Abstract
Nucleoside triphosphate diphosphohydrolases (NTPDases) are a recently described family of ectonucleotidases that differentially hydrolyze the γ and β phosphate residues of extracellular nucleotides. Expression of this enzymatic activity has the potential to influence nucleotide P2 receptor signaling within the vasculature. We and others have documented that NTPDase1 (CD39, 78 kd) hydrolyzes both triphosphonucleosides and diphosphonucleosides and thereby terminates platelet aggregation responses to adenosine diphosphate (ADP). In contrast, we now show that NTPDase2 (CD39L1, 75 kd), a preferential nucleoside triphosphatase, activates platelet aggregation by converting adenosine triphosphate (ATP) to ADP, the specific agonist of P2Y1 and P2Y12 receptors. We developed specific antibodies to murine NTPDase1 and NTPDase2 and observed that both enzymes are present in the cardiac vasculature; NTPDase1 is expressed by endothelium, endocardium, and to a lesser extent by vascular smooth muscle, while NTPDase2 is associated with the adventitia of muscularized vessels, microvascular pericytes, and other cell populations in the subendocardial space. Moreover, NTPDase2 represents a novel marker for microvascular pericytes. Differential expression of NTPDases in the vasculature suggests spatial regulation of nucleotide-mediated signaling. In this context, NTPDase1 should abrogate platelet aggregation and recruitment in intact vessels by the conversion of ADP to adenosine monophosphate, while NTPDase2 expression would promote platelet microthrombus formation at sites of extravasation following vessel injury. Our data suggest that specific NTPDases, in tandem with ecto-5′-nucleotidase, not only terminate P2 receptor activation and trigger adenosine receptors but may also allow preferential activation of specific subsets of P2 receptors sensitive to ADP (eg, P2Y1, P2Y3, P2Y12) and uridine diphosphate (P2Y6).
Introduction
Nucleoside triphosphate diphosphohydrolases (NTPDases) are a family of ectonucleotidases, previously classified as E-type ATPases, ATPDases, ecto-ATPases, or ecto-apyrases.1-3 These enzymes differentially hydrolyze the terminal γ and β phosphate residues of nucleotides, resulting in the rapid formation of the respective diphosphonucleosides and/or monophosphonucleosides. To date, 6 members of this NTPDase family have been identified.3-13 NTPDase1, NTPDase2, and NTPDase3 are transmembrane proteins associated with the plasma membrane with an active site facing the extracellular space.4,5,7,9,14These NTPDase members have been demonstrated to differ in their substrate specificity. For example, NTPDase1 (CD39 [human] or cd39 [murine]) hydrolyzes both nucleoside triphosphates and diphosphates—eg, adenosine triphosphate (ATP) and adenosine diphosphate (ADP),4,5—whereas NTPDase2 (CD39L1) is a preferential nucleoside triphosphatase or ATPase.7 14-16
Extracellular nucleotides in various forms, and at different concentrations, activate multiple P2 receptors: ionotropic P2X and metabotropic P2Y receptors.17-20 While NTPDases would be anticipated to generally terminate P2 receptor agonist signaling, we have also observed that NTPDase1 may prevent receptor desensitization by the catalysis of extracellular nucleotides.21 NTPDase1 may then also facilitate subsequent responses to “pulses” of nucleotides. In addition, NTPDases in tandem with ecto-5′-nucleotidase may facilitate the salvage of nucleotides by the ultimate generation of dephosphorylated forms that are taken up by cells via specific transporters.1 22
NTPDase1 is the major ectonucleotidase at the luminal surface of blood vessels.5,21,23,24 By converting the ADP released from activated platelets, NTPDase1 modulates platelet aggregation in vitro.5,25-28 The hypothesis that NTPDase1 is a key thromboregulatory factor has been further supported by in vivo experiments with NTPDase1-null (or cd39−/−) mice. This model has demonstrated important functions of the ectoenzyme in both regulating platelet aggregation and controlling the expression of procoagulants by the endothelium.21
Whether NTPDase2 is expressed in the vasculature has not been established to date. Furthermore, nothing is known about the functional integration of either NTPDase1 or NTPDase2 with each other or other ectonucleotidases/NTPDases. Biochemical analyses of murine and porcine cardiac tissues have demonstrated ATPase/ADPase ratios of 10, suggesting expression of enzymes with preferential ATPase activity (this paper and Lemmens et al29). Given the levels of NTPDase2 messenger RNA (mRNA) expression in murine and human hearts, this enzyme would be a likely candidate responsible for such an activity.6-8 NTPDase3 is another potential ectoenzyme that may modulate cardiac extracellular nucleotide levels. Although little is known about the distribution of human NTPDase3, cardiac mRNA expression appears low when compared with the signals observed for NTPDase1 and NTPDase2.8
In this paper, we show that catalytic differences between NTPDase1 and NTPDase2 are associated with the differential ability to regulate platelet function. This observation is consistent with the distinct localization of these ectoenzymes in the vasculature of the murine heart that affords yet another level of complexity by which these biologic activities may be regulated.
Materials and methods
Transient transfection and protein preparation
COS-7 cells were transfected using lipofectamine, as previously described.5 Forty-four hours after transfection, the cells were washed twice with Tris-saline, harvested by scraping, and washed twice by centrifugation at 300g for 5 minutes at 4°C. Prior to sonication, pellets were resuspended in 45 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, 10 μg/mL aprotinin, and 45 mM Tris, pH 7.6. Nucleus and cellular debris were discarded by another centrifugation as outlined above. Supernatants containing about 10 mg proteins per milliliter were kept frozen at −80°C. Homogenates and particulate fractions of murine tissues were prepared as previously described.24
NTPDase activity measurement
Enzyme activity in the protein fractions was determined as previously described.24 Briefly, enzyme activity was tested in 1 mL of 8 mM CaCl2, 50 mM Tris, and 50 mM imidazole, pH 7.4. After the addition of protein sample to the buffer, the mixture was preincubated at 37°C for 3 minutes and reaction was started by the addition of 0.3 mM substrate (ATP or ADP as indicated). This was terminated at 5 to 15 minutes with 0.25 mL of the malachite green reagent, and inorganic phosphate released from exogenous nucleotide measured.30 To determine specific activities, the protein content of the enzyme preparations was measured using the technique of Bradford.31
Analysis of platelet activation in vitro
Platelet-rich plasma (PRP) was prepared from human donors according to published methods.5 Platelet aggregation was measured in a Lumi-aggregometor apparatus (Chrono-log, Havertown, PA). Samples of 0.33 mL PRP were incubated at 37°C, and percent light transmission was measured and compared with platelet-poor plasma. Final concentrations of 1 to 20 μM ADP (Chrono-log) were used for platelet activation. In select experiments, aliquots to achieve final concentrations of 1 to 40 μM ATP (Sigma, St Louis, MO) were also added to PRP. Where indicated, varying amounts of recombinant protein samples (1-230 μg total protein) derived from NTPDase1- or NTPDase2-transfected COS cell lysates, diluted in 0.9% saline, were added to PRP.
Antibodies, plasmids, and other reagents
Antimurine NTPDase1 and NTPDase2 polyclonal antibodies (pAbs) were raised in rabbits by direct intramuscular and subcutaneous injection of complementary DNA (cDNA) encoding the whole gene ligated into pcDNA3.21,32 The plasmids expressing mouse NTPDase121 and the open reading frame of rat NTPDase2 have been described previously.7 Serum titers were determined by standard Western blot analysis under nonreducing conditions in the screening protein lysates from COS-7 cells expressing recombinant murine NTPDase1 or NTPDase2. The antibody specifically detected either NTPDase1 or NTPDase2 of both mouse and rat tissues in immunohistochemistry.
For selected double- and triple-labeling experiments, immunoglobulin G (IgG) fractions from NTPDase2 antisera, purified using a protein A–Sepharose column, were biotinylated using the EZ-Link Sulfo-NHS-LC-Biotinylation kit, according to the manufacturer's instructions (Pierce, Rockford, IL). The monoclonal antibody (mAb) anti–PECAM-1 (CD31),33 recognizing murine endothelial cells, and phycocyanin avidin were purchased from Pharmingen (San Diego, CA). The mAb anti-NG2 34,35 corresponding to the human high-molecular-weight melanoma-associated antigen, an epitope expressed on pericytes and smooth muscle cells,36 and the mAb antilaminin β2 chain37 used to detect basal lamina were purchased from Chemicon (Temecula, CA). The mAb antismooth muscle α-actin (clone 1A4)38 used as a marker for smooth muscle cells (and pericytes in other tissue than the heart) was purchased from Sigma. The mAb F4/80 recognizing blood monocytes and macrophages was purchased from Serotec (Raleigh, NC). Biotinylated rabbit antimouse IgG F(ab′)2 fragments and biotinylated swine antirabbit IgG F(ab′)2 were purchased from DAKO (Glostrup, Denmark). Normal serum (rabbit, mouse, goat, and swine) and IgG (rat, rabbit, and mouse) were purchased from Sigma. The biotinylated rabbit antirat IgG, Texas Red Avidin D, goat antirabbit IgG Texas Red conjugate, and rabbit antirat IgG fluorescein conjugate were purchased from Vector Laboratories (Burlingame, CA).
Immunoblotting procedures
Proteins were fractionated by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis according to Laemmli.39 Protein samples were boiled in sample buffer (2% [wt/vol] SDS, 10% [vol/vol] glycerin, 0.001% bromophenol blue in 65 mM Tris, pH 6.8) under nonreducing conditions. The proteins were separated on a 10% acrylamide SDS-gel and transferred to Immobilon-P membrane (Millipore, Bedford, MA) by semidry electroblotting (Bio-Rad, Hercules, CA).40 After incubation with the rabbit anti-NTPDase pAbs, the bands were visualized using horseradish peroxidase–conjugated goat antirabbit IgG (Pierce), at a dilution of 1:4000, and the Renaissance Chemiluminescence Reagent Plus, according to the manufacturer's instructions (NEN Life Science Products, Boston, MA).
Immunohistochemistry
Cardiac and other tissues from C57BL/6 and 129SVEV×C57BL/6 mice were harvested, embedded in Triangle Biomedical Sciences (TBS) tissue freezing medium (American Master*tech Scientific, Lodi, CA) and snap-frozen in isopentane cooled on liquid nitrogen, and stored at −80°C. Six-micrometer serial cryostat sections were fixed in ice-cold acetone for 10 minutes and rinsed in phosphate-buffered saline (PBS). IgG binding sites were blocked with appropriate control serum diluted 1:5 in solution 9 (PBS, pH 7.4, supplemented with 0.1% bovine serum albumin, 150 mM tranexamic acid, 20 μg/mL aprotinin [3-7 trypsin inhibitory units per milligram], 1.8 mM ethylenediaminetetraacetic acid, and 2 mM iodoacetic acid) further supplemented with 2% 3-omega fatty acid (Sigma) for 1 hour at room temperature. Sections were then incubated with primary antibody for 1 hour at room temperature (biotinylated NTPDase2-purified IgGs were incubated for 2 hours at 37°C), rinsed in PBS, and incubated with 3% H2O2 in methanol for 5 minutes to deplete endogenous peroxidase. After incubation with the appropriate biotinylated IgG or F(ab′)2 fragment of IgG for 30 minutes, staining was performed with the Vectastain ABC elite kit (Vector Laboratories) with diaminobenzidine as the peroxidase substrate. Sections were counterstained lightly with Mayer's hematoxylin, dehydrated, cleared in xylene, and mounted in Permount. Detection of mouse antigens with mouse mAbs was performed as previously described.41 All antibodies were diluted in solution 9 unless otherwise indicated. Optimal antibody concentrations were determined by serial dilution.
Triple staining and confocal imaging
Cryostat sections 6 μm thick were fixed in 100% acetone at 4°C for 20 minutes and rehydrated in PBS. Sections were blocked with 20% normal goat serum and 20% normal mouse serum containing 2% 3-omega fatty acid for 1 hour at room temperature. Sections were then incubated for 1 hour at room temperature with rabbit anti-NTPDase1 IgG, rinsed, incubated with goat antirabbit IgG fluorescein isothiocyanate conjugate or biotinylated goat antirabbit IgG for 30 minutes, and then rinsed. Sections were blocked with 5% normal rabbit serum for 30 minutes, rinsed, incubated with biotinylated NTPDase2 IgGs or avidin-coupled phycocyanin for 2 hours at 37°C and 30 minutes at room temperature, respectively, and rinsed. The sections were then incubated with Texas Red Avidin D or antismooth muscle actin fluorescein isothiocyanate conjugate for 1 hour at room temperature, rinsed, and blocked for residual binding sites to avidin using the avidin-biotin blocking kit (Vector Laboratories). These sections were then incubated with biotinylated anti-CD31 or biotinylated NTPDase2 IgG for 2 hours at room temperature and 37°C, respectively. After rinsing, the sections were incubated with avidin-coupled phycocyanin or Texas Red Avidin D for 30 minutes at room temperature and rinsed 5 times in PBS and once with distilled water. Sections were then mounted in fluoromount G and analyzed using an MRC-1024 confocal microscope equipped with an argon/krypton laser (Bio-Rad). Series of sequential optical sections were digitalized, filtered with edge definition and median filters, and viewed as compiled images.41
Statistics
Where appropriate, data are expressed as the mean ± SD. Groups were compared using the Student t test. Differences between experimental groups and controls were considered significant forP < .05.
Results
NTPDase activity in mouse hearts and transfected cells
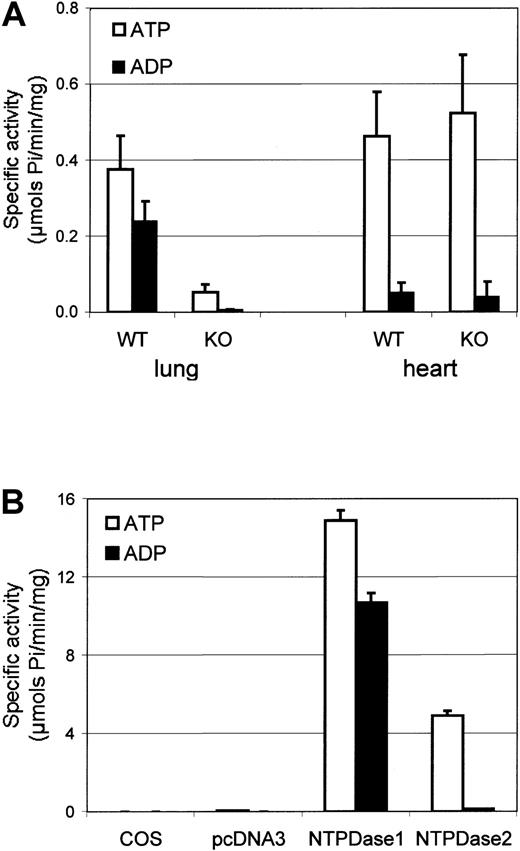
We determined levels of ATPase and ADPase activity in homogenates derived from a variety of wild-type mouse tissues (data not shown). Whereas relative ATPase and ADPase activities were similar in most tissues studied, the ATP-to-ADP hydrolysis ratios in the mouse hearts were strikingly higher, to the order of 10 (Figure1A). These data are comparable to prior observations in pig tissues.29
Biochemical activity of murine tissues and recombinant NTPDase1 and NTPDase2.
NTPDase activities of protein preparations were determined by measurement of phosphate release from the respective substrates. Data are expressed as the mean ± SD for specific ATPase (□) or ADPase (■) activities. (A) NTPDase activities ofcd39+/+ (n = 5) andcd39−/− (n = 5) mouse lung and heart preparations were measured. (B) NTPDase activity of protein extracts from NTPDase1- or NTPDase2-transfected cells were compared with control cells—untransfected (COS) or those transfected with pcDNA3 empty vector (n = 3 different transfections).
Biochemical activity of murine tissues and recombinant NTPDase1 and NTPDase2.
NTPDase activities of protein preparations were determined by measurement of phosphate release from the respective substrates. Data are expressed as the mean ± SD for specific ATPase (□) or ADPase (■) activities. (A) NTPDase activities ofcd39+/+ (n = 5) andcd39−/− (n = 5) mouse lung and heart preparations were measured. (B) NTPDase activity of protein extracts from NTPDase1- or NTPDase2-transfected cells were compared with control cells—untransfected (COS) or those transfected with pcDNA3 empty vector (n = 3 different transfections).
To identify the relative contribution of NTPDase1 to the total NTPDase activities in heart tissues, we then contrasted the activity of homogenates derived from cd39+/+ andcd39−/− tissues. Both ATPase and ADPase activities in cardiac tissue preparations from NTPDase1-null mice were not significantly different in wild-type tissues. Lung, a highly vascularized organ known to express high levels of NTPDase1, was used for comparison (Figure 1A). From the known distribution of NTPDases by Northern blots,7,8 the activity detected in the heart could likely be explained by expression of NTPDase2, a preferential ATPase.7
COS-7 cells were transfected by an expression vector (pcDNA3) containing the cDNA of murine NTPDase1 or NTPDase2. Biochemical activity of protein extracts derived from transfected cells exhibited significant increases in the hydrolysis of both substrates, ATP and ADP (Figure 1B). While the ratios of ATPase and ADPase activities in NTPDase1-transfected cells were similar, the ratio of ATP/ADP hydrolysis in protein extracts from NTPDase2-transfected cells was of the order of 40, in keeping with prior data derived from intact Chinese hamster ovary cells transfected with rat NTPDase2.7
NTPDase1 and NTPDase2 differentially regulate platelet function
The differential substrate specificity of NTPDase1 and NTPDase2 may explain functional effects of these enzymes. That NTPDase1 activity modulates platelet function in vivo has been suggested by studies in mice deficient in NTPDase1.21 Therefore, we contrasted the ability of both enzymes to influence platelet aggregation in the presence of the nucleotides ATP (a competitive antagonist of this process) and ADP (an agonist).18 19
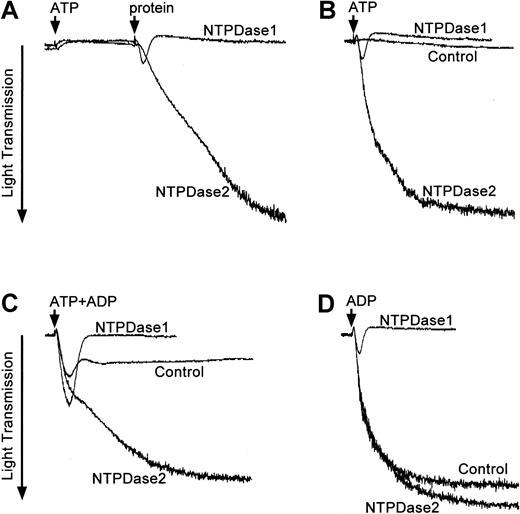
Consistent with the literature, no aggregation occurred when 20 μM ATP was added to PRP (Figure 2A). Addition of protein extracts from NTPDase1-transfected COS cells, in the presence of ATP, resulted in low levels of reversible aggregation that appeared related to conversion of ATP to ADP. However, because NTPDase1 efficiently hydrolyzed both nucleotides, induced platelet aggregation was rapidly and completely reversed (n = 6). In contrast, high levels of platelet aggregation were obtained when NTPDase2 protein extracts were added to PRP and ATP (n = 4). Similar data were obtained when protein extracts were preincubated with the PRP samples prior to the addition of ATP. Experiments with control protein extracts from COS cells transfected with pcDNA3 empty vector revealed no alterations in either platelet shape change or aggregation responses (Figure 2B).
Opposing effects of NTPDase1 and NTPDase2 on platelet aggregation.
PRP prepared from human donors was tested for platelet activation in the presence of exogenous nucleotides (ATP and/or ADP) and protein extracts from COS cells transfected with pcDNA3 encoding NTPDase1 or NTPDase2. Light transmittance (%) was recorded over a period of 8 (B-D) or 10 minutes (A). Protein extracts from COS cells transfected with vector DNA were added as controls at the required protein level. Representative aggregation profiles from 3 to 6 experiments are shown. (A) An arrow indicates the addition of 20 μM ATP to PRP followed 3 minutes later by the addition of 20 μg NTPDase1 or NTPDase2 protein preparation (second arrow). (B) PRP was preincubated for 25 seconds with 20 μg NTPDase1 (0.3 U ATPase) or NTPDase2 (0.1 U ATPase) protein extracts before the addition of 40 μM ATP. (C) PRP was preincubated for 25 seconds with 0.06 units of ATPase activity of NTPDase1 (4 μg) or NTPDase2 (12 μg) prior to the addition of a mixture of nucleotides (4 μM ATP plus 1 μM ADP). (D) PRP was preincubated for 25 seconds with 0.06 units of ATPase activity with either NTPDase1 (4 μg = 0.043 U ADPase) or NTPDase2 (12 μg) prior to the addition of 5 μM ADP.
Opposing effects of NTPDase1 and NTPDase2 on platelet aggregation.
PRP prepared from human donors was tested for platelet activation in the presence of exogenous nucleotides (ATP and/or ADP) and protein extracts from COS cells transfected with pcDNA3 encoding NTPDase1 or NTPDase2. Light transmittance (%) was recorded over a period of 8 (B-D) or 10 minutes (A). Protein extracts from COS cells transfected with vector DNA were added as controls at the required protein level. Representative aggregation profiles from 3 to 6 experiments are shown. (A) An arrow indicates the addition of 20 μM ATP to PRP followed 3 minutes later by the addition of 20 μg NTPDase1 or NTPDase2 protein preparation (second arrow). (B) PRP was preincubated for 25 seconds with 20 μg NTPDase1 (0.3 U ATPase) or NTPDase2 (0.1 U ATPase) protein extracts before the addition of 40 μM ATP. (C) PRP was preincubated for 25 seconds with 0.06 units of ATPase activity of NTPDase1 (4 μg) or NTPDase2 (12 μg) prior to the addition of a mixture of nucleotides (4 μM ATP plus 1 μM ADP). (D) PRP was preincubated for 25 seconds with 0.06 units of ATPase activity with either NTPDase1 (4 μg = 0.043 U ADPase) or NTPDase2 (12 μg) prior to the addition of 5 μM ADP.
Lower concentrations of nucleotides were also tested in various combinations. Mixtures of 1 μM ADP and 4 μM ATP added to PRP preincubated with control protein extracts resulted in low levels of platelet aggregation (Figure 2C). In the presence of low levels of NTPDase1 (4 μg per assay), the initial rates of platelet aggregation were increased (90 arbitrary units/min ± 4) when compared with controls (63 arbitrary units/min ± 4; P = .02, n = 3) but were transient and rapidly reversed. Higher concentrations of NTPDase1 COS cell extracts (> 20 μg per assay) totally prevented aggregation (data not shown). In contrast, in the presence of NTPDase2 samples, marked and sustained platelet aggregation always occurred.
Experiments in the presence of 5 μM ADP induced high levels of platelet aggregation in control samples (amplitude of 73% ± 2%, n = 3); this could be reversed by the addition of NTPDase1 (Figure2D). The initial extent of aggregation was slightly increased by the addition of NTPDase2 (78% ± 3%, P = .03, n = 3). Notably, NTPDase2 did not reverse the low levels of platelet aggregation induced by 1 μM ADP, even when larger amounts of NTPDase2 were added (700 μg transfected cell lysate per milliliter of PRP; data not shown). These data confirmed that ADPase activities of NTPDase2 were minimal and could not compete with platelet P2 receptor binding of the agonist ADP.
Experiments were also conducted in the simultaneous presence of both enzymes. Results obtained with the addition of equivalent units of ATPase activity from NTPDase1 and NTPDase2 protein extracts gave similar results to NTPDase1 alone. These data suggested that the effects of NTPDase1 on platelet aggregation were dominant over NTPDase2 (data not shown).
All aggregation data presented above were confirmed with PRP from different donors and using different batches of recombinant proteins. In all experiments conducted here, protein extracts from COS cells transfected with control DNA gave similar aggregation profiles to the assays performed without protein extracts (data not shown).
In summary, NTPDase1 largely inhibits and reverses platelet aggregation in the presence of ADP and/or ATP. In contrast, NTPDase2 promotes platelet aggregation in the presence of ATP and further facilitates it in the presence of ADP.
Immunolocalization of NTPDase1 and NTPDase2 in mouse heart
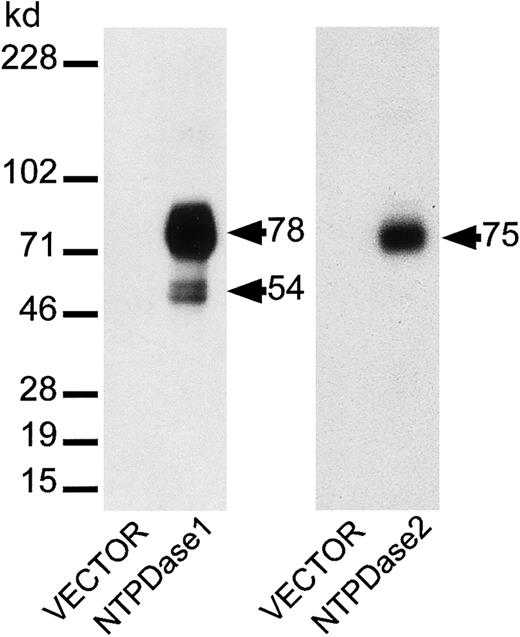
NTPDase1 and NTPDase2 pAbs were generated against the murine proteins and specificity established by Western blot analysis (Figure3). As expected, NTPDase1 antiserum detected bands of about 78 and 54 kd (proteolytic product of the 78-kd form) in the protein extracts of COS cells transfected with the NTPDase1 construct, as we previously observed for bovine, porcine, and human NTPDase1.24,29,42 No bands were detected in control protein extracts (Figure 3). NTPDase2 antibodies detected a band evaluated at 75 kd in the protein extracts from cells transfected with NTPDase2 construct only. Specificity of the antibodies was also analyzed for the ability of the anti-NTPDase pAbs to bind the native protein at the cell surface. Cross-reactivity of NTPDase1 pAb with NTPDase2 was excluded and vice versa.43 These antisera were used to elucidate the distribution of NTPDase1 and NTPDase2 in vivo by immunohistology.
Specificity of antisera for murine NTPDase1 and NTPDase2.
Protein samples (5 μg) from COS cells, transiently transfected with NTPDase1 or NTPDase2 cDNA constructs or from cells transfected with empty vector DNA (as a negative control), were fractionated on 10% acrylamide SDS–polyacrylamide gel electrophoresis under nonreducing conditions. Separated proteins were then transferred to an Immobilon-P membrane and incubated with pAbs directed against NTPDase1 or NTPDase2. Each antiserum detected protein bands only in the appropriately transfected cells.
Specificity of antisera for murine NTPDase1 and NTPDase2.
Protein samples (5 μg) from COS cells, transiently transfected with NTPDase1 or NTPDase2 cDNA constructs or from cells transfected with empty vector DNA (as a negative control), were fractionated on 10% acrylamide SDS–polyacrylamide gel electrophoresis under nonreducing conditions. Separated proteins were then transferred to an Immobilon-P membrane and incubated with pAbs directed against NTPDase1 or NTPDase2. Each antiserum detected protein bands only in the appropriately transfected cells.
In most organs studied, immunolocalization of both NTPDase1 and NTPDase2 was predominantly confined to vascular structures (Figures4 and 5and data not shown). However, NTPDase1 and NTPDase2 were expressed on different cell types within the vascular wall of both microvessels and larger muscularized vessels (Figures 4 and 5 and data not shown).
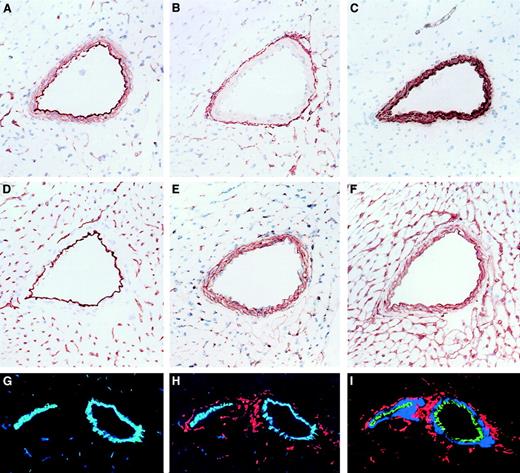
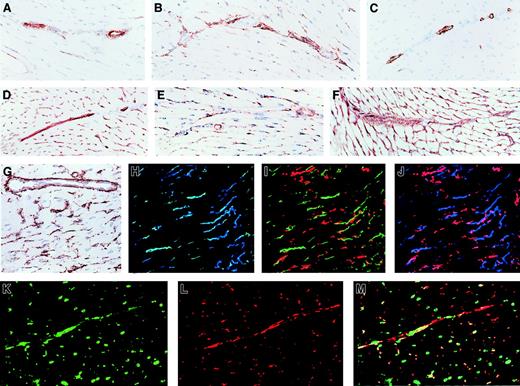
Immunohistologic localization of NTPDases in cardiac vasculature.
Serial sections of a mouse heart were stained by immunohistochemistry with various antibodies detecting (A) NTPDase1, (B) NTPDase2, (C) α-actin (a marker of smooth muscle), (D) CD31 (a marker of endothelium), (E) NG2 (a marker of pericytes and smooth muscle cells), and (F) laminin (a marker of basement membranes). Positive reactions were seen as rust color (A-F), and sections were counterstained with hematoxylin (blue). In panels G-I, immunofluorescence and confocal analysis were performed: (G) CD31 in blue and NTPDase1 in green (colocalization as aquamarine); (H) as for panel G with addition of NTPDase2 in red; (I) NTPDase1 in green, smooth muscle α-actin in blue, and NTPDase2 in red (no colocalization). In medium-sized vessels, CD31 and NTPDase1 were found to colocalize while NTPDase2 did not colocalize with these 2 markers or the smooth muscle α-actin–expressing cells. Original magnifications, ×200.
Immunohistologic localization of NTPDases in cardiac vasculature.
Serial sections of a mouse heart were stained by immunohistochemistry with various antibodies detecting (A) NTPDase1, (B) NTPDase2, (C) α-actin (a marker of smooth muscle), (D) CD31 (a marker of endothelium), (E) NG2 (a marker of pericytes and smooth muscle cells), and (F) laminin (a marker of basement membranes). Positive reactions were seen as rust color (A-F), and sections were counterstained with hematoxylin (blue). In panels G-I, immunofluorescence and confocal analysis were performed: (G) CD31 in blue and NTPDase1 in green (colocalization as aquamarine); (H) as for panel G with addition of NTPDase2 in red; (I) NTPDase1 in green, smooth muscle α-actin in blue, and NTPDase2 in red (no colocalization). In medium-sized vessels, CD31 and NTPDase1 were found to colocalize while NTPDase2 did not colocalize with these 2 markers or the smooth muscle α-actin–expressing cells. Original magnifications, ×200.
Immunohistologic localization of NTPDases in the cardiac microvasculature.
Immunohistochemistry of mouse heart sections (A-G) are shown in the same order as in Figure 4. Antibody specificity: (A) NTPDase1, (B) NTPDase2, (C) α-actin (a marker of smooth muscle), (D) CD31 (a marker of endothelium), (E) NG2 (a marker of pericytes and smooth muscle cells), (F) laminin (a marker of basement membranes), and (G) a higher magnification of NTPDase2. Sections were counterstained with hematoxylin. In panels H-M, immunofluorescence and confocal analysis were performed: (H) NTPDase1 in green and CD31 in blue (colocalization is aquamarine); (I) NTPDase1 in green and NTPDase2 in red; (J) CD31 in blue and NTPDase2 in red (no colocalization); (K) pericyte marker NG2 in green; (L) NTPDase2 in red; (M) panels K and L combined (colocalization is yellow). In the microvasculature, NTPDase1 colocalized with an endothelial cell marker (CD31) while NTPDase2 colocalized with a pericyte marker (NG2). Original magnifications A-F and H-M, ×200; G, ×400.
Immunohistologic localization of NTPDases in the cardiac microvasculature.
Immunohistochemistry of mouse heart sections (A-G) are shown in the same order as in Figure 4. Antibody specificity: (A) NTPDase1, (B) NTPDase2, (C) α-actin (a marker of smooth muscle), (D) CD31 (a marker of endothelium), (E) NG2 (a marker of pericytes and smooth muscle cells), (F) laminin (a marker of basement membranes), and (G) a higher magnification of NTPDase2. Sections were counterstained with hematoxylin. In panels H-M, immunofluorescence and confocal analysis were performed: (H) NTPDase1 in green and CD31 in blue (colocalization is aquamarine); (I) NTPDase1 in green and NTPDase2 in red; (J) CD31 in blue and NTPDase2 in red (no colocalization); (K) pericyte marker NG2 in green; (L) NTPDase2 in red; (M) panels K and L combined (colocalization is yellow). In the microvasculature, NTPDase1 colocalized with an endothelial cell marker (CD31) while NTPDase2 colocalized with a pericyte marker (NG2). Original magnifications A-F and H-M, ×200; G, ×400.
Serial sections of muscularized vessels stained with smooth muscle cell and endothelial cell markers suggested that NTPDase1 was predominantly expressed on endothelium and to a lesser extent in the smooth muscle layer. In contrast, NTPDase2 was expressed in cells present in the vascular adventia, coinciding with staining for the basal lamina component, laminin (Figure 4A-F). Triple immunofluorescent staining (using antibodies recognizing NTPDase1, NTPDase2, CD31 [marker of endothelial cells], and α-actin [marker of smooth muscle cells], analyzed by confocal microscopy) confirmed that NTPDase1 was predominantly localized to the endothelium, whereas NTPDase2 was expressed in the advential layer of muscularised vessels (Figure 4G-I). Standard markers recognizing endothelium, pericytes, smooth muscle cells, and macrophages did not stain cells in the advential layer of medium-sized muscularized vessels, indicating absence of vasa vasora in these vessels (Figure 4 and data not shown). The cellular component of the vascular adventia in smaller muscularized vessels consists predominately of nerves and fibroblasts embedded in a connective tissue matrix.44
Standard immunohistochemical techniques also demonstrated that NTPDase1 and NTPDase2 were expressed in microvascular structures (Figure 5A-G). To identify the cell types expressing NTPDase1 and NTPDase2, double and triple immunofluorescent staining was performed (using antibodies recognizing pericytes and endothelial cells in conjunction with antibodies to NTPDase1 and NTPDase2). Samples were then analyzed using confocal laser microscopy. Expression of NTPDase1 was localized to the endothelium, whereas NTPDase2 was expressed in cells juxtapositioned to the endothelium and enveloped in the basal membrane, suggesting that NTPDase2 was expressed on pericytes (Figure 5). Double immunofluorescent staining with biotinylated IgG recognizing NTPDase2 in conjunction with NG2 confirmed the predominant expression of NTPDase2 on microvascular pericytes (Figure 5K-M). Anti-NTPDase2 and anti-NG2 antibodies colocalized with variable intensities within these individual structures, suggesting that the distribution of these markers on the pericyte cell surface might differ somewhat. Also of interest, NTPDase1 was expressed in cells in the endocardium, whereas NTPDase2 was expressed in a population of cells located immediately adjacent to the endocardium, in the subendocardial space (data not shown).
Alpha-actin is widely used as a marker for pericytes and smooth muscle cells. However, pericyte expression of α-actin varies between different tissues and is dependent upon cellular activation.36,41,45,46 In the mouse heart, α-actin was found to be present on smooth muscle cells of larger vessels but was consistently absent on microvascular pericytes (Figures 4C and 5C). In this organ, no coexpression of NTPDase2 and α-actin was observed. Partially muscularized vessels contained supporting cells that were either positive for both NG2 and smooth muscle α-actin or for NG2 only. These data suggest that such vessels contain intermediate cells, a transitional cell with a phenotype that shares characteristics with both pericytes and smooth muscle cells.44 In these vessels, NTPDase2 expression was confined to NG2-positive and smooth muscle α-actin–negative cells, supporting the initial findings that NTPDase2 is exclusively expressed on microvascular pericytes (data not shown).
In other tissues analyzed, NTPDase2 was also consistently absent from the smooth muscle layer of medium-sized vessels. In these tissues, including striated muscle and large intestine, smooth muscle α-actin expression was present in microvessels. In the latter tissues, NTPDase2 colocalized with smooth muscle α-actin, suggesting that the expression of NTPDase2 on pericytes was not confined to the heart muscle but applies to other organ systems as well (data not shown). Qualitatively, similar results with respect to the cellular distribution of NTPDase1 and NTPDase2 were observed in murine striated muscle, large intestine, and heart (Figures 4 and 5 and data not shown).
Discussion
We and others have previously identified the major vascular endothelial cell NTPDase to be CD39 and demonstrated thomboregulatory properties of this ectoenzyme.5,21,28 We have now identified a second NTPDase that is expressed in vasculature—namely, NTPDase2 (or CD39L1). We observed that NTPDase1 and NTPDase2 have similar molecular masses of 78 kd and 75 kd, respectively (Figure 3), and we have contrasted the individual biochemical properties of the vascular NTPDase1 and NTPDase2. Specifically, NTPDase1 hydrolyses both ATP and ADP while NTPDase2 is a preferential ecto-ATPase7,14 (Figure 1). These differential biochemical properties of NTPDase1 and NTPDase2 may have important implications with respect to platelet function. By hydrolyzing ADP, NTPDase1 inhibited platelet aggregation; in contrast, NTPDase2 promoted platelet aggregation in the presence of ATP and facilitated this process in the presence of ADP (Figure 2). The more pronounced aggregation observed in the latter experiments could be explained by the hydrolysis of ATP released from the platelet granules in an autocrine and paracrine manner. This process would increase the local concentrations of ADP because platelets concentrate both ATP and ADP in dense granules to the order of 1 M.47 Interestingly, our data suggest also that NTPDase2 facilitates ADP-specific P2 receptor activation by the conversion of ATP. Indeed, ATP is generally considered a competitive antagonist of platelet P2Y1 and P2Y12 while ADP triggers their activation and promotes aggregation,18-20 as seen in Figure 2. Thus, NTPDase1 and NTPDase2 activities have opposing effects on platelet aggregation in vitro.
NTPDase2 did not influence platelet aggregation in the presence of low micromolar ADP. On the other hand, NTPDase2 efficiently converted low micromolar concentrations of ATP to ADP, triggering platelet aggregation in vitro. These data demonstrate that the rate of ATP hydrolysis by NTPDase2 with the consequent generation of ADP is potentially relevant to the regulation of platelet activation in vivo. ATP appears to be released by injured cells while platelets release both ATP and ADP in similar concentrations.47 Also of interest, the NTPDase1 effects on nucleotide-mediated platelet aggregation appear dominant over NTPDase2 activity; both enzymes in combination inhibited platelet aggregation as efficiently as did NTPDase1 alone (data not shown).
Consistent with the opposing actions on platelet aggregation, NTPDase1 and NTPDase2 were expressed within different strata and by different cell types within the vasculature. In mouse heart, we observed that NTPDase1 was expressed on vascular endothelium and endocardium and to a lesser extent on vascular smooth muscle cells—in keeping with published descriptions of other vascular beds as carried out by standard immunohistochemistry.24 48-50 These data, and other unpublished observations (J.S., E.C., S.C.R., 1999), demonstrate that NTPDase1 is consistently expressed within the vasculature of mammals.
To date, little has been determined about the expression and function of NTPDase2.6,7,14,51 NTPDase2 mRNA has been detected in homogenates of several human and rat tissues.6,7 In vitro, NTPDase2 mRNA expression in mouse hepatoma cells has been shown to be induced by dioxin.52 NTPDase2 mRNA has also been detected in the PC12 cell line.7 By immunohistologic techniques, we demonstrate here that NTPDase2 is expressed in the vasculature, mainly by microvascular pericytes, adventitial cells in muscularised vessels, and by distinct cell populations in the subendocardial space.
This differential localization of NTPDase1 and NTPDase2 may have direct implications for the control of platelet activation and coagulation responses in vivo. The expression of NTPDase1 on the endothelium and endocardium allows clearance of ADP (and ATP) from the blood plasma, thus blocking and/or modulating platelet aggregation under resting conditions.21,53 NTPDase1 may represent an important thromboregulatory factor, together with nitric oxide and prostaglandins PGI2 and D2, in the modulation of platelet activation.21,53 Targeting the endothelium with recombinant adenoviral-expressed NTPDase1 to increase levels of expression of this ectoenzyme has been shown to have beneficial thrombomodulatory effects in transplantation models.21,54In addition, both cd39+/− andcd39−/− mice express a prothrombotic phenotype (Enjyoji et al21; M.I., O.G., J.S., S.C.R., unpublished observation, November 2000).
Upon damage to the vessel, supporting cells that express NTPDase2 are exposed to increased levels of ATP released from injured cells, red blood cells, activated platelets, smooth muscles, and endothelium.47 Subsequent conversion to ADP by NTPDase2 will then promote platelet plug formation. Notably, strong effects on platelet aggregation were observed with minimal amounts of recombinant NTPDase2 in vitro. Comparable levels of recombinant NTPDase1 and NTPDase2 (at either units of ATPase activity or total protein levels) both influence platelet aggregation in vitro (Figure 2 and data not shown). This specific localization of NTPDase1 and NTPDase2 spatially compartmentalizes the distinct catalytic activities of these 2 enzymes within different elements of the vasculature. The functional and structural integrity of the endothelium dictates that NTPDase1 activity is dominant under quiescent conditions. This scenario would facilitate basal antithrombotic activity of NTPDases (NTPDase1) that could rapidly shift to a prothrombotic activity upon vascular damage and consequent exposure of elements of the outer vessel wall to blood (NTPDase2).
It has been proposed that pericytes and adventitial cells may be important in the process of clot formation through effects on the coagulation cascade.55-58 Our results suggest that these cell types also promote clot formation in a previously unrecognized fashion via direct effects on platelet function. Pericytes and adventitial cells may serve therefore as a hemostatic envelope capable of initiation and/or promotion of thrombus formation when vascular integrity is disrupted.
NTPDase2 may serve as a novel marker to discriminate between pericytes and smooth muscle cells in situ. A limited number of markers for pericytes have recently become available that have facilitated purification and research on the functional significance of these cells. However, the interpretation of the expression of such “specific” markers reveals several limitations. Expression of different pericyte markers depends largely on the activation state of the tissue and has proven less useful in identifying pericytes in resting tissues. No antibodies can discriminate between pericytes, smooth muscle cells, and myofibroblasts. Importantly, we showed that NTPDase2 is not expressed on smooth muscle cells or on transitional supporting cells in partially muscularized vessels that share characteristics between smooth muscle cells and pericytes,44 thus allowing discrimination between these cell types. NTPDase2 was expressed on microvascular pericytes in resting tissues from several different organs (Figures 4 and 5 and data not shown). Our data suggest that NTPDase2 may serve as a useful marker for defining and characterizing pericytes in vivo.
Biochemical properties and specific localization of NTPDase2 in the heart suggest that the enzyme may regulate functions other than platelet activation. Extracellular ATP and its degradation product adenosine exert pronounce inotropic and chronotropic actions and influence electrical impulse conduction as well as metabolic processes.59-61 Adenosine has also been shown to have important cardioprotective effects following ischemic injury to the heart.62,63 These effects are triggered through the activation of various adenosine60 and P2 receptors.64-66
The hydrolysis of ATP by NTPDase2 at the surface of adventitial cells and pericytes may also be important in the control of vascular tone—a function that could be coordinated further by NTPDase1 expressed on vascular smooth muscle cells. Indeed P2X receptors,67predominantly P2X1, are expressed on vascular smooth muscle cells as well as P2Y1, P2Y2, P2Y4, and P2Y6 receptors.68 Much of the vasomotor tonic action of ATP can be shown to be related to the activation of P2X1, but P2Y-mediated signaling also plays a role in this process.68 To our knowledge, there are no published studies that document P2 receptor expression in pericytes. However, there are similarities between pericytes and smooth muscle cells, known to express various P2 receptors. P2Y receptors are also involved in vascular smooth muscle cell proliferation.68 69 It is unclear whether NTPDase1 and NTPDase2 play regulatory roles in these events.
In summary, differential biochemical properties and expression patterns of NTPDase1 and NTPDase2 correlate with the distinct functions of these enzymes on platelet aggregation. Importantly, NTPDase1 faces the blood circulation and abrogates platelet aggregation. In contrast, NTPDase2 is expressed in supporting cells of the vasculature and facilitates platelet aggregation. Our data further support the view that NTPDases may not only directly terminate P2 receptor signaling but under certain circumstances may also mediate the activation of specific ADP receptors such as platelet P2Y170 and P2Y12 by the generation of the specific agonists.
We thank Dr R. Neal Smith of the Immunopathology Unit, Department of Pathology, Massachusetts General Hospital, Boston, MA, for valuable discussion.
Supported by NIH grants RO1HL57307 and RO1HL63972-01 (to S.C.R.), the American Liver Foundation and the Canadian Institutes of Health Research (to J.S.), the Deutsche Forschungsgemeinschaft (SFB 269, A4) and the Fonds der Chemischen Industrie (to H.Z.), and a grant from the Swedish Cancer Foundation and the King Gustaf the Fifth's 80 year fund (to C.S.). O.G. was a recipient of a fellowship from the Deutsche Forschungsgemeinschaft (GU 490/1-2).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jean Sévigny, Centre de Recherche en Rhumatologie et Immunologie, 2705 Blvd Laurier, Local T1-49, Sainte-Foy, Québec, G1V 4G2, Canada; e-mail:jean.sevigny@crchul.ulaval.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal