During the past few years an avalanche of papers describing the conversion of transplanted bone marrow cells into skeletal muscle, heart, liver, brain, and various epithelial tissues suggested a surprising degree of hematopoietic stem cell plasticity. For the clinician they raised the expectation that adult hematopoietic stem cells might one day become useful for tissue replacement therapies and in ethically less controversial ways than embryonic stem cells. For the developmental hematologist these findings were more incremental in nature, because evidence accumulated over the last 2 decades indicated that hematopoietic cells can be induced to convert from one lineage into another. Although a number of excellent reviews have covered the general topic of stem cell plasticity1-5 (also seehttp://www.nih.gov/news/stemcell/scireport.html), none has primarily focused on hematopoietic cells, and little has been written about intrahematopoietic or “lineage” plasticity. The present review aims to fill this gap. In the first part I will discuss the capacity of cultured blood cells to transdifferentiate from one lineage to another and present a model of how this might happen. In the second part I will review experiments that show that bone marrow cells can apparently turn into almost any tissue and discuss the question whether or not the cells that “switch” represent bona fide hematopoietic cells.
Lineage switches within the hematopoietic system
Current models of hematopoietic differentiation state that stem cells become gradually restricted in their differentiation potential by a succession of symmetric and asymmetric divisions. Each intermediate progenitor and each monopotent precursor has a distinct gene expression program that is established and maintained by specific combinations of transcription factors and chromatin remodeling components. It is generally assumed that once a cell commits to a given lineage and acquires a defined phenotype, it can no longer change its fate. However, as summarized in Table 1 and Figure 1, and as will be elaborated in the following section, there are numerous examples where seemingly committed hematopoietic cells can be induced to convert into cells of another lineage.
Induced hematopoietic lineage switches
| Cell type of origin . | Resulting cell type . | Type of manipulation . | Reference . |
|---|---|---|---|
| Pre–B lymphoma cell line | Macrophages | 5-azacytidine treatment | 6 |
| Eu-Myc B lymphoma lines | Macrophages | v-raf expr | 7 |
| Pre–B cell lines | Macrophages | v-fms, CSF1-R plus CSF1 | 8 |
| Pre–B lymphoma | Neutrophil granulocytes | Transplantation | 9 |
| Pax-5−/− pre–B cells | Mac/Gr, dendrites, NK cells | Different cytokines | 11 |
| Pax-5−/− pre–B cells | As above, plus T cells | Transplanted into Rag2−/− | 10 |
| Lymphoid prog. (CLPs) | GM-CSF | IL-2 Rα expr, IL-2 | 13 |
| CD19+ B220−cells | Macrophages | Myeloid culture conditions | 14 |
| Pro–T cells | Macrophages | Conditioned medium | 22 |
| Macrophage cell line (70Z/3) | B-lineage cells | E2A, EBF expr | 15 |
| MEP cells | Myeloid cells | Inducible PU.1 expr | 30 |
| MEP cells | Eosinophils | Inducible C/EBP expr | 31,41 |
| Myeloid cell line (HD50M) | Eosinophils | Inducible GATA-1 expr | 32 |
| Myeloid cell line (HD50M) | MEPs; erythrocytes | High GATA-1 expr; ts Ets | 32 |
| Eosinophil cell line (1A1) | MEP cells | FOG-1 expr | 33 |
| Erythroid cell line (MEL) | Macrophagelike cells | Inducible PU.1 expr | 34 |
| Myeloid cell line (416B) | Megakaryocytic cells | GATA-1, GATA-2 expr | 35 |
| Myeloid cell line (FDCW2) | Erythroid cells | GATA-1 expr | 36 |
| Myeloid cell line (M1) | Megakar, erythroid cells | GATA-1 expr | 37 |
| GM precursors | Ery, eosin, basoph cells | Inducible GATA-1 | T. Enver pers comm., September 2001 |
| Cell type of origin . | Resulting cell type . | Type of manipulation . | Reference . |
|---|---|---|---|
| Pre–B lymphoma cell line | Macrophages | 5-azacytidine treatment | 6 |
| Eu-Myc B lymphoma lines | Macrophages | v-raf expr | 7 |
| Pre–B cell lines | Macrophages | v-fms, CSF1-R plus CSF1 | 8 |
| Pre–B lymphoma | Neutrophil granulocytes | Transplantation | 9 |
| Pax-5−/− pre–B cells | Mac/Gr, dendrites, NK cells | Different cytokines | 11 |
| Pax-5−/− pre–B cells | As above, plus T cells | Transplanted into Rag2−/− | 10 |
| Lymphoid prog. (CLPs) | GM-CSF | IL-2 Rα expr, IL-2 | 13 |
| CD19+ B220−cells | Macrophages | Myeloid culture conditions | 14 |
| Pro–T cells | Macrophages | Conditioned medium | 22 |
| Macrophage cell line (70Z/3) | B-lineage cells | E2A, EBF expr | 15 |
| MEP cells | Myeloid cells | Inducible PU.1 expr | 30 |
| MEP cells | Eosinophils | Inducible C/EBP expr | 31,41 |
| Myeloid cell line (HD50M) | Eosinophils | Inducible GATA-1 expr | 32 |
| Myeloid cell line (HD50M) | MEPs; erythrocytes | High GATA-1 expr; ts Ets | 32 |
| Eosinophil cell line (1A1) | MEP cells | FOG-1 expr | 33 |
| Erythroid cell line (MEL) | Macrophagelike cells | Inducible PU.1 expr | 34 |
| Myeloid cell line (416B) | Megakaryocytic cells | GATA-1, GATA-2 expr | 35 |
| Myeloid cell line (FDCW2) | Erythroid cells | GATA-1 expr | 36 |
| Myeloid cell line (M1) | Megakar, erythroid cells | GATA-1 expr | 37 |
| GM precursors | Ery, eosin, basoph cells | Inducible GATA-1 | T. Enver pers comm., September 2001 |
Prog indicates, progenitor; expr, expression; megakary, megakaryocyte; ery, erythrocyte; eosin, eosinophil; basoph, basophil; and pers comm, personal communication.
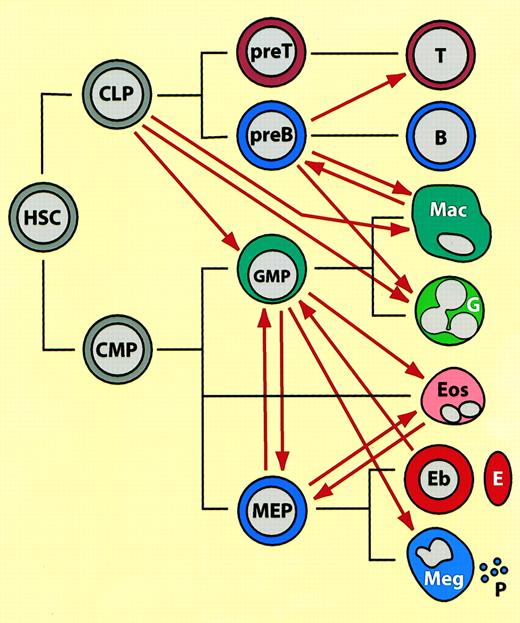
Summary of lineage switches observed between hematopoietic cells, placed in the context of normal hematopoietic differentiation.44 45
(See text for details). The black lines indicate normal lineage relationships, the thick red arrows represent induced switches. (These do not necessarily imply direct transitions.) HSC indicates hematopoietic stem cell; CLP, common lymphoid progenitor; CMP, common multipotent progenitor; GMP, granulocyte/macrophage progenitor (in the text also called “myeloblasts”); MEP, megakaryocyte erythrocyte progenitor; T, T lymphocyte; B, B lymphocyte; Mac, macrophage; G, neutrophil granulocyte; Eos, eosinophil, Eb, erythroblast; E, erythrocyte; Meg, megakaryocyte; P, platelet. Mast cells, NK cells, and dendritic cells have been omitted from the scheme and placement of eosinophils is speculative. Note that most of the switches were observed with transformed cell lines in culture and that the cell type designations (both before and after the switch) may not accurately reflect the phenotype of the normal counterparts.
Summary of lineage switches observed between hematopoietic cells, placed in the context of normal hematopoietic differentiation.44 45
(See text for details). The black lines indicate normal lineage relationships, the thick red arrows represent induced switches. (These do not necessarily imply direct transitions.) HSC indicates hematopoietic stem cell; CLP, common lymphoid progenitor; CMP, common multipotent progenitor; GMP, granulocyte/macrophage progenitor (in the text also called “myeloblasts”); MEP, megakaryocyte erythrocyte progenitor; T, T lymphocyte; B, B lymphocyte; Mac, macrophage; G, neutrophil granulocyte; Eos, eosinophil, Eb, erythroblast; E, erythrocyte; Meg, megakaryocyte; P, platelet. Mast cells, NK cells, and dendritic cells have been omitted from the scheme and placement of eosinophils is speculative. Note that most of the switches were observed with transformed cell lines in culture and that the cell type designations (both before and after the switch) may not accurately reflect the phenotype of the normal counterparts.
Switches between lymphoid and myeloid cells
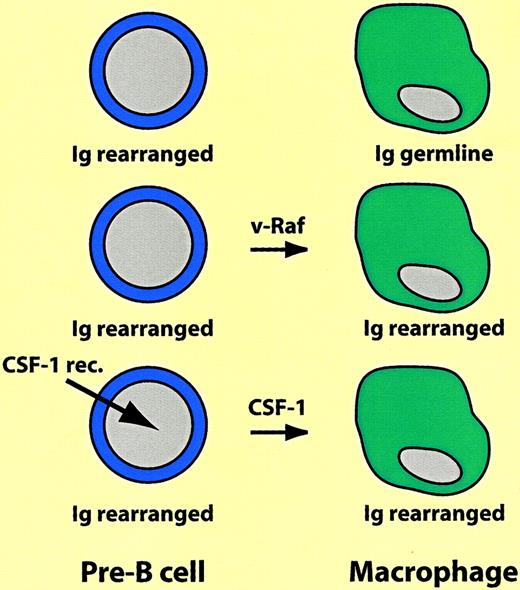
The first experiments demonstrating that differentiated hematopoietic cells are not cast in stone were reported almost 2 decades ago. Following reports that 3T3 fibroblasts treated with the demethylating agent 5-azacytidine can differentiate into striated muscle cells, chondrocytes, and adipocytes, Boyd and Schrader tested the effects of the drug on Abelson virus–transformed pre–B lymphoma cell lines. They found that a subset of the cells acquired properties of macrophages, including the ability to adhere to plastic as well as phagocytic and esterase activity.6 Later, using pre-B and B-cell lines immortalized with Eu-myc, Adams and colleagues showed that v-raf oncogene overexpression had a similar effect. The v-raf–transfected macrophagelike cells not only expressed a plethora of myelomonocytic markers (such as the colony-stimulating factor [CSF]-1 receptor and lysozyme), not seen before the switch, but also retained immunoglobulin rearrangements characteristic of the original cells.7 Similarly, a proportion of early B-cell lines ectopically expressing thev-fms oncogene (encoding a constitutively active form of the CSF-1 receptor) switched into macrophages, as did murine cells expressing the human CSF-1 receptor gene after treatment with human CSF-1.8 It is difficult to assess the frequency of the B lineage-to-macrophage transitions, but it does not appear to be high because in the various studies only a fraction of the cells expressing the transgene switched lineage. Most likely the outgrowth of the reprogrammed cells involved selection (see also below), but the immunoglobulin rearrangements in the resulting macrophages leave no doubts about their origin. These experiments are summarized in Figure 2.
Switch of B-lineage cells into macrophages.
Ig rearranged and Ig germline indicate immunoglobulin heavy chain gene in rearranged or germline configuration, respectively. For explanation, see text.
Switch of B-lineage cells into macrophages.
Ig rearranged and Ig germline indicate immunoglobulin heavy chain gene in rearranged or germline configuration, respectively. For explanation, see text.
In addition to the observed transition of B-lineage cells into macrophages, one study reported a switch of B-lymphoid cells to neutrophil granulocytes.9 While creating transgenic mouse lines involving the max gene, the authors obtained one line (called max 41) that contained dramatically increased numbers of granulocytes at the expense of B lymphocytes. Although this unbalance per se does not imply an in vivo lineage switch, the following experiments suggest that this is indeed the case. When max41 was crossed with the Eu myc mouse, which has a propensity to form pre–B cell lymphomas, animals were obtained that likewise developed pre–B cell lymphomas but, in addition, contained elevated levels of granulocytes. These lymphomas were transplantable and generated in the recipient not only new lymphoma cells but also granulocytes of donor origin. Transplantation experiments with cloned pre–B-lymphoma cell lines derived from this cross showed that its granulocytes contained the same pattern of immunoglobulin rearrangements.9 The gene whose altered activity in the max 41 mice facilitates the switch has not been reported so far. It is also unclear whether or not a B lymphoid-to-granulocyte switch occurs normally (see also below).
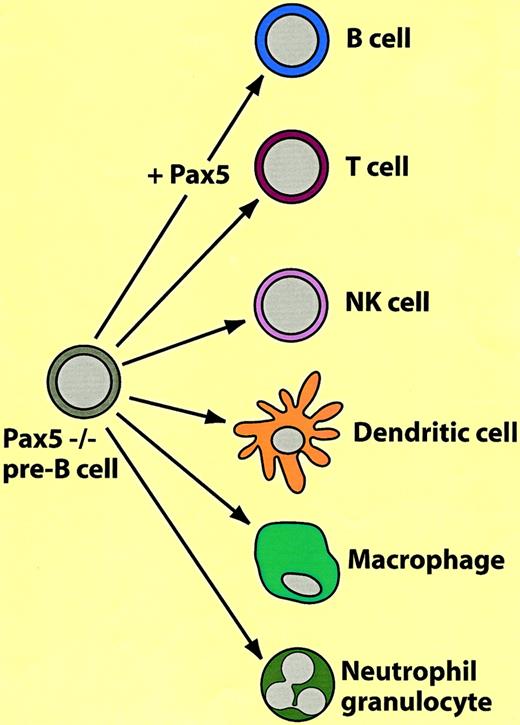
A surprising degree of plasticity was discovered in B-lineage cells derived from Pax-5 knockout mice: on the one hand these cells exhibit characteristics of pre–BI cells (B220+, AA4.1+, c-Kitlow, SL chain+ but CD19−) but, on the other hand, have characteristics of pluripotent stem cells. In contrast to wild-type pre–BI cells, they are capable of continuous-to-unlimited self-renewal on stromal cells and interleukin-7 (IL-7), home to the bone marrow after transplantation, and can differentiate into most hematopoietic cell types in vitro and in vivo. As summarized in Figure3, they can be induced to differentiate into B cells by restoration of Pax-5 expression and culturing on stromal cells in the presence of IL-7. However, when transplanted into RAG2 knockout mice they differentiate into T cells10; when grown in the presence of IL-2 they differentiate into natural killer (NK) cells, dendritic cells with CSF-1 and granulocyte-macrophage colony-stimulating factor (GM-CSF), macrophages with CSF-1, and neutrophils with IL-3, IL-6, stem cell factor (SCF), and granulocyte colony-stimulating factor (G-CSF).11 Subclones from immortalized Pax-5−/− lines retain the capacity for multilineage differentiation, suggesting that the pluripotency of the original culture is not due to heterogeneity. It appears that the main role of Pax-5 in the establishment of B-cell commitment is the repression of lineage inappropriate genes, such as the CSF-1 receptor gene (c-fms), which is expressed in the pre–BI Pax-5 knockout cells.12 It needs to be added that cells with a phenotype comparable to the Pax-5 knockout pre–B cells probably do not exist in wild-type animals (eg, pre–B cells from wild-type mice cannot easily be established in culture).
Another series of experiments demonstrated that common lymphoid progenitors (CLPs), can be reprogrammed to become myelomonocytic.13 These experiments involved the analysis of a transgenic mouse line that ectopically expresses the human IL-2 receptor β chain. When CLPs from this mouse were cocultured with OP9 stromal cells plus IL-7 they differentiated into B cells and NK cells. However, additional treatment with human IL-2 induced the formation of granulocytes and macrophages (no such cells developed when CLPs from control mice were grown under the same conditions). The signaling pathways required for the establishment of the 2 myelomonocytic lineages seem to be separable: When introduced into CLPs by retroviral infection, a mutant β chain of the receptor lacking the Shc/Lck-binding domain yielded predominantly macrophages, whereas a Stat5-binding site mutant yielded mainly granulocytes. Because the IL-2 receptor is not normally expressed in CLPs, how likely is it that the observed cytokine-induced reprogramming reflects a process that goes on during normal lineage commitment? Although the answer is not in, 2 observations suggest that this treatment mimics the effects of GM-CSF receptor signaling: activation of the IL-2 receptor in CLPs leads to an up-regulation of the GM-CSF receptor, and ectopic expression of the GM-CSF receptor followed by stimulation with GM-CSF likewise leads to a lymphoid-to-myeloid conversion.13 Together, these results indicate that common lymphoid progenitors are highly plastic and that cytokine receptor signaling may play a role during normal lineage commitment.
All of the experiments discussed so far were performed with cells that were either derived from lymphomas, transformed by an oncogene, or altered by mutations. This raises the possibility that the observed plasticity represents a phenomenon restricted to cells with altered growth properties. However, recent work suggests it can also happen with normal B-lineage cells, at least in culture. Thus, CD19 antigen, DJ rearrangement–positive, B220− cells from adult mouse bone marrow can develop into adherent cells that resemble macrophages when cultured in the presence of IL-3, IL-6, and GM-CSF. They can also differentiate into B cells but not into T nor NK cells.14
If B cells can be switched to become macrophages, can such a switch also be observed in the opposite direction? In the cell system where this was studied the answer seems to be yes. The experiment consisted in characterizing the pattern of gene expression of macrophagelike variants obtained from the pre–B lymphoma cell line 70Z/3, and restore the down-regulation of transcription factors that are suspected to play a role in commitment. Of the 3 transcription factors known to be required for progression through the early stages of B-cell development, E2A, EBF, and Pax5,12 E2A and EBF were completely absent in the macrophage variant, whereas expression of Pax-5 was reduced. Enforced expression of the E12 encodingE2A gene caused a decreased adherence of the cells, down-regulation of Mac-1 and of c-fms, and induction of a number B lineage–specific genes, that included the IL7R, EBF,and RAG1 genes, and the ability to form κ light chain in response to mitogens. Similar, but less dramatic, changes were observed after enforced expression of the EBF gene, suggesting that it acts downstream of E2A.15 Experiments in which E2A and EBF were expressed in an embryonic kidney cell line in the presence or absence of RAG1 showed that combinations of these genes are capable of inducing light-chain immunoglobulin rearrangements in nonhematopoietic cells. The fact that the cells did not acquire a full B-lineage phenotype suggests that the combination of E2A, EBF,and RAG gene expression is not sufficient for lineage specification and that additional genes must be involved.16
The examples of lineage plasticity discussed between B cells and myelomonocytic cells suggest a close developmental relationship. This might reflect the fact that they arise from a common progenitor in fetal liver (together with T cells, NK cells, and dendritic cells)17,18 and that they both require the transcription factor PU.1 for commitment (for reviews, see Singh et al19and Orkin20). In addition, lin− fetal liver cells from PU.1 knockout mice infected with a PU.1-containing retrovirus convert into B-lineage cells when expressing low concentrations of the factor and into macrophages PU.1 at about 4-fold higher concentrations.21 Whether commitment to these 2 lineages depends on the graded expression of PU.1 at the level of a common progenitor, or whether the factor is needed to maintain lineage identity after commitment, is not known. It will be interesting to determine whether increasing the level of PU.1 expression in B-lineage cells reprograms them to become myeloid and, conversely, whether reducing the levels of the factor in myeloid cells induces them to become B lymphoid.
There is also an example of a possible developmental connection between T-lineage cells and macrophages. The thymus is known to harbor macrophages that act as scavengers to remove dead cell bodies generated during T-cell development. Although the developmental origin of these macrophages is obscure, a recent study suggests that they arise locally through the differentiation of immature T cells. Thus, culturing of a subset of purified pro–T cells in the presence of conditioned medium from a thymic stromal cell line generated functional macrophages. This lymphoid-to-myeloid transition could also be observed in the presence of a combination of IL-6, IL-7, and macrophage colony-stimulating factor (M-CSF; CSF-1), but at a lower frequency.22
Lineage switches within the myeloid/erythroid compartment
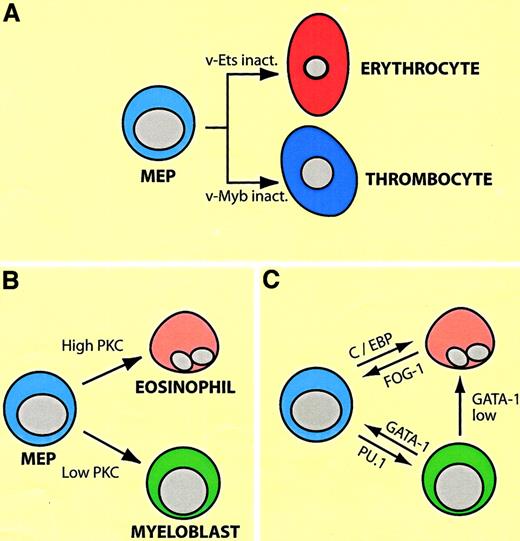
Most experiments showing lineage plasticity within the myeloid/erythroid compartment are based on enforced transcription factor expression. Perhaps, surprisingly, the factors capable of inducing commitment are all activators of lineage-specific gene expression, paralleling observations made with myogenic transcription factors. In the following I will describe an experimental system that has allowed investigators to direct the differentiation of cultured cell lines from one lineage into another in reproducible and predictable ways, fitting into a logical framework. (For the interested reader the biology of the relevant factors in a nutshell: GATA-1 and FOG-1 are required for erythroid and megakaryocytic differentiation and synergize in the induction of genes specific for these lineages. GATA-1 seems to have a wider role than FOG-1 in that it is also expressed in eosinophils and mast cells. Importantly, however, both factors are down-regulated in myelomonocytic cells [for a review, see Orkin20]. In contrast, PU.1 and C/EBP family members are required for myeloid cell differentiation and synergize in the induction of myelomonocytic genes. The inactivation of PU.1 leads to the abrogation of myeloid [as well as lymphoid] cells; that of C/EBPα to a lack of neutrophil and eosinophil granulocytes; and ablation of C/EBPβ to an impairment of macrophage function. All 3 factors are down-regulated during erythroid/megakaryocytic differentiation [for a review see Tenen et al23]). The system consists of erythroid/megakaryocytic progenitors transformed by the Myb-Ets–encoding E26 leukemia virus, dubbed “MEPs” for Myb-Ets-progenitors (illustrated in Figure4A). E26-MEP cells express a number of megakaryocytic/stem cell surface markers (such as MEP21/thrombomucin/PCLP1 and GPIIaIIIb/CD4124,25), as well as GATA-1 and FOG-1, but no myelomonocytic cell surface markers and no or low levels of PU.1, C/EBPα, and C/EBPβ. Inactivating v-Ets in these cells, either by using a temperature sensitive (ts) mutant of E26 with a point mutation in the DNA binding domain of Ets, or by deleting it with a FLP recombinase approach, induces erythroid differentiation.26,27 Conversely, inactivation of v-Myb induces the differentiation of MEP cells into thrombocytes, the avian equivalents of megakaryocytes and platelets.28 In contrast, introducing into MEP cells retroviruses encoding oncogenes of the ras pathway, or treating them with phorbolesters, thus activating protein kinase C (PKC), induces them to become either eosinophils or myeloblasts, depending on the strength of the signal (Figure 4B).24,29 These lineage conversions can be mimicked by enforced transcription factor expression (Figure 4C): PU.1 converts MEPs into cells that resemble myeloblasts (macrophage/granulocyte precursors),30 whereas C/EBPα converts them into eosinophils.31 They can also move backwards and “sideways”: forced expression of GATA-1 in myeloid cells induces the formation of either MEP cells or eosinophils, depending on the concentration of the factor. (The transition into MEP cells requires relatively high levels of ectopic GATA-1, whereas eosinophils require 2- to 3-fold lower levels, reflecting the relative proportions of endogenous GATA-1 expressed in these cell types32). The myeloid cells can be reprogrammed all the way into erythrocytes, using a 2-step protocol. If the myeloid-to-MEP cell switch induced by GATA-1 is performed in a background of ts E26 cells, inactivation of v-Ets induces erythroid differentiation.32 Finally, enforced expression of FOG-1 in eosinophils causes these cells (but not myeloblasts) to acquire an MEP phenotype.33 Again, in all of these conversions, the phenotypic changes involve a complete down-regulation of MEP markers concomitant with an up-regulation of lineage-specific markers.
Plasticity of MEP cells transformed by E26 leukemia virus.
(A) Differentiation potential of MEP cells, as demonstrated by the use of ts mutants of E26 virus with point mutations in eitherv-ets or v-myb. (B) Differentiation of MEP cells into eosinophils and myeloblasts (granulocyte/macrophage precursors) by activation of the Ras or PKC pathways. Different phorbolester concentrations preferentially lead to the differentiation of either eosinophils or myeloblasts (high and low PKC levels). (C) Arrows indicate induction of eosinophil, myeloblast, and MEP cell differentiation by enforced expression of transcription factors shown. GATA-1 low indicates that lower levels of the factor are required for eosinophil differentiation than for MEP cell differentiation.
Plasticity of MEP cells transformed by E26 leukemia virus.
(A) Differentiation potential of MEP cells, as demonstrated by the use of ts mutants of E26 virus with point mutations in eitherv-ets or v-myb. (B) Differentiation of MEP cells into eosinophils and myeloblasts (granulocyte/macrophage precursors) by activation of the Ras or PKC pathways. Different phorbolester concentrations preferentially lead to the differentiation of either eosinophils or myeloblasts (high and low PKC levels). (C) Arrows indicate induction of eosinophil, myeloblast, and MEP cell differentiation by enforced expression of transcription factors shown. GATA-1 low indicates that lower levels of the factor are required for eosinophil differentiation than for MEP cell differentiation.
Similar observations were made in other systems and in normal, cultured cells. PU.1 has been reported to induce the expression of macrophage markers in a murine erythroid cell line.34 Conversely, GATA-1 confers an erythroid/megakaryocytic phenotype in myeloid cells.32,35,36 These cells up-regulate a number of erythroid/megakaryocytic genes, such as c-mpl, EPO receptor, a-globin, EKLF, NF-E2, and ets-1, while down-regulating myelomonocytic genes, such as mac-1and c-fms.36 37 Finally, a recent study suggests that primary myelomonocytic progenitors can be reprogrammed into erythroid cells by enforced expression of GATA-1. As in the MEP system, eosinophils (and basophils) were also observed (C. Heyworth and T. Enver, personal communication, September 2001).
As can be seen from the summary in Figure 1, not all possible transitions between lineages have been described. Whether this simply reflects limited number of attempts or lack of appropriate conditions, or whether certain switches are not possible, remains to be seen. Probably, however, the facility with which cells from one lineage can transdifferentiate into another depends on how closely related they are to each other and how much their transcription factor machinery overlaps.
Selection of a few or instruction of many?
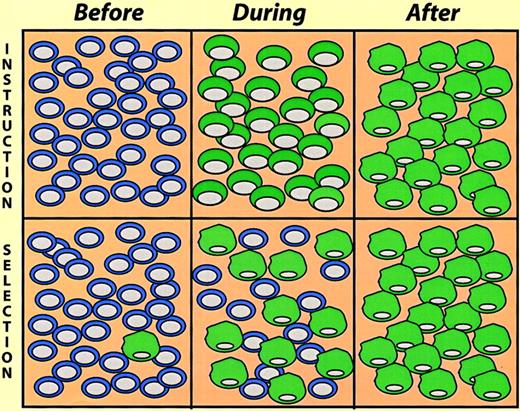
Induced lymphoid-to-nonlymphoid conversions, because of the irreversibility of immunoglobulin gene rearrangements, leave little doubt about the occurrence of true reprogramming events. However, in the case of conversions involving myeloid/erythroid cells this becomes a more difficult question. Here an alternative explanation of the observations made is the selection of pre-existing cells in the population studied. These instruction-versus-selection alternatives (Figure 5) reflect a problem formally akin to the question whether cytokine receptor signaling induces cell fate commitment or merely promotes selective cell survival (for a discussion of this issue, see Kaushansky,38Metcalf,39 and Enver et al40). In the E26 leukemia virus system the selection alternative could be ruled out by the use of inducible transcription factors. This was achieved by fusion to the estrogen receptor (ER) hormone-binding domain, making their activity dependent on estrogen or tamoxifen treatment. Thus, β-estradiol treatment of MEP cells expressing PU.1-ER led to a change, over a period of 2 to 4 days, into cells resembling myeloblasts. This involved the concomitant down-regulation of MEP markers and the up-regulation of myeloid markers via cells that expressboth sets of markers, although at reduced levels.30 Likewise, β-estradiol treatment of MEP cells containing C/EBPβ-ER induces them to switch, in this case into eosinophils.31,41 Finally, β-estradiol induces myeloblasts expressing GATA-1-ER to convert into eosinophils.32 In these experiments a limited exposure to β-estradiol (2-3 days), activating the transcription factor temporally, led to an irreversible commitment. This observation, together with the fact that the conversion of one population into another occurred gradually via cells that express both markers (as opposed to the sudden outgrowth of cells with a distinct phenotype) strongly argues against the hypothesis of selection of pre-existing cells.
Instruction versus selection models of lineage switching.
The cells illustrate a hypothetical experiment where cultures were exposed to switch-promoting conditions, and the cell phenotypes determined at different times thereafter (before, during, and after). In the instruction model the cells gradually change phenotype, going through a phase where they exhibit mixed traits. In the selection model a pre-existing minor cell population becomes dominant in the “after” population.
Instruction versus selection models of lineage switching.
The cells illustrate a hypothetical experiment where cultures were exposed to switch-promoting conditions, and the cell phenotypes determined at different times thereafter (before, during, and after). In the instruction model the cells gradually change phenotype, going through a phase where they exhibit mixed traits. In the selection model a pre-existing minor cell population becomes dominant in the “after” population.
Does transdifferentiation imply a reversal of differentiation?
The arrows in Figure 1 seem to imply that different cell types can directly convert into one another by transdifferentiation. However, it is also possible that these conversions involve retrodifferentiation to an earlier progenitor. What is the evidence that blood cells can retrodifferentiate? An example in case are chicken myelomonocytic cells transformed by a ts mutant of v-myb. These cells exhibit an immature phenotype, resembling myeloblasts, at the permissive temperature. If shifted to the nonpermissive temperature, they differentiate into adherent, phagocytic, macrophagelike cells and cease dividing. Time-lapse experiments showed that this process is reversible, with most adherent cells acquiring a blastlike morphology and re-entering the cell cycle within 2 to 3 days after shift to the permissive temperature.42 Another example is the possible formation of primitive erythroid cells after injection of transgenic bone marrow cells into blastocysts. Here, the resulting embryos contained small amounts of donor type α-globin, as revealed by polymerase chain reaction (PCR) analysis.43
Despite their apparent capacity to retrodifferentiate, there is no evidence that any of the lineage switches described involve a detour via an earlier progenitor. For example, if it is assumed that a B cell-to-myeloid switch involves the formation of a lymphohematopoietic progenitor, then cells from lineages other than macrophages, such as T cells or erythroid cells, should be generated. However, this has not been described. Likewise, the GATA-1–induced expression of eosinophil markers in myeloid cells is not preceded by the up-regulation of MEP cell markers.32,33 Finally, the CSF-1–induced transition of Pax-5−/− pre–B cells into macrophages also does not seem to involve the formation of an earlier progenitor.11
Do intrahematopoietic lineage switches occur in vivo?
Can lineage switches also be shown to occur in vivo, under physiologic conditions, or are they restricted to experimental manipulations in cell culture? The existing knowledge, accumulated over many years of research, would suggest that the answer is no. Thus the concept that cells may “change their mind” during differentiation flies in the face of the belief that hematopoiesis is a linear process with multilineage progenitors becoming restricted in a stepwise but irreversible fashion. The following examples illustrate this point: (1) The fact that lethally irradiated mice receiving transplants of short-term repopulating (STR) cells do not survive suggests that these cells cannot convert into long-term repopulating (LTR) cells in vivo. (2) The existence of progenitors that are apparently exclusively committed to the production of restricted progeny (CLPs, common multipotent progenitors [CMPs], granulocyte/macrophage progenitors [GMPs], MEPs, B- and T-cell progenitors44,45) would also argue that, if lineage switching occurs, it must be at low frequencies. (3) A similar argument can be made for the high degree of congruency in the phenotype of individual colonies observed after replating single cells from colony “starts.”46 However, none of these approaches rule out the possibility that lineage switching occurs in vivo, but at frequencies too low to be detected with technologies used so far. Finally, even if lineage switching does not occur in unperturbed animals, it is still possible that it happens under conditions of stress, such as during immunization, inflammation, or pathogen infection.
How can the question of in vivo plasticity best be approached experimentally? Lymphoid-to-myeloid cell transitions offer a seemingly straightforward approach because immunoglobulin and T-cell receptor gene rearrangements are irreversible and nonlymphoid cells of this origin should be irreversibly marked. For example, the transition from CD19+ cells into macrophages (and B cells),14if it happens in vivo, should result in a fraction (if small) of resident monocyte/macrophages that contain DJ rearrangements. However, this has (not yet) been reported. In practice, Southern blots may not be sensitive enough to detect minor fractions of cells with rearrangements, and PCR techniques require cell populations that are 100% pure, which is not always easy to achieve. Because no DNA rearrangements occur in cells of other lineages, it is even more difficult to demonstrate transitions between nonlymphoid cells and other cell types. Here a recently developed approach might be informative. It uses bacterial recombinase (Cre-LoxP or FLP-FRT) to mark the DNA of cells that express a given lineage marker. Briefly, mice are created in which the expression of the recombinase gene, sayCre, is driven by a cell type–specific marker, say the B cell–specific CD19 gene. These mice are then crossed to a reporter mouse that contains a DNA cassette framed by LoxP sites, sequences that are recognized by the bacterial recombinase. In this cross, Cre removes the cassette in all CD19-expressing B cells and this change is retained in all non–B-cell derivatives. Conveniently, removal of the cassette can be visualized at the single cell level by β-galactosidase or green fluorescent protein (GFP) expression, using reporter strains in which the indicator is driven by the enhancer/promoter of a ubiquitously expressed gene such asROSA2647-49 or β-actin.50 The method has successfully been used to trace the ancestry ofwnt-1 or protein O expressing neuroectodermal progenitors, using either Cre50,51 or FLP recombinase.52However, it also has its pitfalls because any infidelity of gene expression, if not properly controlled, can lead to incorrect lineage assignments.
There may be an example of an in vivo lineage switch after all, involving the formation of dendritic cells. Transplantation experiments showed that CMPs give rise to myeloid type (Mac-1+, CD8α−) as well as to lymphoid-type (Mac-1−, CD8α+) dendritic cells.53 This was also the case, to a lesser extent, for granulocyte/macrophage precursors, whereas megakaryocyte/erythroid precursors could not be reprogrammed.54 Conversely, common lymphoid progenitors can also generate both types of dendritic cells, with T-cell precursors having low and B-cell precursors having no dendritic cell forming potential.54 Although these experiments suggest plasticity of immature myeloid and lymphoid cells, another study indicates that functional macrophages can differentiate into dendritic cells in vivo. In these experiments microspheres were injected subcutaneously, attracting inflammatory monocytes, which subsequently became phagocytic (a hallmark of macrophages). Some of these phagocytic cells could be shown to migrate to draining lymph nodes, where they localized to regions within the nodes that are known to contain dendritic cells but not macrophages. The bead-containing cells also displayed cell surface and functional markers of dendritic cells.55 A caveat in the interpretation of these data as an example of an in vivo switch is that myelomonocytic and myeloid dendritic cells might be too closely related as to represent separate lineages. The comparison of gene expression profiles by microarray technologies might help to clarify the issue.
A model of how hematopoietic cells might switch lineage
How can the process of lineage switching be explained at the molecular level? Before speculating about possible mechanisms, it is useful to briefly recapitulate current ideas about how blood cell lineages become specified during the differentiation of multipotent progenitors. One popular model proposes that hematopoietic stem cells “sneak preview” a large range of lineage-restricted genes and specific gene expression programs become dominant after that in a random process of gene fluctuation.40,56 This concept is based on the finding that hematopoietic multipotent progenitors express low levels of genes characteristic for different lineages, such as globin and myeloperoxidase as well as diverse cytokine receptor genes,57 and that Pax-5−/− pre–BI cells express genes from nonlymphoid lineages.11 However, extrinsic events such as cytokine signaling might also play a role in commitment,39 with both intrinsic and extrinsic processes probably reinforcing each other's effects. Ultimately, the phenotype of a cell is established and maintained by combinations of transcription factors that regulate lineage-specific gene expression programs.
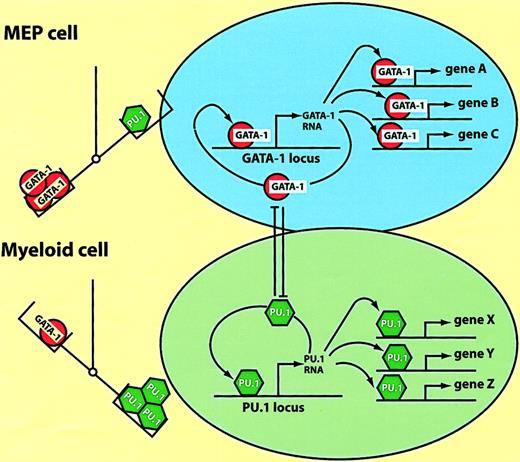
How do ectopically expressed transcription factors manage to reprogram cells in a reversible fashion? Here I will discuss a model, derived from studies with the E26 system described earlier, postulating that the relative excess of a key transcription factor over another can lead to the “controlled collapse” of a cell's phenotype. This involves both the suppression of the cell's gene expression program and the activation of novel genes, in a fashion that can be compared to a reversible chemical reaction, with different phenotypes representing “energy sinks.” As summarized in Figure 4C, MEPs, eosinophils, and myeloblasts can be converted into one another by inappropriate transcription factor expression. Lineage transitions can be induced according to a simple code that defines cell identity: high GATA-1 plus FOG-1 specifies MEPs; moderate GATA-1 plus C/EBPα/β specifies eosinophils; and PU.1 plus C/EBPβ specifies myeloblasts. The observed phenotypic changes involve both the up-regulation of lineage-specific genes as well as the extinction of lineage markers characteristic of the starting cell population. Whereas activation occurs through the combinatorial action of the above transcription factors on specific promoter/enhancer elements (megakaryocyte-specific genes,58-60 myeloid-specific genes,23 and eosinophil-specific genes61,62), suppression appears to be mediated by a completely different mechanism. Thus, GATA-1 suppresses PU.1 activity and PU.1 suppresses GATA-1 activity through direct protein interactions.63-66Therefore, as illustrated in Figure 6, the stoichiometry between the 2 factors determines the cell's phenotype, with an excess of GATA-1 resulting in MEP cells and an excess of PU.1 resulting in myeloid cells. (The balance between GATA-1 and PU.1 is also decisive in determining the differentiation of murine erythroleukemia cells, with an excess of PU.1 maintaining an immature phenotype, and an excess of GATA-1 inducing terminal differentiation.63) The cross-antagonism model in Figure 6can also explain why the transcription factor–induced phenotypic changes become irreversible, resulting in “commitment.”20,67,68 Expression of high levels of GATA-1 in myeloid cells would interrupt the PU.1 autoregulatory loop,69 activate MEP cell-specific genes, and induce the expression of endogenous GATA-1, thus establishing its own autoregulatory loop.70 In turn, an excess of PU.1 in MEP cells would induce them to acquire a myeloid phenotype, following the same principles but in reverse order. Thus, once an exogenous transcription factor has turned on the corresponding endogenous gene the cells become “committed,” no longer requiring the initial stimulus.
Model of hematopoietic lineage switching.
The scheme is based on the reciprocal lineage switches between MEP cells and myeloblasts induced by GATA-1 and PU.1 (see text). The left part of the figure illustrates that the balance between the 2 factors determines the cell's phenotype; MEPs are specified by an excess of GATA-1, myeloblasts by an excess of PU.1. The right part of the figure illustrates what might be going on in the nucleus. In MEP cells GATA-1 activates multiple target genes (A, B, C), as well as possiblyGATA-1 itself. In myeloid cells PU.1 activates another set of target genes (X, Y, Z), including PU.1 itself. Enforced expression of PU.1 in MEP cells leads to the inactivation of GATA-1 through direct protein interactions. GATA-1, when overexpressed in myeloid cells, inactivates PU.1 through a similar mechanism (the cross-antagonism between the 2 factors is illustrated by the T bars).
Model of hematopoietic lineage switching.
The scheme is based on the reciprocal lineage switches between MEP cells and myeloblasts induced by GATA-1 and PU.1 (see text). The left part of the figure illustrates that the balance between the 2 factors determines the cell's phenotype; MEPs are specified by an excess of GATA-1, myeloblasts by an excess of PU.1. The right part of the figure illustrates what might be going on in the nucleus. In MEP cells GATA-1 activates multiple target genes (A, B, C), as well as possiblyGATA-1 itself. In myeloid cells PU.1 activates another set of target genes (X, Y, Z), including PU.1 itself. Enforced expression of PU.1 in MEP cells leads to the inactivation of GATA-1 through direct protein interactions. GATA-1, when overexpressed in myeloid cells, inactivates PU.1 through a similar mechanism (the cross-antagonism between the 2 factors is illustrated by the T bars).
What is the ground state of transcription factor expression in erythroid/myeloid progenitors and how does commitment get started? The idea is that multipotent progenitors express both GATA-1 and PU.1 and that the latter is expressed at relatively low levels.45External signaling events or intrinsic fluctuations or both may tip the balance between the 2 transcription factors, thus allowing the progenitors to differentiate along one or another pathway. This interpretation is supported by the observation that MEP cell clones can differentiate spontaneously into myeloblasts at low frequencies and that this transition can be accelerated by treatments that mimic growth factor receptor stimulation.24 29
Is the transcription factor cross-antagonism model more generally applicable? Although a definitive answer is not yet in, a few more examples can be cited. Within the E26 system, FOG-1 antagonizes C/EBP expression, driving an eosinophil to MEP transition, and vice versa, although a direct interaction of the 2 factors remains to be demonstrated.20,33 The latter process may involve the interruption by FOG-1 of an autoregulatory loop of C/EBPα.71,72 Another example of transcription factor cross-antagonism has been described for Ets-1 and Maf-B. Here overexpression of the macrophage-specific factor Maf-B in erythroid cells leads to a block of differentiation through a direct interaction with Ets-1 and its subsequent inactivation, although the relevance of this observation in the context of normal differentiation is unclear. It is possible that the cross-antagonism principle is also operative within the lymphoid compartment. Thus, a direct protein interaction between PU.1 and Pax-5 has been reported and this might play a role in the decision of whether a cell acquires a myeloid cell or a B-cell fate, respectively.73
The handful of transcription factors covered in this review are not the only players involved, and readers are referred to other reviews discussing additional factors (for myeloid/erythroid lineages, see Tenen et al,23 Yamanaka,74 Nagamura-Inoue et al75; for lymphoid lineages, see Rothenberg et al,76 Glimcher and Singh,77 Glimcher and Murphy78). In addition, transcription factors involved in lineage specification almost certainly do not act in simple 2-way combinations but are part of cell-specific transcription factor networks, where they participate in positive and negative cross-regulations at transcriptional and posttranscriptional levels. One can imagine that these networks have a 3-dimensional shape that changes as the cell's transcription factor composition is altered. Such “conformational changes,” rather than being merely consequences of differentiation, might constitute a driving force during commitment.79
Other mechanisms, not involving direct transcription factor interactions, also play a role during hematopoietic lineage specification and transdifferentiation. Examples include competition for the general coactivators CBP/p30080-82 and histone-modifying factors that are involved in chromatin remodeling. These processes help to establish and stabilize the differentiated state.83
Conversions between hematopoietic and nonhematopoietic cells
Until recently it has been widely assumed that there are 2 major classes of stem cells: embryonic stem cells derived from early embryos, able to replenish all cell types in the animal, and stem cells located in various organs of the body, dedicated to the replenishment of specific tissues, such as blood, muscle, and brain (for a review, see Fuchs and Segre84). This assumption was shaken a few years ago by the observation that bone marrow cells can differentiate into muscle cells after transplantation,85 followed by reports that cells derived from brain and muscle can fully reconstitute the hematopoietic system of lethally irradiated mice.86-88 Are all of these conversions due to the plasticity of hematopoietic cells? It is particularly difficult to assess whether true conversions occur from nonhematopoietic to hematopoietic tissues because hematopoietic stem cells (HSCs), entering these tissues via the circulation, might act as “contaminants.” Indeed, recent experiments revealed that skeletal muscle tissue contains multilineage hematopoietic progenitors and LTR-HSCs. These cells were clearly derived from bone marrow because HSCs recovered from the muscle of mice that were previously reconstituted with marked donor cells were of donor origin.89 In addition, cells in the skeletal muscle capable of generating hematopoietic colonies and of hematopoietic engraftment are CD45+, Sca-1+, whereas cells that contribute to muscle cell formation after transplantation are CD45− (M. Goodell, personal communication). These findings suggest that the muscle-to-hematopoietic cell switch originally described can be attributed to hematopoietic progenitors homing to muscle tissue. The reported switch of neuronal precursors to blood cells has also come under question. Thus, extensive experiments involving the transplantation of neural stem cell cultures (“neurospheres”) into irradiated mice, following the protocol of Bjornson et al,86 failed to reconstitute the hematopoietic system (C. Morshead, personal communication, October 2001). In addition, experiments in which neurospheres were injected into mouse blastocysts revealed the presence of donor-type cells in a wide variety of tissues in the resulting embryos with the notable exception of hematopoietic tissues.90
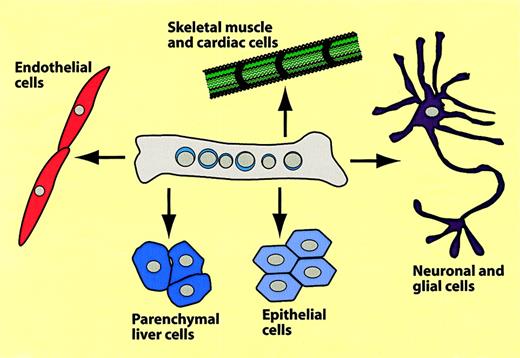
The remainder of this review will focus on conversions from hematopoietic to nonhematopoietic tissues. The different types of conversions that have been reported are summarized in Figure7 and Table2. Using mice transgenic for the muscle-specific myosin light chain 3F promoter driving LacZ (MLC3F-lacZ mice) as donors, Ferrari and coworkers injected bone marrow cells into leg muscles regenerating after chemical injury. When they analyzed muscle sections 2 to 5 weeks later they detected β-galactosidase–positive muscle fibers.85 These authors also described that muscle cells can be recruited from the circulation. For this, lethally irradiated mice received transplants of MLC3F-lacZ bone marrow; a muscle injury was induced after 5 weeks and muscles were analyzed 2 to 3 weeks later. Most animals again contained labeled myocytes, albeit at lower frequencies than after direct injection.85 Several groups reported that transplantation of bone marrow cells into mice with a defect in the dystrophin-relatedmdx gene induced the formation of Y chromosome–positive, Mdx-expressing muscle fibers.88,91 In one of these studies, as few as 2000 “side population (SP) cells,” purified from bone marrow after Hoechst 3042 staining, were able to generate muscle cells.88 However, the efficiency of the tissue replacement was exceedingly low and no ameliorating effects on muscular dystrophy could be observed.92 In addition to donor-origin skeletal muscle cells, the animals that received transplants had cardiomyocytes, particularly when previously infarcted by arterial occlusion. The new cardiomyocytes located preferentially to the ischemic region.91,93,94 Lin−Kit+ cells, but not Lin−Kit− cells, exhibited cardiomyogenic potential.93 The observation that cardiomyocyte-generating cells, which alleviate myocardial infarction, can be generated by mobilizing blood-borne cells by treatment with G-CSF and SCF caused great excitement among clinicians.95Whether the relevant cells are of hematopoietic origin has yet to be established.
Summary of hematopoietic-to-nonhematopoietic cell conversions.
The scheme summarizes experiments in which bone marrow cells were transplanted into mice, which were then analyzed several months later for hematopoietic and nonhematopoietic donor type cells.
Summary of hematopoietic-to-nonhematopoietic cell conversions.
The scheme summarizes experiments in which bone marrow cells were transplanted into mice, which were then analyzed several months later for hematopoietic and nonhematopoietic donor type cells.
Bone marrow cells yielding nonhematopoietic tissues after transplantation
| Donor cells, fraction . | Nonhematopoietic derivative . | Recipients . | Reference . |
|---|---|---|---|
| BM, adherent cells | Skeletal muscle | Scid/bg mice | 85 |
| BM, SP cells | Skeletal muscle | Mdx mice | 88 |
| BM | Skeletal/cardiac muscle | Mdx mice | 91 |
| Lin−Kit+ cells | Endothelial cells, cardiac muscle | Irrad, infarct mice | 93 |
| CD34lo, Sca-1+Kit+cells | Endothelial cells, cardiac muscle | Irrad, infarct mice | 94 |
| CD34+ lin−cells | Hepatocytes | Irradiated mice | 96 |
| Lin−Sca-1+Kit+cells | Hepatocytes | FAH−/−, irrad mice | 97 |
| BM | Astrocytes, microglia | Irradiated mice | 101 |
| BM | Immature neuronal cells | PU.1−/− mice | 102 |
| BM | Immature neuronal cells | Irrad mice | 103 |
| CD34 high, Kit+ cells | Various epithelial cells | Serial transplants | 104 |
| Cultured CD34+ cells | Vascular endothelial cells | Canine model | 113 |
| CD34+Kit+Flk1+AC133+cells | Vascular endothelial cells | Irrad mice | 114 |
| BM | Liver oval cells, hepatocytes | Rats, liver injuries | 137 |
| Donor cells, fraction . | Nonhematopoietic derivative . | Recipients . | Reference . |
|---|---|---|---|
| BM, adherent cells | Skeletal muscle | Scid/bg mice | 85 |
| BM, SP cells | Skeletal muscle | Mdx mice | 88 |
| BM | Skeletal/cardiac muscle | Mdx mice | 91 |
| Lin−Kit+ cells | Endothelial cells, cardiac muscle | Irrad, infarct mice | 93 |
| CD34lo, Sca-1+Kit+cells | Endothelial cells, cardiac muscle | Irrad, infarct mice | 94 |
| CD34+ lin−cells | Hepatocytes | Irradiated mice | 96 |
| Lin−Sca-1+Kit+cells | Hepatocytes | FAH−/−, irrad mice | 97 |
| BM | Astrocytes, microglia | Irradiated mice | 101 |
| BM | Immature neuronal cells | PU.1−/− mice | 102 |
| BM | Immature neuronal cells | Irrad mice | 103 |
| CD34 high, Kit+ cells | Various epithelial cells | Serial transplants | 104 |
| Cultured CD34+ cells | Vascular endothelial cells | Canine model | 113 |
| CD34+Kit+Flk1+AC133+cells | Vascular endothelial cells | Irrad mice | 114 |
| BM | Liver oval cells, hepatocytes | Rats, liver injuries | 137 |
BM indicates bone marrow; SP, side population; irrad, irradiated; and infarct, infarcted.
Although the reported transitions of bone marrow–derived cells into muscle cells were unexpected, a series of papers describing conversions into epithelial cells were even more surprising. Several studies reported the formation of liver cells from transplanted bone marrow in mice,96,97 rats,98 and humans.99Others suggested that bone marrow cells can turn into neuronal cells. Following publications describing donor-type microglia and astrocytes in mice that received bone marrow transplants,100,101 Mezey et al102and Brazelton et al103 reported the detection of cells with neural markers in the brain PU.1 defective or irradiated recipients, respectively. However, these studies failed to demonstrate the formation of mature/functional neurons and their hematopoietic origin is also unclear. Finally, a recent paper described the in vivo conversion of bone marrow cells into epithelial cells of various organs in mice that underwent transplantation.104 This paper will be discussed in more detail below.
Which of the bone marrow-to-nonblood cell conversions can be attributed to bona fide hematopoietic cells?
If it is already difficult to rigorously establish hematopoietic cell plasticity using cultured cell lines, it is even more difficult to conclude that hematopoietic cells are plastic based on transplantation experiments. There is always the possibility that the bone marrow hosts a variety of dedicated tissue-specific stem cells, such as muscle stem cells, neuronal stem cells, and hepatic progenitors. The following requirements have to be met before concluding that a hematopoietic-to-nonhematopoietic transdifferentiation has been demonstrated (“the gold standard”): (1) Show that bone marrow cells purified on the basis of established cell surface markers cannot only reconstitute irradiated mice but can also participate in the formation of nonhematopoietic tissues. (2) Demonstrate that this can be done with a single cell or else clonal reconstitution of both types of tissues. (3) Demonstrate that the bone marrow–derived nonhematopoietic cells are differentiated and functional.
Although there is (as yet) no evidence for the presence of dedicated muscle, neuronal, or liver cell progenitors in the bone marrow, this tissue is certainly heterogeneous and appears to contain various types of hematopoietic as well as nonhematopoietic progenitors (Figure8). Based on their long-term or short-term repopulation capacity of lethally irradiated hosts, 2 types of hematopoietic stem cells can be distinguished: LTR-HSCs and STR-HSCs (eg, see Lanzkron et al105). Both types of HSCs in the mouse are Lin−, CD45+, Kit+, Sca-1+, but LTR-HSCs are mostly CD34−CD38+, whereas STR-HSCs are CD34+CD38−.106 However, definition of a cell's potential by its phenotype is complicated by the fact that expression of a given marker does not necessarily indicate a cell's developmental potential. Thus, mice treated with 5-fluorouracil (to deplete cycling progenitors) develop LTR-HSCs that transiently down-regulate c-Kit and up-regulate Mac-1107and reversibly modulate CD34 expression.108 In addition, CD34 expression levels of murine, and possibly human, HSCs is age dependent, whereas the percentage of CD34+ HSCs is high in fetal liver and in bone marrow of young mice it decreases dramatically in adult animals, with only about 20% of HSCs expressing CD34.109 110
The bone marrow cavity contains different types of hematopoietic and nonhematopoietic progenitors.
HSC indicates hematopoietic stem cells; MSC, mesenchymal stem cells; AnBl, angioblasts. The boxes represent “drawers” that indicate differentiation potential. Colors within the drawers indicate tissue conversions that have been described or postulated (question marks). The presence of angioblasts in the bone marrow is speculative. Additional types of precursors may also exist.
The bone marrow cavity contains different types of hematopoietic and nonhematopoietic progenitors.
HSC indicates hematopoietic stem cells; MSC, mesenchymal stem cells; AnBl, angioblasts. The boxes represent “drawers” that indicate differentiation potential. Colors within the drawers indicate tissue conversions that have been described or postulated (question marks). The presence of angioblasts in the bone marrow is speculative. Additional types of precursors may also exist.
In addition to hematopoietic cells, the bone marrow cavity probably contains angiogenic precursors (also called endothelial progenitor cells or angioblasts), which in humans are CD34+, Kithigh, Flk-1+, Tie-2+, AC133. Endothelial progenitor cells have been found in the circulation111-113 and can be mobilized by G-CSF.114 These cells, when transplanted, give rise to mature endothelial cells in vessels and can be selected by adherence and culture under endothelial conditions.114 However, some of the selected cells might be of hematopoietic origin because human monocytes cultured in the presence of angiogenic growth factors have been shown to acquire properties of endothelial cells.115-117 A third class of stem cells contained in the bone marrow are mesenchymal stem cells (MSCs, also called stromal cells) which are CD45−, CD34−, Mac-1−. Like macrophages and angiogenic cells, they can be selected by plastic adherence.118 Cultured human MSCs have been shown to differentiate into adipocytes, chondrocytes, osteocytes, and muscle cells.119,120 Perhaps surprisingly, they were also found to differentiate into cardiomyocytes following in utero transplantation in a sheep model.121 The amazing differentiation potential of MSCs does not stop here. Cultures of rat, human, and mouse MSCs can differentiate into neuronlike cells in culture,122,123 and injection of mouse MSCs into neonatal mouse brains gives rise to astrocytes and possibly neurons.124
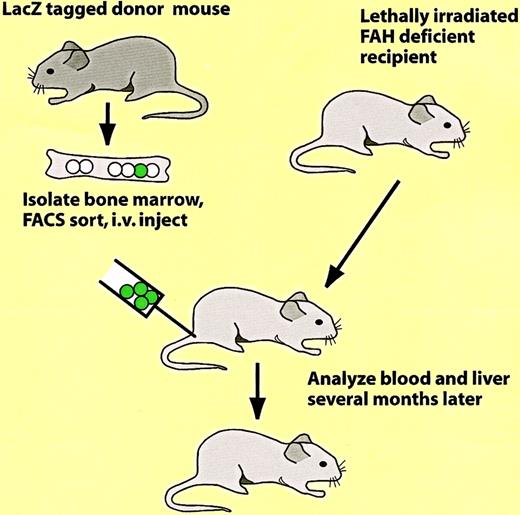
Which of the reported conversions of bone marrow to nonhematopoietic cells involve bona fide hematopoietic progenitors, rather than reflecting developmental potentials of other types of progenitors? Here 2 papers will discussed in some detail. In an elegant study, Lagasse et al97 transplanted bone marrow cells from a mouse ubiquitously labeled with β-galactosidase (ROSA26 LacZ mouse125) into lethally irradiated mice with a congenital liver defect (the protocol used is illustrated in Figure9). This resulted in a rescue of the animals from disease onset and death through the restoration of the hematopoietic system and the formation of large β-galactosidase–positive colonies of parenchymal liver cells. Sorting experiments revealed that effective cell fractions were Kit+, Lin− and Sca1+, thus exhibiting a phenotype corresponding to that of HSCs. As little as 50 Lin−Sca-1+Kit+ cells were able to both reconstitute hematopoiesis and to produce donor-derived liver cells. Although it cannot be excluded that the bone marrow contains, in addition to HSCs, contaminating liver progenitors, this appears unlikely because both biologic activities correlated in limiting dilution experiments. However, liver stem cells (“oval cells”) are known to share a number of markers with HSCs, such as CD34 and Kit (eg, see Crosby et al126).
Protocol used to show the in vivo conversion of hematopoietic cells into parenchymal liver cells.97
Protocol used to show the in vivo conversion of hematopoietic cells into parenchymal liver cells.97
That a single bone marrow–derived stem cell can differentiate not only into blood cells but also into a wide variety of epithelial cells is indicated by work of Krause et al.104 In this study, the authors enriched Lin−, quiescent cells from the bone marrow, labeled them with a vital dye, transplanted them into a lethally irradiated primary recipient, and recovered donor type cells from the bone marrow 2 days later. (The dye-positive cells were 500- to 1000-fold enriched for LTR-HSC activity and contained approximately 10 times more CD34+Sca-1+ cells than the starting population.) Then, single cells obtained by limited dilution were injected into secondary recipients and the animals receiving transplants were analyzed several months later for donor contribution, using fluorescent in situ hybridization for the identification of Y chromosome–positive cells. A sixth of these animals were fully reconstituted in their hematopoietic system and showed, in addition, significant levels of donor engraftment in alveoli, bronchi, esophagus, stomach, small and large bowel, skin, and bile duct (as well as in liver hepatocytes, D. Krause, personal communication, July 2001). Some of these cells, such as lung type II pneumocysts, were shown to be functional through the detection of messenger RNA encoding a surfactant protein characteristic of these cells. Because of the high percentages of donor-derived epithelial cells observed (up to 20%), there must have been a considerable expansion of the originally transplanted cell, although perhaps oddly, no distinct y-chromosome–positive colonies were found. These results raise a number of questions: Can donor-type epithelial cells also be observed in animals that received primary transplantations or is the serial transplantation protocol used a prerequisite? If the latter is true, what happens to the HSCs in the bone marrow of the primary recipients? Do the nearly totipotent cells described still represent classical HSCs or are they more akin embryonic stem cells? Where does the cell expansion and the mesenchymal-to-epithelial cells transition occur—in the bone marrow or after the cells home to epithelial tissues? Is tissue injury necessary for engraftment in epithelial tissues (see also below), and what are the homing clues?
Although experiments discussed strongly indicated the potential of bona fide hematopoietic cells to convert into various types of epithelial cells, this is less clear for the reported conversions into muscle, endothelial cells, and brain cells. Considering that the bone marrow contains various types of stem cells (Figure 8), it is possible that the donor-derived muscle and endothelial cells observed after bone marrow transplantation85,93,94 originate from mesenchymal stem cells and angiogenic precursors, respectively. Likewise, the neuronal cells observed in the transplantation experiments of Mezey et al102 and Brazelton et al103 might have arisen from mesenchymal stem cells. As an alternative to this more conventional interpretation, it has recently been postulated that a universal adult stem cell exists that comes in different coats.5 In this extreme view the various types of stem cells residing in the bone marrow, brain, heart, and muscles are considered to represent different states of a universal adult progenitor whose phenotype is defined by its local environment. These stem cells can move from one tissue into another via the circulation and are more plastic in early than in more differentiated stages.5 Accepting this view, which might be called the “Heisenberg principle of stem cell biology,” the issue of stem cell plasticity becomes almost semantic. Taking it to the limit, the only way to prove plasticity would be in instances where already differentiated cells can be shown to transdifferentiate into other, well-defined cell types. That this is a possibility is illustrated by examples from the world outside Blood. Myocytes have been shown to “transdifferentiate” into proliferating stem cells during tissue regeneration in amphibians127 or following expression of the msx-1 gene,128; C/EBPβ has been shown to induce a conversion of pancreas to liver cells129; and oligodendrocyte precursors have been induced to retrodifferentiate into stem cells that can go on to become astrocytes and neuronal cells.130
Do hematopoietic-to-nonhematopoietic conversions occur normally during embryonic development and in adult animals?
Almost all reported conversions between hematopoietic and nonhematopoietic cells in vivo were carried out by forced tissue relocations in irradiated or injured animals. For example, the experiments demonstrating a conversion from hematopoietic to epithelial cells were performed with irradiated animals. Irradiation, on the other hand, has been shown to destroy not only hematopoietic but also endothelial and epithelial tissues.131 It is therefore important to know whether such tissue conversions also occur under physiologic conditions. This question seems irrelevant if the ultimate purpose is to use somatic stem cells in tissue replacement therapies. However, knowledge about existing cellular processes might well be useful in guiding the development of tissue replacement protocols. In principle, tissue conversions might occur during embryonic development, involving the migration of hematopoietic cells from the yolk sac, aorta/gonad/mesonephros (AGM), or fetal liver to the anlagen of nonhematopoietic organs. For example, it has been reported that macrophages migrate from the yolk sac into the neural fold area,132 possibly seeding the brain with microglia (strictly speaking, however, this would not represent a “switch,” because microglia are phagocytic cells that express macrophage markers). So far, there are no reports showing a reprogramming of hematopoietic cells nonhematopoietic tissues during development, but some evidence suggests a developmental flow in the opposite direction, from endothelial to hematopoietic cells. Thus, early blood cell progenitors share a number of markers with endothelial cells133 and colony assays indicate the existence of a common endothelial/blood cell progenitor, the “hemangioblast.”134 In addition, endothelial cells within the chick embryonic aorta labeled with a vital stain or with a LacZ-encoding retrovirus were shown to develop into blood cells.135,136 These experiments were interpreted as evidence for cells with a dual hematopoietic and endothelial differentiation potential. However, it is also conceivable that hemangioblasts/hematopoietic cells can originate from fully functional endothelial cells,133 such as through local signaling events that induce their transdifferentiation. This issue might be hard to resolve. Clearly, there is a close relationship between endothelial cells and cells from other mesodermal lineages, a concept strengthened by the recently described “meso-angioblast” discovered in birds. This embryonic aorta-derived stem cell, which expresses CD34, c-Kit, and Flk-1, is capable of giving rise to vessels, blood, cartilage, bone, as well as to smooth, skeletal, and cardiac muscle cells (G. Cossu, personal communication, February 2002).
Conclusions
The cell culture experiments reviewed here leave little doubt about the intrinsic plasticity of hematopoietic cells, with cells from almost any lineage being capable of switching into cells of another lineage. The switches appear to represent true transdifferentiation events, with cells moving backwards as well as sideways within the differentiation tree. Reprogramming is initiated either by extrinsic or intrinsic events that result in changes in transcription factors active in commitment and maintenance of lineage-specific gene expression programs. The transcription factors involved in commitment appear to have a dual role; they not only set up novel gene expression programs through transcriptional activation but also extinguish old programs through inactivating interactions with an antagonistic transcription factor. Whether committed hematopoietic cells can “change their mind” during normal development and in adult animals, perhaps in situations of stress, is an area that needs to be explored. Knowledge of such mechanisms may shed new light on the evolution of the hematopoietic system, its homeostasis, and its adaptation to pathogen invasion during innate immunity.
The interpretation of the hematopoietic-to-nonhematopoietic cell conversions described is complicated by the fact that the bone marrow cavity not only hosts bona fide hematopoietic precursors but 2 or more additional types of stem cells. In most studies it is therefore difficult to conclude with certainty that a blood-to-nonblood cell switch has actually occurred. Thus, although it is clear that hepatocytes and other epithelial cell types can derive from transplanted hematopoietic stem cells, the conversion to skeletal and cardiac muscle, neuronal cells, macroglia, and vessels is less clear and could involve mesenchymal stem cells as well as angiogenic precursors. And, as for the intrahematopoietic lineage conversions, it remains to be seen whether blood-to-nonblood conversions occur during normal embryonic development or even as an ongoing process in adults. Another interesting area for future investigations is to define stem cell niches, and how different microenvironments influence the developmental potential of bone marrow–derived stem cells.
Some of the reports covered in this review have suggested that hematopoietic stem cells can serve as an alternative to embryonic stem cells for tissue replacement after injury or disease, such as stroke and heart infarct, Parkinson disease, cystic fibrosis, muscular dystrophy, and congenital liver diseases. Because of the vast amount of knowledge accumulated in their handling for the purpose of transplantation and their relatively easy isolation, they would indeed be an ideal source for organ repair and reconstitution. The clinical prospects for the use of adult hematopoietic stem cells for these purposes largely depends on whether it will be possible to develop protocols that increase the tissue conversion frequencies and noninvasive strategies that involve the mobilization of bone marrow progenitors. Clearly, the field is still in its infancy and basic research will have to go hand in hand with clinical research to explore the suitability of hematopoietic stem cells for tissue replacement therapies.
I would like to thank Todd Evans, Gordon Keller, Kelly McNagny, Fabio Rossi, Gwen Randolph, and members of the Graf lab for discussions and suggestions.
Supported by NIH grants 1 RO1 CA89590-01 and 1 RO1 N543881-01.
References
Author notes
Thomas Graf, 1300 Morris Park Ave, Bronx, NY 10461; e-mail: graf@aecom.yu.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal