Abstract
Mice deficient for the transcription factor interferon consensus sequence binding protein (ICSBP) are immunodeficient and develop granulocytic leukemia. Further analyses indicated that ICSBP is a molecular switch factor directing the differentiation of bipotential myeloid precursors to the monocytic lineage. To reveal the molecular mechanisms responsible for the deregulation of myelopoiesis, we examined the signaling of the colony-stimulating factor 1 receptor (CSF-1R) in bone marrow–derived macrophages (BMMs) from ICSBP−/− mice. We found that in the absence of ICSBP, CSF-1R signaling is attenuated as seen from an accelerated termination of Erk phosphorylation and reduced cell growth. This finding coincides with an increased CSF-1R ubiquitination and an enhanced accumulation of c-Cbl. c-Cbl is an ubiquitin-ligase known to down-regulate activated CSF-1R by targeting it to the endocytic pathway. Our results indicate that upon CSF-1R activation, c-Cbl itself is partly proteolytically degraded in ICSBP+/+ but not in ICSBP−/−BMMs. Congruently, the expression of a major endosomal/lysosomal protease, cathepsin B, is strongly reduced in ICSBP−/− BMMs.
Introduction
During myelopoiesis, mature granulocytes and macrophages are generated from a common bipotential progenitor.1,2 The fate of a granulocytic/macrophage progenitor cell is determined by transcription factors that activate the expression of lineage-specific genes.3,4Identification of the critical transcriptional regulators is fundamental for our understanding of normal myeloid, as well as pathologic (eg, leukemogenic) cell development. So far, PU.1,5,6 C/EBPα,7,8 and MafB9have been defined as essential factors for myeloid differentiation. Recently, the transcription factor interferon consensus sequence binding protein (ICSBP), also termed interferon regulatory factor 8 (IRF-8), emerged as a novel myeloid regulator.10-12
Mice made deficient for ICSBP (ICSBP−/−) by gene targeting are immunodeficient and develop a syndrome similar to human chronic myelogenous leukemia (CML).10,13 These results revealed a novel role for ICSBP as a tumor suppressor that regulates proliferation of hematopoietic cells. Lack of or strongly reduced expression of ICSBP in patients with CML or acute myelogenous leukemia (AML) indicate a possible relevance of ICSBP also in human leukemias.14 This notion was supported by analyses of cytokine responses and extracellular matrix adhesion of myeloid progenitors from ICSBP−/− mice that exhibit several features in common with CML progenitors, and inferred the possibility of overlapping signaling pathways in these 2 disorders.10,11 This view was further enforced by the finding of reduced ICSBP expression in Bcr-Abl–induced CML-like disease in a mouse model. Importantly, the myeloproliferative disease was inhibited by enforced coexpression of ICSBP, thus providing direct evidence for ICSBP tumor suppressor activity.15 This result was in agreement with the most recent finding of Deng and Daley16 who reported that ectopic expression of ICSBP generated immunity against Bcr-Abl–induced leukemia in mice.
We have shown previously that the myeloid progenitors from ICSBP−/− mice have a strongly reduced growth response to colony-stimulating factor 1 (CSF-1) in vitro.11The colonies grown in the presence of CSF-1 consist mainly of granulocytes instead of monocytes. This asymmetric differentiation toward cells of the granulocytic lineage in vitro is paralleled by the expansion of granulocytes in leukemia of ICSBP−/− mice and indicates that ICSBP is a molecular switch factor acting on bipotential precursors, directing their differentiation to the monocytic pathway. Further evidence for the role of ICSBP in myeloid cell lineage selection was provided by Tamura et al,12 who showed that retrovirally transduced ICSBP directed macrophage differentiation of a myeloid progenitor cell line established from ICSBP−/− mice. Nevertheless, the molecular mechanisms of ICSBP regulatory functions remain unknown.
The results described by Scheller et al11 suggested that the hematopoietic alteration in ICSBP−/− mice could result from altered CSF-1 receptor (CSF-1R) expression or signaling. CSF-1, also known as M-CSF, regulates the survival, proliferation, and differentiation of mononuclear phagocytic cells by signaling through the CSF-1R,17 a tyrosine kinase encoded by the c-fms proto-oncogene.18 Ligand binding leads to autophosphorylation of the receptor, followed by an initiation of intracellular signal transduction cascades including the Ras/Erk and STAT pathways.19 One signaling protein known to interact with activated CSF-1R is p120 c-Cbl. Recently, ample evidence was provided for c-Cbl acting as an ubiquitin ligase (reviewed in Joazeiro and Weissman20) that terminates signaling of receptor tyrosine kinases (RTKs), including the CSF-1R.21-24
Here, we have investigated if the deregulated myelopoiesis observed in ICSBP−/− mice and the proposed function of ICSBP as a molecular switch of myeloid differentiation reflect an alteration in CSF-1R expression or CSF-1R signaling. The results of our study provide evidence for attenuated CSF-1R signaling in bone marrow–derived macrophages (BMMs) from ICSBP−/− mice, which results from a reduced proteolytic degradation of c-Cbl.
Materials and methods
Mice and cell cultures
ICSBP deficient mice were generated as described.10 All experiments were performed with 6-week-old to 7-week-old mice on a C57Bl/6×129/Sv F2 background. BMMs were prepared as described by Meraz et al25 with some modifications. Briefly, bone marrow cells were plated on 100-mm nontissue culture plates (Falcon, BD Biosciences, San Jose, CA) and cultivated in Dulbecco modified Eagle medium in the presence of 20 ng/mL recombinant murine CSF-1 for 7 days. Growth curves were generated as described by Lee et al.22 For inhibition of ubiquitin-dependent proteolysis, BMMs were maintained in 10 ng/mL CSF-1 for 16 hours, treated with protease inhibitors for an additional 6 hours, and harvested. Total cell extracts were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and western blot analysis. Inhibitors were used as follows: 25 μM N-acetyl-leucinyl-leucinylnorleucinal-H (LLnL) (Sigma, St. Louis, MO); 10 μM lactacystin (Calbiochem, San Diego, CA); 25 μM cathepsin B inhibitor 1 (Z-Phe-Ala-CH2F; Calbiochem); 25 μM calpain inhibitor V (Mu-Val-HPh-CH2F; Calbiochem). For stimulation of BMMs, recombinant murine interferon γ (IFN-γ) (R&D Systems, Minneapolis, MN) and lipopolysaccharide (LPS) (Sigma) were used.
Reagents and antibodies
Purified recombinant murine IFN-γ was purchased from Sigma and purified recombinant murine CSF-1 (rmCSF-1) was provided by R&D Systems. The anti-ICSBP antiserum was described by Rosenbauer et al.26 Anti–CSF-1R (rabbit polyclonal, C-20), antiubiquitin (mouse monoclonal, clone P4D1), and anti–c-Cbl (rabbit polyclonal, C-15) antibodies were obtained from Santa Cruz; anti–mitogen-activated protein (MAP) kinase antibodies were from Biolabs; anti–murine CSF-1R antiserum (rabbit polyclonal), anti–rat cathepsin B (rabbit polyclonal), and antiphosphotyrosine (clone 4G10) antibodies were from Upstate (Lake Placid, NY). Preabsorbed horseradish peroxidase–conjugated secondary antibodies and chemiluminescence Luminol Reagent were purchased from Santa Cruz (Santa Cruz, CA); Protein A– and Protein G–Agarose were from Roche (Mannheim, Germany).
Protein extracts
BMMs were washed with warm endotoxin-free phosphate buffered saline (PBS), trypsinized, subsequently washed again with medium and PBS, and pelleted. For western blot analysis, cells were lysed on ice in RIPA buffer (50 mM Tris-HCL, pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 50 mM NaCl, 10 mM ethylenediaminetetraacetic acid [EDTA], 1 mg/ml aprotinin, leupeptin, pepstatin, 1 mM Na3(VO)4, 1 mM phenylmethanesulfonyl fluoride [PMSF]).
Subcellular fractionation
BMMs were fractionated as described by Wang et al.27 Cells were harvested by trypsin as described above, suspended in 5 vol buffer C (10 mM HEPES-KOH, pH 7.6, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 1 mM EDTA, 1 mM ethyleneglycotetraacetic acid [EGTA]) supplemented with proteinase inhibitors (see above) and incubated on ice for 1 hour before passaging 20 times through a 22-gauge needle. Lysats were centrifuged in a microcentrifuge for 1 minute at 16 000gand the resulting crude nuclear pellets were extracted with an equal volume of buffer D (20 mM HEPES-KOH, pH 7.6, 25% glycerol, 0.5 M NaCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA) supplemented with proteinase inhibitors (see above) followed by a 30-minute centrifugation at 16 000g. The supernatants were designated as nuclear extracts. The supernatants from the initial centrifugation were further spun at 100 000g for 1 hour to obtain the cytosolic fractions (supernatant) and membrane fraction (pellet). The crude membrane pellets were washed in buffer D supplemented with proteinase inhibitors, recovered by another centrifugation step at 100 000g, and solubilized in RIPA buffer. Membrane fractions may contain insoluble cytoskeletal-associated proteins, whereas nuclear fractions may include perinuclear-associated structures.
Immunoprecipitations and Western blot analysis
For immunoprecipitations, cell extracts were precleared by incubation with 20 μL Protein A–Agarose for 1 hour. Immunoprecipitations were performed with 4 mg of the polyclonal IgG or a 1:100 dilution of the anti–CSF-1R antiserum and 25 μL Protein A–Agarose for polyclonal IgG or Protein G–Agarose for the antiserum for 4 hours. Precipitates were washed, and boiled in SDS sample buffer. Cell extracts or immunoprecipitates were analyzed by 7% to 10% SDS-PAGE. Proteins were transferred to nitrocellulose membranes and immunoblotting was performed with the antibodies as indicated in the figure legends. Quantitative analyses of blot scans were performed with EASYWin32 from Herolab (Wiesloch, Germany). The measured values were normalized for amounts of the precipitated protein.
Synthetic oligonucleotides, polymerase chain reaction, and RNA analyses
For reverse transcriptase–polymerase chain reaction (RT-PCR), the following primers were used: for c-Cbl, 5′CCTGGGGAGCAAGGGGAAAG 3′ (upper strand) and 5TGAGAGCTGCGGTGAGGGTG3′ (lower strand); for cathepsin B, 5′GCCCGACCATTGGACAGATTAGAG3′ (upper strand) and 5′TCCACTGGGCCATTTTTGTAGATT3′ (lower strand); for β-actin, 5′TGGAATCCTGTGGCATCCATGAAAC3′ (upper strand), 5′TAAAACGCAGCTCAGTAACAGTCC3′ (lower strand). For PCR analysis, 26 cycles of RT-PCR were used. For β-actin, 19 cycles of the following parameters were used: 24 seconds at 94°C; 22 seconds at 60°C; 36 seconds at 72°C. For c-Cbl and cathepsin B, 21 cycles of the following parameters were used: 30 seconds at 94°C; 30 seconds at 60°C; 35 seconds at 72°C.
Total RNA was extracted with Trizol reagent (Sigma). For northern blot analysis, 10 μg RNA per lane was loaded on a 1.2% agarose gel containing 0.4 M formaldehyde. The probe was generated using c-Cbl cDNA primers described above, and hybridization was carried out using ExpressHyb Hybridization Solution (Clontech, Palo Alto, CA) according to the manufacturer's recommendation. After stripping, the blot was rehybridized with a β-actin probe.
Results
Reduced growth of ICSBP−/− BMMs in response to CSF-1
To study the role of ICSBP in growth and differentiation of monocytic cells, we used BMMs that are responsive to CSF-1 and were employed recently to study CSF-1R signaling.22,28 BMMs from ICSBP−/− and control mice were prepared and their growth in the presence of CSF-1 was compared. Despite the higher cellularity of bone marrow from ICSBP−/−mice,11 the yield of BMMs per femur from ICSBP−/− mice was constantly lower (1.2 ± 0.2 × 106 BMMs per 23 ± 4 × 106 cells plated) in comparison to control mice (3.0 ± 0.8 × 106 BMMs per 14 ± 4 × 106 cells plated). However, BMMs derived from ICSBP−/− and ICSBP+/+ mice are similar regarding general morphology and expression of the representative monocytic markers FcγRIII and F4/80 as revealed by fluorescence-assisted cell sorting (data not shown). To analyze the cell growth, differentiated BMMs were cultured for 8 days with CSF-1 and cell counts were monitored (Figure1). The growth curves demonstrate approximately 3-fold lower counts of BMMs from ICSBP−/−as compared to ICSBP+/+. Thus, similarly to previous studies in myeloid progenitor cells,11 CSF-1–dependent growth of BMMs is impaired in the absence of ICSBP. BMMs therefore provide an attractive system to analyze the role of ICSBP in CSF-1R signaling.
Reduced growth of ICSBP−/− BMMs in response to CSF-1.
Day 5 BMMs were seeded at 1 × 105 cells per plate and cultured in the presence of 20 ng/mL CSF-1. Cells were fed every 2 days for 8 days. At the indicated time points cells from triplicate plates were trypsinized and counted.
Reduced growth of ICSBP−/− BMMs in response to CSF-1.
Day 5 BMMs were seeded at 1 × 105 cells per plate and cultured in the presence of 20 ng/mL CSF-1. Cells were fed every 2 days for 8 days. At the indicated time points cells from triplicate plates were trypsinized and counted.
Unaltered expression and phosphorylation but increased ubiquitination of CSF-1R in ICSBP−/− BMMs
To distinguish whether the reduced CSF-1 response of ICSBP−/− BMMs is due to reduced receptor expression or ligand/receptor interaction, CSF-1R expression and phosphorylation were analyzed by western blotting. A comparable expression of p165, the mature receptor form, is seen in both ICSBP+/+ and ICSBP−/− BMMs. However, a degradation product of 105 kd, which has been described before,22 is more prominent in ICSBP−/− BMMs, suggesting a higher receptor turnover (Figure 2A).
Expression, autophosphorylation, and ubiquitination of CSF-1R in BMMs from ICSBP+/+ and ICSBP−/− mice.
(A) Total cell extracts from 5 × 105 BMMs cultured in the presence of 20 ng/mL CSF-1 (as described in “Materials and methods”) were loaded per lane and probed with an anti–CSF-1R antiserum by western blotting (upper panel). Equal loading of the lanes was confirmed by detection of an unspecific band, p80, with the ICSBP antibody (lower panel). (B) BMMs were deprived of CSF-1 for 16 hours, followed by stimulation with 100 ng/mL CSF-1 at 37°C for the indicated times. After incubation, cells were lysed and immunoprecipitated with an anti–CSF-1R antiserum. Immuncomplexes were analyzed by western blotting either with antiphosphotyrosine antibody (upper panel), antiubiquitin antibody (middle panel), or anti–CSF-1R antiserum (lower panel). In the CSF-1 western blot the position of the band corresponding to the 150-kd isoform of CSF-1 is indicated as a loading control.
Expression, autophosphorylation, and ubiquitination of CSF-1R in BMMs from ICSBP+/+ and ICSBP−/− mice.
(A) Total cell extracts from 5 × 105 BMMs cultured in the presence of 20 ng/mL CSF-1 (as described in “Materials and methods”) were loaded per lane and probed with an anti–CSF-1R antiserum by western blotting (upper panel). Equal loading of the lanes was confirmed by detection of an unspecific band, p80, with the ICSBP antibody (lower panel). (B) BMMs were deprived of CSF-1 for 16 hours, followed by stimulation with 100 ng/mL CSF-1 at 37°C for the indicated times. After incubation, cells were lysed and immunoprecipitated with an anti–CSF-1R antiserum. Immuncomplexes were analyzed by western blotting either with antiphosphotyrosine antibody (upper panel), antiubiquitin antibody (middle panel), or anti–CSF-1R antiserum (lower panel). In the CSF-1 western blot the position of the band corresponding to the 150-kd isoform of CSF-1 is indicated as a loading control.
We next investigated the autophosphorylation of the CSF-1R after stimulation of the cells with CSF-1. As seen in the upper panel of Figure 2B, phosphorylated forms of CSF-1R appear with similar kinetics in both types of cells, although the intensity of phosphorylated p165 in ICSBP−/− BMMs is reduced. Additionally, antiphosphotyrosine (anti-pY)–stained bands migrating slower than pp165 are more prominent in ICSBP−/− BMMs. However, quantitative analysis did not reveal significant differences in the total amount of phosphorylated CSF-1R forms between ICSBP+/+ and ICSBP−/− BMMs.
It has been shown that CSF-1 stimulation results not only in rapid tyrosine phosphorylation but is followed by multiubiquitination of the CSF-1R.29 Reprobing of the blot with an antiubiquitin antibody (Figure 2B, middle panel) confirmed that the slower migrating bands represent multiubiquitinated forms of CSF-1R. A quantitative evaluation of the blot scan indicated a 2- to 3-fold enhanced ubiquitination of CSF-1R in ICSBP−/− BMMs.
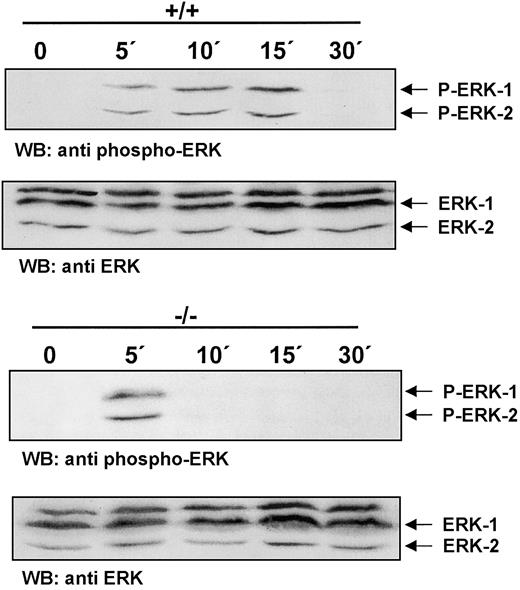
Rapid termination of CSF-1R signaling in ICSBP−/− BMMs
Ligand binding by CSF-1R triggers a signaling cascade that leads to the activation of the Ras/Erk pathway.28 To test whether CSF-1R signaling in ICSBP-deficient BMMs proceeds to the distal end of the Ras/Erk signaling pathway, activation of Erk-1/Erk-2 was measured. Striking differences in the kinetics of Erk phosphorylation in ICSBP+/+ and ICSBP−/− BMMs in response to CSF-1 are documented in Figure 3. Both types of cells respond to CSF-1 by initial phosphorylation of Erk-1/Erk-2 seen within 5 minutes of stimulation. Erk-1/Erk-2 phosphorylated forms persist unchanged in ICSBP+/+ BMMs for up to 15 minutes. However, in ICSBP−/− BMMs a strong reduction of Erk-1/Erk-2 phosphorylation can be observed already at 10 minutes after stimulation. Thus, our results demonstrate a rapid termination of CSF-1R signaling in ICSBP−/−cells.
Rapid termination of CSF-1R signaling in ICSBP−/− BMMs.
BMMs were deprived of CSF-1 for 16 hours following stimulation with 200 ng/mL CSF-1 at 37°C for the indicated time points. Total extracts were subjected to SDS-PAGE and western blot analyses with an antiphospho-Erk-1/2 antibody (upper panel). To confirm equal protein loading the membrane was stripped and blotted with an anti–Erk-1/2 antibody (lower panel).
Rapid termination of CSF-1R signaling in ICSBP−/− BMMs.
BMMs were deprived of CSF-1 for 16 hours following stimulation with 200 ng/mL CSF-1 at 37°C for the indicated time points. Total extracts were subjected to SDS-PAGE and western blot analyses with an antiphospho-Erk-1/2 antibody (upper panel). To confirm equal protein loading the membrane was stripped and blotted with an anti–Erk-1/2 antibody (lower panel).
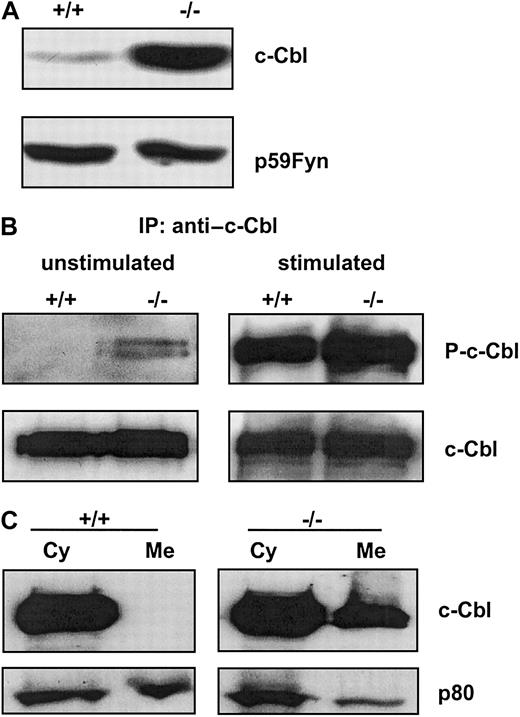
Constitutive overexpression and membrane association of c-Cbl in BMMs from ICSBP−/− mice
The results in Figure 2 suggest that enhanced ubiquitination is a possible mechanism for the impaired CSF-1R signaling in ICSBP−/− BMMs. Therefore, we analyzed the expression of c-Cbl, an ubiquitin ligase, and a well-defined negative regulator of RTKs.21-24 In total cell extracts of BMMs from ICSBP+/+ mice, only a low expression level of c-Cbl is detected whereas BMMs from ICSBP−/− mice display constitutively high expression levels of c-Cbl protein (Figure4A).
Expression, phosphorylation, and subcellular distribution of c-Cbl in ICSBP+/+ and ICSBP−/− BMMs.
Cells were deprived of CSF-1 for 16 hours, harvested, and lysed in RIPA buffer. (A) Total cell extracts of 5 × 105 cells were subjected to western blotting with anti–c-Cbl antibody. Anti-Fyn antibody was used as control. (B) BMMs deprived of CSF-1 for 16 hours were left untreated or were stimulated for 4 minutes with 200 ng/mL CSF-1 at 37°C. Total cell extracts (1.5 mg) were used for immunoprecipitation with a c-Cbl antibody. The precipitates were subjected to western blot analyses with an antiphospho-tyrosine antibody (upper panels). After stripping the membrane was probed with an anti–c-Cbl antibody (lower panels). (C) BMMs continuously grown in the presence of 20 ng/mL CSF-1 were fractionated as described in “Materials and methods.” A quantity of 25 μg of membrane (Me) and cytosolic fraction (Cy) were subjected to western blotting with an anti–c-Cbl antibody (upper panel). Equal protein loading was controlled by the detection of p80 with the anti-ICSBP antiserum (lower panel).
Expression, phosphorylation, and subcellular distribution of c-Cbl in ICSBP+/+ and ICSBP−/− BMMs.
Cells were deprived of CSF-1 for 16 hours, harvested, and lysed in RIPA buffer. (A) Total cell extracts of 5 × 105 cells were subjected to western blotting with anti–c-Cbl antibody. Anti-Fyn antibody was used as control. (B) BMMs deprived of CSF-1 for 16 hours were left untreated or were stimulated for 4 minutes with 200 ng/mL CSF-1 at 37°C. Total cell extracts (1.5 mg) were used for immunoprecipitation with a c-Cbl antibody. The precipitates were subjected to western blot analyses with an antiphospho-tyrosine antibody (upper panels). After stripping the membrane was probed with an anti–c-Cbl antibody (lower panels). (C) BMMs continuously grown in the presence of 20 ng/mL CSF-1 were fractionated as described in “Materials and methods.” A quantity of 25 μg of membrane (Me) and cytosolic fraction (Cy) were subjected to western blotting with an anti–c-Cbl antibody (upper panel). Equal protein loading was controlled by the detection of p80 with the anti-ICSBP antiserum (lower panel).
Since tyrosine phosphorylation of c-Cbl was shown to promote its ubiquitin ligase activity and in turn to contribute to the degradation of CSF-1R,22 we analyzed the phosphorylation status of c-Cbl in ICSBP−/− BMMs as well as control cells. Figure4B demonstrates that c-Cbl was tyrosine-phosphorylated in response to CSF-1 stimulation in BMMs of both ICSBP−/− and control mice. Remarkably, a fraction of c-Cbl in ICSBP−/− BMMs was found to be phosphorylated prior to CSF-1 stimulation, suggesting a constitutively higher basal c-Cbl activity in these cells.
c-Cbl associates with activated RTKs and promotes their ubiquitination, during which process cytoplasmic c-Cbl is relocated to the plasma membrane and to early endosomes.22 24 We have therefore fractionated cell extracts and compared the distribution of c-Cbl in cytoplasmic and membrane fractions of ICSBP−/− and ICSBP+/+ BMMs. Figure 4C indicates that the amount of the membrane-bound c-Cbl is increased in ICSBP−/− BMMs as compared with controls, whereas the cytoplasmic level of c-Cbl remains approximately the same. The augmented level of c-Cbl in ICSBP−/− BMMs is also reflected in enhanced ubiquitination of CSF-1R as demonstrated in Figure 2B.
Together these results provide an explanation for the rapid termination of CSF-1R signaling and hence for reduced proliferation of ICSBP−/− BMMs.
c-Cbl is a target of proteolytic degradation in ICSBP+/+ but not in ICSBP−/− BMMs
To reveal the molecular mechanism responsible for accumulation of c-Cbl in ICSBP−/− BMMs, we first asked whether c-Cbl is a direct target of ICSBP. In contrast to the increased protein level no differences in the amount of c-Cbl mRNA transcripts were revealed by northern blot analysis of BMMs from ICSBP−/− and ICSBP+/+ mice (Figure 5A). This result pointed to altered posttranscriptional regulation of c-Cbl in ICSBP−/− BMMs.
Posttranscriptional regulation of c-Cbl.
(A) Unchanged levels of c-Cbl mRNA in BMMs from ICSBP+/+and ICSBP−/− mice. Isolation of RNA and northern blot analysis was performed as described in “Materials and methods.” The positions of the 10.5-kb c-Cbl transcript, 28S rRNA, and β-actin mRNA are indicated. (B-D) Effects of protease inhibitors on the expression of c-Cbl. (B) ICSBP+/+ and ICSBP−/− BMMs were grown in the presence of 20 ng/mL CSF-1 and treated with the protease inhibitor LLnL (25 μM) for 6 hours. Total cell extracts were analyzed by SDS-PAGE and western blotting with the anti–c-cbl antibody. Extracts from 5 × 105 cells were used per lane. Equal loading of the lanes was confirmed by detection of p80 and ICSBP with the anti-ICSBP antiserum. (C) ICSBP+/+ BMMs were treated with different protease inhibitors (25 μM) for 6 hours. Western blot analysis of the total cell extracts was performed with the antibodies as indicated. Lane 1: cathepsin B inhibitor (Z-Phe-Ala-CH2F); lane 2: calpain inhibitor (Mu-Val-HPh-CH2F); lane 3: lactacystin; lane 4: control (dimethyl sulfoxide [DMSO] solvent). Equal loading of the lanes was confirmed by detection of ICSBP. (D) ICSBP+/+ BMMs were treated as in B and subcellular fractionation was performed as described in “Materials and methods.” Protein fractions (50 μg) were analyzed by SDS-PAGE and western blotting with the anti–c-Cbl antibody. Equal loading of the lanes was confirmed with detection of p80 and ICSBP by the anti-ICSBP antiserum.
Posttranscriptional regulation of c-Cbl.
(A) Unchanged levels of c-Cbl mRNA in BMMs from ICSBP+/+and ICSBP−/− mice. Isolation of RNA and northern blot analysis was performed as described in “Materials and methods.” The positions of the 10.5-kb c-Cbl transcript, 28S rRNA, and β-actin mRNA are indicated. (B-D) Effects of protease inhibitors on the expression of c-Cbl. (B) ICSBP+/+ and ICSBP−/− BMMs were grown in the presence of 20 ng/mL CSF-1 and treated with the protease inhibitor LLnL (25 μM) for 6 hours. Total cell extracts were analyzed by SDS-PAGE and western blotting with the anti–c-cbl antibody. Extracts from 5 × 105 cells were used per lane. Equal loading of the lanes was confirmed by detection of p80 and ICSBP with the anti-ICSBP antiserum. (C) ICSBP+/+ BMMs were treated with different protease inhibitors (25 μM) for 6 hours. Western blot analysis of the total cell extracts was performed with the antibodies as indicated. Lane 1: cathepsin B inhibitor (Z-Phe-Ala-CH2F); lane 2: calpain inhibitor (Mu-Val-HPh-CH2F); lane 3: lactacystin; lane 4: control (dimethyl sulfoxide [DMSO] solvent). Equal loading of the lanes was confirmed by detection of ICSBP. (D) ICSBP+/+ BMMs were treated as in B and subcellular fractionation was performed as described in “Materials and methods.” Protein fractions (50 μg) were analyzed by SDS-PAGE and western blotting with the anti–c-Cbl antibody. Equal loading of the lanes was confirmed with detection of p80 and ICSBP by the anti-ICSBP antiserum.
In order to investigate whether c-Cbl is a target of proteolytic regulation different protease inhibitors were used. The stability of c-Cbl from ICSBP+/+ BMMs is unaffected by lactacystin (Figure 5C), an inhibitor that was shown to be highly specific for proteasomal protein degradation.30 However, accumulation of p120 c-Cbl is observed in ICSBP+/+ BMMs treated with LLnL, an inhibitor of calpains and cysteine proteases, including cathepsins (Figure 5B), and methylamine, a specific inhibitor of lysosomal proteases (data not shown). Additionally, accumulation of c-Cbl was found in cells treated with an inhibitor of cathepsin B, a major endosomal/lysosomal cysteine protease, and another calpain-specific inhibitor (Figure 5C). Interestingly, none of these inhibitors affected the elevated level of c-Cbl in ICSBP−/− BMMs (data not shown). Thus, our results indicate that c-Cbl is degraded in BMMs and that the accountable proteolytic activity is reduced in ICSBP−/− cells.
We next tested whether deregulated proteolytical degradation could also account for the increased membrane accumulation of c-Cbl observed in ICSBP−/− BMMs. Subcellular fractionation of ICSBP+/+ BMMs treated with protease inhibitor LLnL reveals that, similar to results obtained in ICSBP−/− BMMs, c-Cbl accumulates in the membrane fraction whereas LLnL has no effect on the cytosolic fraction (Figure 5D).
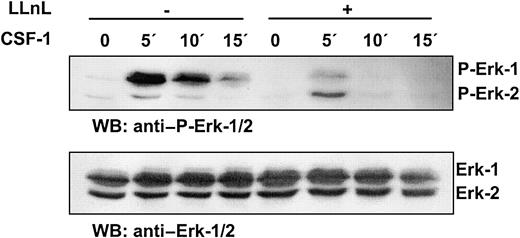
Interestingly, treatment of BMMs with protease inhibitor LLnL directly affected phosphorylation of Erk-1/2, providing further support for the link between accumulation of c-Cbl and reduced CSF-1R signaling (Figure6).
Activation of Erk-1/2 in response to CSF-1 is diminished by protease inhibitor LLnL.
BMMs from ICSBP+/+ mice were incubated in the presence or absence of 25 μM LLnL and deprived of CSF-1 simultaneously for 12 hours. The cells were then stimulated with 100 ng/mL CSF-1 for the times indicated.
Activation of Erk-1/2 in response to CSF-1 is diminished by protease inhibitor LLnL.
BMMs from ICSBP+/+ mice were incubated in the presence or absence of 25 μM LLnL and deprived of CSF-1 simultaneously for 12 hours. The cells were then stimulated with 100 ng/mL CSF-1 for the times indicated.
Expression of cathepsin B is down-regulated in ICSBP−/− BMMs
In order to rationalize the reduced degradation of c-Cbl in ICSBP−/− BMMs, we investigated the expression of endosomal/lysosomal cysteine protease cathepsin B. As revealed by both RT-PCR and western blotting, cathepsin B expression was strongly reduced in BMMs from ICSBP−/− mice at both mRNA and protein levels (Figure 7A-B). It was previously reported that cathepsin B is induced by IFN-γ in human monocytes.31 Since IFN-γ strongly induces ICSBP expression,32 this observation suggested that cathepsin B transcription could be regulated by ICSBP. This notion was further supported by the experiment (shown in Figure 7B) which demonstrated up-regulation of cathepsin B protein levels, following induction of ICSBP expression by IFN-γ and LPS in ICSBP+/+ but not ICSBP−/− BMMs. The fact that this effect was not observed in the absence of ICSBP indicates that the up-regulation of cathepsin B expression by IFN-γ is dependent on ICSBP.
Expression of lysosomal protease cathepsin B is positively regulated by ICSBP.
(A) Cytosolic RNA was isolated from BMMs and analyzed by RT-PCR using specific primers for cathepsin B cDNA. (B) BMMs were cultured with 20 ng/mL CSF-1 and stimulated with 200 U/mL IFN-γ and 100 ng/mL LPS or mock-stimulated for 24 hours. Total cell extracts of 5 × 105 cells per lane were subjected to western blotting with an anti–cathepsin B antibody. Equal protein loading was confirmed by blotting with an anti-ICSBP antiserum detecting ICSBP and p80.
Expression of lysosomal protease cathepsin B is positively regulated by ICSBP.
(A) Cytosolic RNA was isolated from BMMs and analyzed by RT-PCR using specific primers for cathepsin B cDNA. (B) BMMs were cultured with 20 ng/mL CSF-1 and stimulated with 200 U/mL IFN-γ and 100 ng/mL LPS or mock-stimulated for 24 hours. Total cell extracts of 5 × 105 cells per lane were subjected to western blotting with an anti–cathepsin B antibody. Equal protein loading was confirmed by blotting with an anti-ICSBP antiserum detecting ICSBP and p80.
Discussion
In this study we have investigated the molecular mechanisms underlying the proposed function of ICSBP as a molecular switch of myeloid differentiation. Since our previous experiments provided evidence for an altered CSF-1 response of ICSBP-deficient myeloid progenitors,11 we have analyzed CSF-1R expression and signaling in CSF-1-responsive BMMs from ICSBP−/−mice.
Despite unchanged CSF-1R expression or autophosphorylation after ligand engagement, there is a rapid termination of CSF-1R signaling in ICSBP−/− BMMs, as is evident from analysis of cell growth and kinetics of Erk-1/Erk-2 phosphorylation. The molecular explanation for down-regulated CSF-1R signaling is provided by our finding of strongly enhanced membrane accumulation of c-Cbl and accelerated CSF-1R ubiquitination in response to CSF-1 binding in the absence of ICSBP.
Proto-oncogene c-Cbl is the cellular homologue of the retroviral oncogene v-Cbl that is known to induce B-cell lymphoma and myeloid leukemia in mice.33 c-Cbl is expressed mainly in hematopoietic cells, and is associated with negative as well as positive signaling of several growth factors and cytokines (reviewed in Thien and Langdon34). The evidence for a negative regulatory function of c-Cbl in receptor signaling was first shown for the C elegans c-Cbl ortholog, Sli-1.35 The role of c-Cbl in attenuation of tyrosine receptor signaling by epidermal growth factor (EGF), platelet derived growth factor (PDGF), CSF-1, and Her2/Neu was established by further work from several laboratories.22,23,36 c-Cbl has been shown to be an ubiquitin ligase, targeting substrates for ubiquitination (reviewed by Joazeiro and Weissman20). c-Cbl binds to activated receptor tyrosine kinases via its SH2-like domain, and promotes signaling termination by accelerated endocytosis and proteasomal or lysosomal degradation of ligand-receptor complexes.
The negative regulation of cell growth by c-Cbl was also confirmed by targeted mutagenesis in mice.22,37,38 The ubiquitin-ligase activity of c-Cbl has been shown to be directly responsible for down-regulation of the CSF-1R.22Multiubiquitination of the CSF-1R was decreased and the internalization of CSF-1–CSF-1R complexes was significantly delayed in c-Cbl−/− macrophages.22
The enhanced ubiquitination and degradation of the CSF-1R and the rapid termination of ERK phosphorylation in c-Cbl–overexpressing BMMs from ICSBP−/− mice are in accord with previous results using alternative experimental systems. Enforced expression of c-Cbl leads to a faster ligand-induced ubiquitination and degradation of the PDGF-alpha and the Her2/Neu receptor tyrosine kinases.23,36 Consistent with previous observations by Lee et al,22 who showed that BMMs from c-Cbl−/−mice proliferate faster and form denser colonies as compared with those from control mice, ICSBP−/− BMMs that overexpress c-Cbl proliferate more slowly (Figure 1) and form less-dense colonies than ICSBP+/+ BMMs (data not shown).
To elucidate the mechanism leading to enhanced accumulation of c-Cbl in ICSBP−/− BMMs, we examined whether the c-Cbl gene is directly regulated by the transcription factor ICSBP. The observation that c-Cbl transcript levels were not affected by the loss of ICSBP pointed to a deregulated intracellular turnover of c-Cbl in ICSBP−/− BMMs. Experiments using a series of different protease inhibitors showed that the proteolytic degradation of c-Cbl was blocked by inhibitors of both lysosomal proteases (including cathepsin B) and calpains. Subcellular fractionation revealed that in cells treated with protease inhibitors, c-Cbl accumulates in the membrane fraction, which parallels the situation observed in ICSBP−/− BMMs. These results imply that part of the c-Cbl membrane fraction is degraded in the course of CSF-1R signaling, and suggest that the observed accumulation of c-Cbl in ICSBP−/− BMMs is due to reduced protease activity. Thus, the increased accumulation of c-Cbl in ICSBP−/− BMMs leads to the rapid attenuation of CSF-1R signaling and reduced cell growth.
Given the important regulatory function of c-Cbl in receptor signaling, elucidation of the parameters that control cellular levels of c-Cbl is of general interest. Noteably, IFN-γ is known as a potent regulator of immunoproteasomal proteases involved in antigen processing,39 as well as other proteases including cathepsins and calpains.31,40,41 Our results suggest that the down-regulation of cathepsin B in the absence of ICSBP may directly contribute to the increased accumulation of c-Cbl in BMMs. Interestingly, cathepsin B was shown to be involved in endosomal/lysosomal degradation of another receptor tyrosine kinase, epidermal growth factor receptor (EGF-R).42 Furthermore, c-Cbl remains associated with the EGF-R during the endocytic pathway43 and thus is accessible to endosomal/lysosomal proteases. Most recently, evidence was presented for coordinated degradation of the Cbl-b and EGF-R proteins.44
As shown recently by Kondo et al,45 down-regulation of cytokine receptors is an important mechanism in the control of hematolymphopoiesis. Our results strongly suggest that the same molecular mechanism that affects CSF-1R signaling in ICSBP−/− BMMs also contributes to the aberrant myelopoiesis in ICSBP−/− mice. It is likely that disproportional differentiation of bipotential granulocytic/monocytic progenitors toward the granulocytic lineage in the absence of ICSBP10-12 results from attenuated CSF-1R signaling and reduced proliferation of monocytic cells.
We thank Drs S. Feller, C. Huettner, P. Kloetzel, and J. Selfe for stimulating discussions and comments on the manuscript, and M. Wietstruk for excellent technical help.
Supported by the Deutsche Forschungsgemeinschaft (SFB 506) and from Fonds der Chemischen Industrie.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ivan Horak, Department of Molecular Genetics, Institute of Molecular Pharmacology, Krahmerstrasse 6, 12207 Berlin, Germany; e-mail: horak@fmp-berlin.de.



![Fig. 5. Posttranscriptional regulation of c-Cbl. / (A) Unchanged levels of c-Cbl mRNA in BMMs from ICSBP+/+and ICSBP−/− mice. Isolation of RNA and northern blot analysis was performed as described in “Materials and methods.” The positions of the 10.5-kb c-Cbl transcript, 28S rRNA, and β-actin mRNA are indicated. (B-D) Effects of protease inhibitors on the expression of c-Cbl. (B) ICSBP+/+ and ICSBP−/− BMMs were grown in the presence of 20 ng/mL CSF-1 and treated with the protease inhibitor LLnL (25 μM) for 6 hours. Total cell extracts were analyzed by SDS-PAGE and western blotting with the anti–c-cbl antibody. Extracts from 5 × 105 cells were used per lane. Equal loading of the lanes was confirmed by detection of p80 and ICSBP with the anti-ICSBP antiserum. (C) ICSBP+/+ BMMs were treated with different protease inhibitors (25 μM) for 6 hours. Western blot analysis of the total cell extracts was performed with the antibodies as indicated. Lane 1: cathepsin B inhibitor (Z-Phe-Ala-CH2F); lane 2: calpain inhibitor (Mu-Val-HPh-CH2F); lane 3: lactacystin; lane 4: control (dimethyl sulfoxide [DMSO] solvent). Equal loading of the lanes was confirmed by detection of ICSBP. (D) ICSBP+/+ BMMs were treated as in B and subcellular fractionation was performed as described in “Materials and methods.” Protein fractions (50 μg) were analyzed by SDS-PAGE and western blotting with the anti–c-Cbl antibody. Equal loading of the lanes was confirmed with detection of p80 and ICSBP by the anti-ICSBP antiserum.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/9/10.1182_blood.v99.9.3213/6/m_h80922464005.jpeg?Expires=1770743003&Signature=XA0qdiLqJhQMaxbfZsvtPrfTbJPtmu9gt8pTpHMABe0EwOlbsVSl1d4i0~OdGlHVcKnvlYjqME-WP75kPsr~suzT~5m1g~KWaWndLjctEEf~5nDAaXHMa2YtmmtnUghsX4IiFxOIs3D4IZfm2hwhpJt~evOMl8pQkvgBWqCfjJ6laCzBtnHFKamLSLzC8XRsNSsONTBdZBADOwNt1w2NYnWppHhG9rRQNCyN4xKXm8yLmpSg1RD-~zBjW3nvQdc6BiWb3jp9~KQRkiBx3T7~YSpVeQA5qFGxbxCpcy4NnNglP1JOOv8ID-ewtqgceTNQS-UBw97-R~eC2NeSOw9ISg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal