated from all 21 subjects (Figure 2A; supplemental Figure 4). Although all FVIII peptides identified on MoDCs bind with high affinity to 1 or more of their expressed MHC-II molecules, only a subset of peptides that binds with high affinity to an individual subject’s MHC-II variants is identified in the MAPPs assay (Figure 2B). In characterizing the MAPPs assay, we used the FL-rFVIII plus pdVWF. In subsequent studies, we compared the effect of different product classes and protein-protein interactions on peptide processing and presentation by MoDCs.
Affinities of FVIII peptides identified on MoDCs. (A) The distribution of percentile-rank scores predicted by netMHCIIpan3.2 for all peptides found in the MAPPs assay for a single healthy donor, D1098 (black, shaded), compared with the distribution of percentile-rank scores for 1 × 106 random strings of 15 aa (green), 1 × 106 random 15 mers selected from the human proteome (red), and all 15 mers from FVIII and VWF (blue). Peptides found in the MAPPs assay were more likely to have high binding affinity to the patients’ alleles as shown by the greater probability of finding lower percentile rank scores for these peptide/MHC-II binding pairs. The other 3 distributions were close to flat which can be expected from a random distribution of peptides. (B) Peptides identified in the MAPPs assay following incubation with FL-rFVIII plus VWF. Location of the peptides is shown on the primary sequence of FVIII and they are color-coded for the subjects who provided the MoDCs. The gray backdrop shows the predicted number of subjects whose MHC-II variants would bind peptides at each location with high affinity (percentile rank <5%) based on netMHCIIpan predictions. Peptides found in the MAPPs assay were found in regions predicted to be binding regions by netMHCIIpan3.2. Not all regions predicted to be binding regions were found in the assay.
Affinities of FVIII peptides identified on MoDCs. (A) The distribution of percentile-rank scores predicted by netMHCIIpan3.2 for all peptides found in the MAPPs assay for a single healthy donor, D1098 (black, shaded), compared with the distribution of percentile-rank scores for 1 × 106 random strings of 15 aa (green), 1 × 106 random 15 mers selected from the human proteome (red), and all 15 mers from FVIII and VWF (blue). Peptides found in the MAPPs assay were more likely to have high binding affinity to the patients’ alleles as shown by the greater probability of finding lower percentile rank scores for these peptide/MHC-II binding pairs. The other 3 distributions were close to flat which can be expected from a random distribution of peptides. (B) Peptides identified in the MAPPs assay following incubation with FL-rFVIII plus VWF. Location of the peptides is shown on the primary sequence of FVIII and they are color-coded for the subjects who provided the MoDCs. The gray backdrop shows the predicted number of subjects whose MHC-II variants would bind peptides at each location with high affinity (percentile rank <5%) based on netMHCIIpan predictions. Peptides found in the MAPPs assay were found in regions predicted to be binding regions by netMHCIIpan3.2. Not all regions predicted to be binding regions were found in the assay.
FL-rFVIII and BDD-rFVIII; comparison of peptide presentation on MHC-II molecules
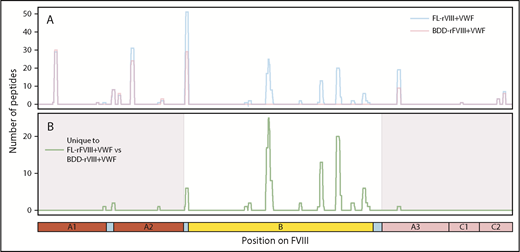
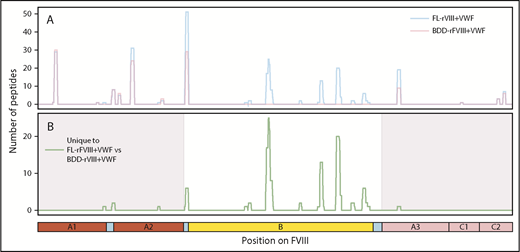
MoDCs from the same individual were incubated with either FL-rFVIII plus pdVWF or BDD-rFVIII plus pdVWF in the MAPPs assay. Fewer unique peptides were identified from the MHC-II proteins when cells were incubated with BDD-rFVIII plus pdVWF (Figure 3). However, all the peptides that were unique to FL-rFVIII were localized to the B domain (Figure 3).
Comparison of FVIII peptides identified in a MAPPs assay following incubation of MoDCs with FL-rFVIII plus pdVWF or BDD-rFVIII plus pdVWF. (A) Pooled data for all subjects showing location and frequencies of peptides identified in the MAPPs assay following incubation of MoDCs with FL-rFVIII plus pdVWF (blue) or BDD-rFVIII plus pdVWF (red). (B) Location and frequencies of peptides identified following incubation with FL-rFVIII plus VWF where no peptides were found after incubation with BDD-rFVIII plus VWF are shown in green. The x-axis represents the primary sequence of FL-FVIII and the different FVIII domains are depicted on the figure.
Comparison of FVIII peptides identified in a MAPPs assay following incubation of MoDCs with FL-rFVIII plus pdVWF or BDD-rFVIII plus pdVWF. (A) Pooled data for all subjects showing location and frequencies of peptides identified in the MAPPs assay following incubation of MoDCs with FL-rFVIII plus pdVWF (blue) or BDD-rFVIII plus pdVWF (red). (B) Location and frequencies of peptides identified following incubation with FL-rFVIII plus VWF where no peptides were found after incubation with BDD-rFVIII plus VWF are shown in green. The x-axis represents the primary sequence of FL-FVIII and the different FVIII domains are depicted on the figure.
Comparing peptide presentation on MHC-II molecules following incubation with rFVIII and pdFVIII products
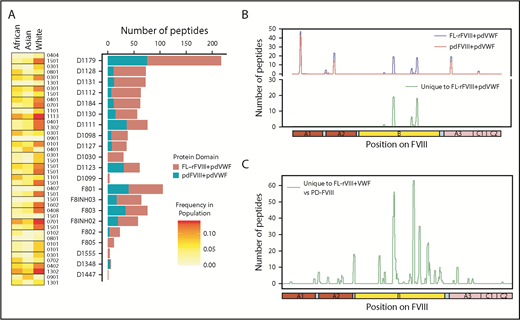
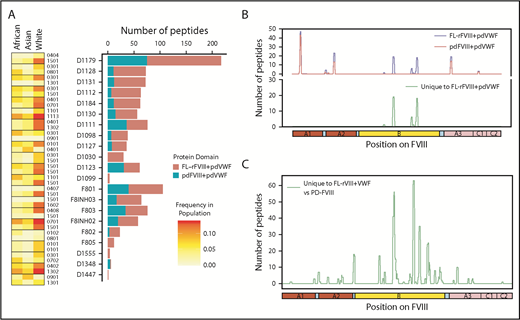
Some clinical studies suggest that specific FVIII products may present a higher immunogenicity risk whereas other studies show that the risks are statistically equivalent.5-8,45-47 The Study on Inhibitors in Plasma-Product Exposed Toddlers (SIPPET), a prospective randomized clinical trial, reported a 1.87-fold increase in inhibitors in previously untreated patients treated with rFVIII compared with pdFVIII.9 We compared FL-rFVIII plus pdVWF and pdFVIII plus pdVWF with respect to presentation of peptides by MHC-II proteins. As pdFVIII products have variable amounts of VWF, we isolated pdFVIII free of pdVWF and reconstituted both FL-rFVIII and pdFVIII with excess pdVWF at a molar ratio of 1:13.4 (see “Methods”). This strategy allows us to compare processing and MHC-II presentation of FL-rFVIII and pdFVIII under identical conditions that mimic the in vivo scenario where, due to the high affinity of the FVIII-VWF interaction, rFVIII products once infused would rapidly associate with VWF. We found that incubation of MoDCs with pdFVIII plus pdVWF results in presentation of fewer peptides than when the same MoDCs are incubated with FL-rFVIII plus pdVWF (Figure 4A). Although there are differences in the number of peptides presented by each donor, cells from all donors present more peptides when incubated with FL-rFVIII plus pdVWF compared with pdFVIII plus pdVWF. The location of the peptides identified following incubation with FL-rFVIII plus pdVWF or pdFVIII plus pdVWF for a typical subject is shown in Figure 4B (data for all patients, individually, is provided in supplemental Figure 5). We also pooled the data to determine the location of all FVIII peptides protected against MHC-II presentation with pdFVIII, that is, not identified when pdFVIII was incubated with the MoDCs (Figure 4C). These results show that across multiple donors there are differences in the processing and presentation of FL-rFVIII and pdFVIII by MoDCs. Differences in the processing and presentation of FL-rFVIII and pdFVIII (even when both are protected by VWF) could contribute to the differences in the reported clinical immunogenicity of these products. However, additional studies are required to understand the molecular mechanisms (see “Discussion”).
Comparison of FVIII peptides identified in a MAPPs assay following incubation of MoDCs with FL-rFVIII plus pdVWF or pdFVIII plus pdVWF. (A) For each donor and HA patient, number of peptides found in the MAPPs assay for FL-rFVIII plus pdVWF (red) and pdFVIII plus pdVWF (blue) are shown. In addition, the MHC-II–DRB1 alleles for each donor are identified as well as their respective frequencies in African, Asian, and white populations. (B) Location and frequencies of peptides for a single donor (D1179) identified in the MAPPs assay following incubation of APCs with FL-rFVIII plus pdVWF (blue) or pdFVIII plus pdVWF (red). (C) Pooled data for all subjects showing location and frequencies of peptides identified following incubation with FL-rFVIII plus pdVWF where no peptides were found after incubation with pdFVIII plus pdVWF.
Comparison of FVIII peptides identified in a MAPPs assay following incubation of MoDCs with FL-rFVIII plus pdVWF or pdFVIII plus pdVWF. (A) For each donor and HA patient, number of peptides found in the MAPPs assay for FL-rFVIII plus pdVWF (red) and pdFVIII plus pdVWF (blue) are shown. In addition, the MHC-II–DRB1 alleles for each donor are identified as well as their respective frequencies in African, Asian, and white populations. (B) Location and frequencies of peptides for a single donor (D1179) identified in the MAPPs assay following incubation of APCs with FL-rFVIII plus pdVWF (blue) or pdFVIII plus pdVWF (red). (C) Pooled data for all subjects showing location and frequencies of peptides identified following incubation with FL-rFVIII plus pdVWF where no peptides were found after incubation with pdFVIII plus pdVWF.
Identification of peptides derived from VWF on MHC-II molecules
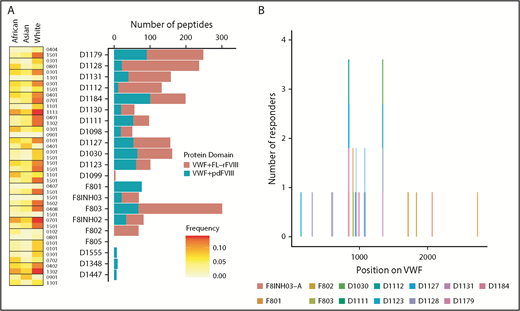
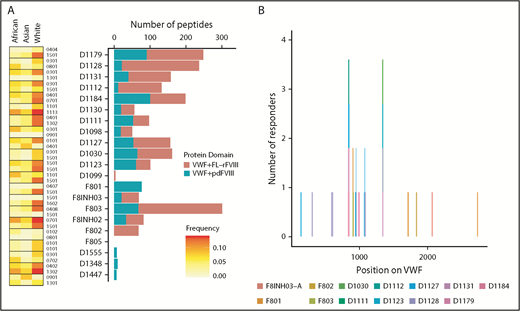
Because in the experiments described previously (and reported in Figure 4) the 3 different FVIII products were incubated with pdVWF, we also identified the pdVWF peptides found to be bound to the MHC-II molecules. Although there are a few exceptions, in general, more VWF peptides are presented when cells are incubated with VWF in the presence of FL-rFVIII (compared with pdFVIII) (Figure 5A). The location, on the primary sequence of VWF, of all peptides identified is shown in Figure 5B.
VWF peptides identified in a MAPPs assay and effect of co-incubation with different FVIII products. (A) For each donor, number of VWF peptides identified in the MAPPs assay is shown by the bars; pdVWF was coincubated with either FL-rFVIII (red) or pdFVIII (blue). Left panel, The MHC-II-DRB1 alleles for each donor and their respective frequencies in African, Asian, and white populations. (B) Location of the VWF peptides identified in the MAPPs assay shown on the VWF primary sequence (x-axis). At each location, the subjects are identified by the color codes.
VWF peptides identified in a MAPPs assay and effect of co-incubation with different FVIII products. (A) For each donor, number of VWF peptides identified in the MAPPs assay is shown by the bars; pdVWF was coincubated with either FL-rFVIII (red) or pdFVIII (blue). Left panel, The MHC-II-DRB1 alleles for each donor and their respective frequencies in African, Asian, and white populations. (B) Location of the VWF peptides identified in the MAPPs assay shown on the VWF primary sequence (x-axis). At each location, the subjects are identified by the color codes.
Mapping the peptides identified using the MAPPs assay on the structure of FVIII
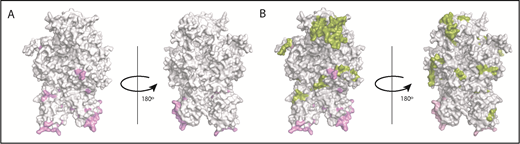
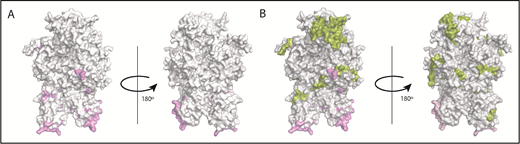
The crystal structure of FVIII (PDB ID 2R7E), generated using PyMOL is depicted in Figure 6. Several studies have used site-directed mutagenesis and antibody competition as well as electron microscopy and mass spectrometry to identify the sites of interaction between VWF and FVIII.35-39 The known locations of FVIII-VWF interactions are depicted in Figure 6A. In the current study, several peptides were identified when cells were incubated with FL-rFVIII plus pdVWF that were not identified following incubation with pdFVIII plus pdVWF (Figure 6B). VWF-mediated protection of FVIII processing would suggest that the peptides that are not presented in the presence of VWF would be located at or near the sites of VWF-FVIII interactions. However, our results show that these peptides do not necessarily locate to the sites of VWF-FVIII interaction.
Mapping of the MAPPs assay derived peptides on FVIII structure. (A) Surface representation of the FVIII structure (PDB ID 2R7E) with the regions identified to be VWF-binding sites10,39 (pink). (B) Structure depicted in panel A with the regions yielding MHC-II–presented FVIII-derived peptides unique to FL-rFVIII (green). Peptides in green (see domain labels for peptide location) were identified on MoDCs incubated with FL-rFVIII plus pdVWF but not identified when MoDCs from the same subject were incubated with pdFVIII plus pdVWF (ie, protected in the pdFVIII protein). The FVIII domains are identified on the figure in bold letters.
Mapping of the MAPPs assay derived peptides on FVIII structure. (A) Surface representation of the FVIII structure (PDB ID 2R7E) with the regions identified to be VWF-binding sites10,39 (pink). (B) Structure depicted in panel A with the regions yielding MHC-II–presented FVIII-derived peptides unique to FL-rFVIII (green). Peptides in green (see domain labels for peptide location) were identified on MoDCs incubated with FL-rFVIII plus pdVWF but not identified when MoDCs from the same subject were incubated with pdFVIII plus pdVWF (ie, protected in the pdFVIII protein). The FVIII domains are identified on the figure in bold letters.
Identification of potential T-cell epitopes on FVIII based on the MAPPs assay
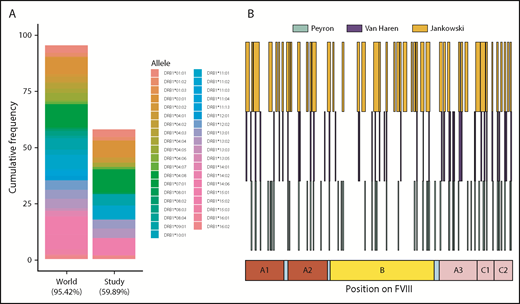
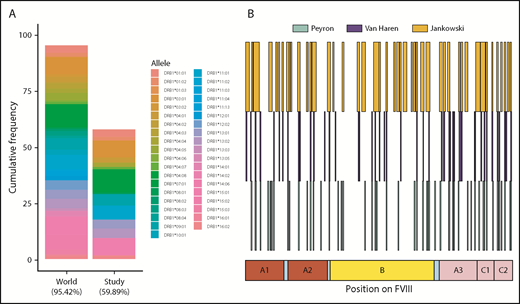
We have shown that there is considerable variation between subjects with respect to the peptides presented on MHC-II molecules (Figures 3-5). Some of these differences may arise due to the distinct MHC-II variants on MoDCs from different subjects. The subjects in this study covered MHC-II variants that occur in ∼50% of the world population (Figure 7A). Moreover, the frequencies of these variants are comparable to those found in this population. The MHC-II repertoire is 1 of the most diverse in the human genome and MAPPs assays are technically challenging, labor intensive, and expensive to carry out. Thus, it may be useful to pool data from different studies to develop a comprehensive list of FVIII peptides that can be processed and presented to T-cell receptors. In addition to this study, 2 previous studies,24,25 using 9 and 4 donors, respectively, have identified FVIII peptides on MHC-II proteins following incubation with FL-rFVIII. We pooled the data for FVIII peptides identified using the MAPPs assays (Figure 7B). Together, the antigen-presenting cells used in these studies represent 57.9% of MHC-II variants in the world population. The peptides identified in the previous studies covered 27.3% of the total length of FVIII. The peptides identified in our study included 84.4% of those positions found in the previous studies and novel peptides covering an additional 19.0% of FVIII.
HLAcII-DRB1 coverage in this study and comparison of FVIII peptides identified in this study and those identified in previous studies. (A) The bar labeled “World” represents a set of 38 MHC-II–DRB1 alleles and their respective frequencies in the world population; together, these alleles represent 95.42% of alleles found in this population. The bar labeled “Study” represents the 16 alleles found in the subjects of the MAPPS assays. This subset represents 59.89% of alleles found in the world population. The relative frequencies of the MHC-II–DRB1 alleles in the study population and the world population are comparable. (B) Previous studies have identified FVIII peptides in MAPPs assays using a BDD-rFVIII, whose primary amino acid sequence corresponds exactly to that of the BDD-rFVIII used in this study; the peptides identified in studies by Peyron et al26 (green), van Haren et al25 (blue), and in the current study are shown for comparison.
HLAcII-DRB1 coverage in this study and comparison of FVIII peptides identified in this study and those identified in previous studies. (A) The bar labeled “World” represents a set of 38 MHC-II–DRB1 alleles and their respective frequencies in the world population; together, these alleles represent 95.42% of alleles found in this population. The bar labeled “Study” represents the 16 alleles found in the subjects of the MAPPS assays. This subset represents 59.89% of alleles found in the world population. The relative frequencies of the MHC-II–DRB1 alleles in the study population and the world population are comparable. (B) Previous studies have identified FVIII peptides in MAPPs assays using a BDD-rFVIII, whose primary amino acid sequence corresponds exactly to that of the BDD-rFVIII used in this study; the peptides identified in studies by Peyron et al26 (green), van Haren et al25 (blue), and in the current study are shown for comparison.
Discussion
We present MAPPs data from MoDCs derived from HA patients and healthy blood donors incubated with different FVIII products. Together, these subjects provide an adequate coverage of MHC-II variants (Figure 1A). The number and location of peptides identified on cells obtained from HA patients or healthy blood donors are similar (Figure 1B-C). This is important as MAPPs assays require large numbers of cells and obtaining these from patients with bleeding disorders can be challenging. In addition, we demonstrate the value of the MAPPs assay as complementary to peptide–MHC-II binding assessments as a risk factor for immunogenicity. The peptides identified on MHC-II proteins are all enriched for high-affinity binders (Figure 2A), providing a mechanistic justification for the positive association between peptide–MHC-II affinity and immune responses.23,48-50 Moreover, peptide-MHC-II affinity is increasingly being used as a surrogate marker in nonclinical studies that seek to estimate the immunogenicity risk during drug development.2,17 The use of peptides in assessing whether immunogenicity can overestimate potential T-cell epitopes as protein processing is not taken into account (see “Introduction”). Only peptides that have successfully undergone protein processing and peptide presentation are identified in the MAPPs assay whereas peptide–MHC-II binding measurements/estimates cannot account for the processing steps that are necessary prior to presentation. We show in Figure 2B that many peptides that can bind with high affinity to donor MHC-II variants are not presented by MHC-II proteins. The MAPPs assay provides a unique data set that cannot be obtained from other in vivo or ex vivo methods used for immunogenicity assessments. We used the MAPPs assay to address specific hypotheses related to the processing and presentation of FVIII-derived peptides of different product classes.
A key issue in the immunogenicity of FVIII products is the role of VWF, which is the chaperone of FVIII in plasma (for review, see Pipe et al10 ). It has been proposed that association with VWF protects some FVIII sites from being processed to explain the clinical observations showing decreased immunogenicity of pdFVIII products.5-8 Studies by Voorberg and colleagues compared FVIII peptides identified in a MAPPs assay following incubation of MoDCs with a BDD-rFVIII.13,26 The studies were conducted with 2 concentrations of BDD-rFVIII, 50 nM and 25 nM, using MoDCs from 3 and 2 donors, respectively. The studies found that VWF modulates the peptides identified in the MAPPs assay. However, only cells from the 2 patients treated with 25 nM showed VWF-mediated protection; that is, fewer peptides are identified following exposure to cells of FVIII plus VWF.
Although the hypothesis that VWF-FVIII interaction protects FVIII processing can be tested in vitro, it is difficult to rationalize under physiological conditions. This is because FVIII has a very high affinity for VWF (0.2-0.5 nM)51 and infused FL-rFVIII would associate very rapidly with endogenous VWF. In this study, we compared FVIII-peptide presentation in the presence of pdVWF for FL-rFVIII, BDD-rFVIII, and pdFVIII. The relative concentrations of VWF and FVIII commercial preparations of pdFVIII vary9 ; we reconstituted all proteins used in this study with the same pdVWF at an identical molar ratio (1:13.4).
As expected, BDD-rFVIII presented fewer total and unique peptides compared with FL-rFVIII (Figure 3) and all the peptides not identified following incubation with BDD-rFVIII mapped to the B domain. This finding shows that although the FL-rFVIII and BDD-rFVIII were manufactured by different companies, the identical peptides were recovered in a MAPPs assay (except for the peptides in the B domain, which is lacking in the BDD-rFVIII product).
The findings in several clinical studies that there is a higher risk of developing inhibitors to rFVIII products than to pdFVIII products6-9 did not provide a mechanistic explanation. Studies have shown that a necessary condition for the immune response to FVIII products used in replacement therapy is the presentation of FVIII-derived peptides by APCs and subsequent stimulation of FVIII-specific CD4 T lymphocytes.52,53 Thus, the MAPPs assay could provide information about a necessary condition for clinically relevant immune responses: MHC-II–mediated peptide presentation. We compared the number and location of peptides identified in the MAPPs assay when MoDCs from the same donor/patient were incubated with either FL-rFVIII plus pdVWF or pdFVIII plus pdVWF. All donors presented larger numbers of peptides when incubated with FL-rFVIII plus pdVWF and several unique peptides were identified following incubation with FL-rFVIII plus pdVWF. Interestingly, these peptides do not necessarily map to the regions of FVIII associated with VWF (Figure 6).
We observed (i) that pdFVIII plus pdVWF presents fewer peptides than FL-rFVIII plus pdVWF and (ii) that the “protected” peptides do not necessarily map onto the regions of VWF-FVIII. These observations were consistent for all subjects and suggest that the folding and/or posttranslational modification of pdFVIII (which is synthesized at physiologic expression levels and in the physiologically relevant human tissue) are less susceptible to proteolytic processing and presentation by MoDCs. It is conceivable that the recombinant proteins overexpressed in a heterologous system exhibit greater microheterogeneity and less optimal folding and/or posttranslational modifications. Additionally, a recent report54 shows different glycosylation patterns in pdFVIII and FL-rFVIII, which could also contribute to the differential enzymatic processing of these 2 product classes. The VWF-mediated mechanisms by which internalization of FVIII is affected11,13 would not account for the differences observed in this study as both FL-rFVIII and pd-FVIII were incubated with dendritic cells in the presence of pdVWF.
Although our results are consistent with some clinical studies5-8 that show that rFVIII products may be more immunogenic than pdFVIII products, due to the small number of HA patients in this study it is not possible to determine whether peptides identified on MoDCs can be used for predicting clinical immunogenicity. It is unlikely that any technique used in isolation will be sufficient to design predictive algorithms. Nonetheless, the MAPPs assay generates a unique data set that provides information about both peptide processing and presentation. It will be necessary to integrate the MAPPs assay into clinical trials to fully understand the mechanistic underpinnings of clinical observations. MAPPs assays performed in conjunction with T-cell–based proliferation assays (using peptides identified in the MAPPs assay) and genotyping of patients (to determine which of the presented peptides are foreign) when carried out on clinical samples would plausibly provide key parameters for predictive algorithms for inhibitor development both at the individual and population levels. The former would be useful in the clinic to determine the best choice of medication and the latter in choosing between candidate drugs during drug development.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Laureen Kelley, Martha Hopewell, and Jodi Weeks of Save One Life for their generous gift of the FL-rFVIII and BDD-rFVIII used in this study.
Research conducted in the laboratory of Z.E.S. was supported by US Food and Drug Administration intramural grants from the Chief Scientist’s Challenge Grant and the Critical Path Initiative. This project was supported in part by an appointment of J.M. to the Research Participation Program at the Office of Tissues and Advanced Therapies, Center for Biologics Evaluation and Research, US Food and Drug Administration, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the US Food and Drug Administration. Research conducted in the laboratory of T.E.H. was supported in part by an investigator-initiated grant from CSL Behring to Haplomics Biotechnology, and relied on the use of University of Texas Rio Grande Valley facilities constructed with the support of National Institutes of Health, National Institute of General Medical Sciences grant C06 RR020537.
The findings and conclusions in this article have not been formally disseminated by the US Food and Drug Administration and should not be construed as representing any Agency determination or policy.
Authorship
Contribution: W.J. and J.M. helped in the design of the study and performed all data analyses; Y.P. and R.K. carried out the clinical studies, including recruitment of patients and preparation of clinical samples; E.M., M.H., and T.E.H. participated in the design of the experiments and writing of the manuscript; V.P.D. participated in the bioinformatics/statistical analyses and in the review and editing of the manuscript; B.W.L. oversaw the collection, storage, and shipping of the therapeutic FVIII proteins used in the MAPPs assay, and assisted in the review and editing of the manuscript; M.H. generated and characterized all protein samples for the MAPPs assay and helped in the drafting and editing of the manuscript; and N.S.K. and Z.E.S. designed the research plan, oversaw the project, and wrote the manuscript.
Conflict-of-interest disclosure: M.H. and E.M. are employees of CSL Behring. T.E.H. is the chief scientific officer for Haplomics Biotechnology Corporation. The remaining authors declare no competing financial interests.
Correspondence: Zuben E. Sauna, Center for Biologics Evaluation and Research, 10903 New Hampshire Ave, Building 52, Room 4120, Silver Spring, MD 20993; e-mail: zuben.sauna@fda.hhs.gov; or Nigel S. Key, Department of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC 27516; e-mail: nigel_key@med.unc.edu.