In 2000, we elucidated the structure of the RHlocus1 by showing that it is an example for a gene cluster; RHD and RHCE face each other by their 3′ tail ends, and a third gene, SMP1, was found to be interspersed between the 2 rhesus genes. Two 9 000 base pair (bp) DNA segments, dubbed “rhesus boxes,” of identical orientation fringed the RHD gene (Figure 1, top).
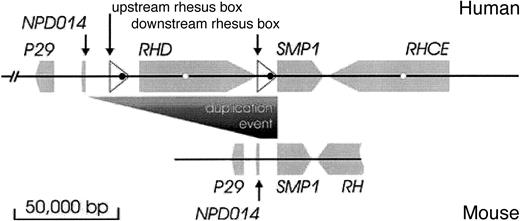
RH locus in human and mouse.
The human RH locus (top) is compared to the mouseRH locus (bottom). The orientations and positions of genes are indicated by gray arrows, the rhesus boxes by white triangles. The fragment of the mouse RH gene shown covers exons 2 to 10. An open reading frame (ORF) in the rhesus box is depicted as a black circle, the succinate dehydrogenase complex, subunit D precursor pseudogene in RH intron 3 as a white circle; no sequences homologous to these ORF were found in the mouse locus. During the duplication event, RHD and its fringing rhesus boxes were inserted between NPD014 and SMP1. The depicted DNA segments are GCIP-binding protein P29 (accession numbers human, NM_015484; mouse, AY033432), NPD014 (AF247168,AK003799), SMP1 (NM_014313, AK014282) and RH(CE)(M34015, NM_011270), and the ORF in the upstream and downstream rhesus boxes (XP_044740 and XP_057505).
RH locus in human and mouse.
The human RH locus (top) is compared to the mouseRH locus (bottom). The orientations and positions of genes are indicated by gray arrows, the rhesus boxes by white triangles. The fragment of the mouse RH gene shown covers exons 2 to 10. An open reading frame (ORF) in the rhesus box is depicted as a black circle, the succinate dehydrogenase complex, subunit D precursor pseudogene in RH intron 3 as a white circle; no sequences homologous to these ORF were found in the mouse locus. During the duplication event, RHD and its fringing rhesus boxes were inserted between NPD014 and SMP1. The depicted DNA segments are GCIP-binding protein P29 (accession numbers human, NM_015484; mouse, AY033432), NPD014 (AF247168,AK003799), SMP1 (NM_014313, AK014282) and RH(CE)(M34015, NM_011270), and the ORF in the upstream and downstream rhesus boxes (XP_044740 and XP_057505).
Based on this structure of the RH locus, the RHDgene deletion was parsimoniously explained by an unequal crossing-over event.1 Furthermore, the inverse orientation of theRH genes may facilitate gene conversion among both rhesus genes, which would explain the high frequency of RHD-CE-D orRHCE-D-CE hybrid alleles.2 However, it remained unknown which rhesus gene, if any, represented the ancestral positioning. The close proximity of the RHCE andSMP1 in humans was startling too.
The duplication of the rhesus gene is known to have occurred during primate evolution,3 giving rise to the RHD andRHCE genes in humans. Hence nonprimate mammals, like mice, may reveal the ancient state of the RH locus. In this context an 89 065 bp genomic DNA segment that was recently deposited in public databases (GenBank entry AL611963), which encompassed the mouse RH locus (Figure 1, bottom), was most disclosing. In order to compare the topology in mouse to the humanRH locus we assembled a 315 242 bp DNA segment that included the human RH locus.
The assembly of this human genomic DNA was complicated by the fact that the current GenBank entry AL139426 contained sequences representative of RHD, SMP1, both rhesus boxes, and parts ofRHCE but did not represent their correct topology. To overcome this limitation we compared the sequence of AL139426 to the sequences of RHD (X63097) and RHCE (M34015) cDNA, of RHD (AB035192) and RHCE (AB035191) intron 3, of RHD (AB035185) and RHCE (AB035184) intron 9, and of the upstream (AJ252311) and downstream (AJ252312) rhesus boxes. We determined multiple misassemblies occurring in long regions between almost identical paralogous sequences (join ofRHD exon 3 to RHCE exon 4, RHCE exon 3 to RHD exon 4, 5′ upstream rhesus box to 3′ downstream rhesus box, 3′ upstream rhesus box to 5′ upstream rhesus box, and failed assembly of RHCE intron 9). We compiled the 315 242 bp human genomic DNA contig (Figure 1, upper panel) including both rhesus genes and a stretch of surrounding DNA comprising more than 100 000 bp using AL031432 (5′ of RHD), AL031284 (RHCE), AB035185 (RHD intron 9), AB035184 (RHCE intron 9) and a corrected version of AL139326. This third party annotated human DNA segment was deposited under GenBank accession number BN000065.
The position and orientation of proteins in the human and mouse DNA segments were determined by a homology search against the nonredundant protein database of the GenBank (Tblastx) utilizing the NCBI Blast page. Then, each possible match was manually evaluated after a 2-sequence alignment (Blast 2 sequences).4 Both genomic DNA segments contained the RH gene(s), SMP1, and the 2 additional genes, GCIP-interacting protein P29 andNPD014. In addition, the human DNA segment, but not the mouse DNA segment, contained the 2 rhesus boxes carrying one open reading frame each and a succinate dehydrogenase pseudogene located in the introns 3 of RHD and RHCE (Figure 1). The 3′ ends of the rhesus boxes carry GC-rich regions that are typical for some strong promoters. The juxtapositioning of this structure right in front of the SMP1 start codon may modify the expression of smp1 in primates compared to nonprimate species.
Based on the gene positions and orientations, RHCE was determined to represent the ancestral state. The close proximity ofSMP1 and RH known in humans1 was also observed in the mouse RH locus (Figure2). In the mouse, there were 8 639 bp between NPD014 and SMP1. This size of a DNA stretch corresponded to the 11 437 bp between NPD014 and the upstream rhesus box rather than to the 91 136 bp betweenNPD014 and SMP1. The limited conservation of the noncoding regions did not allow a more detailed analysis of theRH duplication site in the moment.
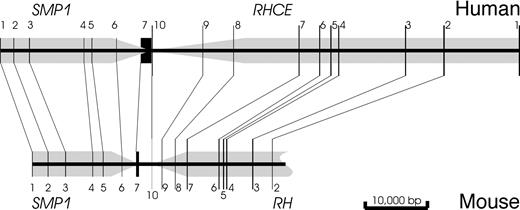
Topology of SMP1 and RHCE.
The orientations and positions of the genes are indicated by gray arrows and the localization of the exons by black lines at their approximate scale. The relative position and orientation ofRHCE and SMP1 is conserved. Human and mouseSMP1 have a very similar topology. Both human rhesus genes are much enlarged relative to the mouse RH gene because of considerably elongated introns.
Topology of SMP1 and RHCE.
The orientations and positions of the genes are indicated by gray arrows and the localization of the exons by black lines at their approximate scale. The relative position and orientation ofRHCE and SMP1 is conserved. Human and mouseSMP1 have a very similar topology. Both human rhesus genes are much enlarged relative to the mouse RH gene because of considerably elongated introns.
Among the 4 proteins, smp1 was most conserved and Rh was least conserved (Table 1). There are 2 human smp1–analogous proteins, smp1 (accession number AAD17754) located in chromosome 1 and c21orf4 (P56557) located in chromosome 21. These 2 human proteins corresponded to 2 different mouse proteins, BAB29242 andBAB32266, that had 94% and 98% homology to the human genes, respectively.
Conservation of proteins at the RH gene locus between mouse and human
| Protein . | Protein database accession numbers . | Length (amino acids) . | Amino acid positions . | |||
|---|---|---|---|---|---|---|
| Human . | Mouse . | Human . | Mouse . | Identical (%) . | Conserved (%) . | |
| RhD | Q02161 | NP_035400 | 417 | 418 | 239 (56) | 296 (69) |
| RhCE | CAA44811 | NP_035400 | 417 | 418 | 240 (56) | 296 (69) |
| npd014 | AAG44543 | BAB23002 | 242 | 248 | 177 (75) | 187 (79) |
| p29 | NP_056299 | AAK54459 | 243 | 242 | 221 (90) | 229 (93) |
| smp1 | AAD17754 | BAB29242 | 157 | 157 | 149 (94) | 152 (96) |
| Protein . | Protein database accession numbers . | Length (amino acids) . | Amino acid positions . | |||
|---|---|---|---|---|---|---|
| Human . | Mouse . | Human . | Mouse . | Identical (%) . | Conserved (%) . | |
| RhD | Q02161 | NP_035400 | 417 | 418 | 239 (56) | 296 (69) |
| RhCE | CAA44811 | NP_035400 | 417 | 418 | 240 (56) | 296 (69) |
| npd014 | AAG44543 | BAB23002 | 242 | 248 | 177 (75) | 187 (79) |
| p29 | NP_056299 | AAK54459 | 243 | 242 | 221 (90) | 229 (93) |
| smp1 | AAD17754 | BAB29242 | 157 | 157 | 149 (94) | 152 (96) |
In conclusion, RHD arose by a duplication ofRHCE. It is likely that the orientation of RHDwas inverted during this event. We propose that the rhesus boxes were instrumental for the duplication. SMP1 is a highly conserved gene located in the immediate proximity of RH during much of the mammalian evolution. An understanding of the events shaping the rhesus polymorphism and the underlying mechanisms will contribute to improving genotyping strategies for rhesus as well as possibly for a host of other loci with clustered genes in the genome.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal