Abstract
CCAAT/enhancer-binding proteins (C/EBPs) are critical transcriptional regulators of differentiation of hematopoietic cells. Previous studies have shown that targeted disruption of theC/ebpα gene results in a lack of granulocytic differentiation with an arrest at the stage of immature myeloblasts. By using a gene replacement strategy in which C/EBPβ was expressed from the C/ebpα gene locus of C/EBPα-null mice, we have evaluated the ability of C/EBPβ to function for C/EBPα in directing differentiation along the granulocytic pathway. We show that the morphology and the differential cell counts of the bone marrow and peripheral blood cells from C/EBPβ knockin mice are indistinguishable from those of their wild-type littermates, indicating that hematopoiesis occurs normally in these animals. Additionally, we analyzed expression of 21 myeloid-specific genes, including markers for distinct stages of granulocytic differentiation, and found no significant differences in their levels of expression in the bone marrow of C/EBPβ knockin and wild-type mice. These results imply that C/EBPβ can substitute for C/EBPα during hematopoiesis when expressed from the C/ebpα gene locus.
Introduction
CCAAT/enhancer-binding proteins (C/EBPs) are a family of structurally related transcription factors made up of 6 members (C/EBPα, -β, -γ, -δ, -ε, and -ζ).1-7All C/EBPs share conserved C-terminal regions that contain leucine-zipper dimerization motifs adjacent to basic DNA-binding domains.8 Their N-terminal regions are more diverse and contain transcriptional activation domains. Dimerization within the C/EBP family or with other transcription factors is a prerequisite for DNA binding and subsequent transactivation. With the exception of C/EBPε, proteins in the C/EBP family are expressed in partially overlapping patterns in multiple tissues.9 However, targeted inactivation of C/EBP family genes in mice has demonstrated their individual contributions to cellular differentiation.
Knockout mice models have defined a critical role for C/EBPs in hematopoietic tissues.10-14 Two members, C/EBPα and C/EBPβ, play key roles in determining the fate of differentiating hematopoietic cells. For example, C/EBPα is expressed in early myeloid cells,9 and its absence in C/EBPα−/− mice leads to a complete lack of granulocytic differentiation with an arrest at the stage of immature myeloblasts.10 Although mature neutrophils and eosinophils are absent in C/EBPα-null mice, other hematopoietic lineages are not affected. C/EBPβ, however, appears to be a critical signaling molecule for more mature myeloid cells as well as for B lymphocytes because its expression is dramatically induced during macrophage differentiation9,15 and lymphopoiesis.12Targeted deletion of C/EBPβ in mice results in impaired macrophage function, lymphoproliferative disorders, and defective B lymphopoiesis. Differential expression of these 2 C/EBPs in hematopoietic tissues underscores their individual roles in the development of mature blood cells.
In addition to their roles in the hematopoietic system, C/EBPα and C/EBPβ are important for normal development of liver16,17and adipose tissue.4,18 Mice lacking C/EBPα die within 8 hours of birth because of a severe loss of liver function. However, a gene replacement approach in which C/EBPβ is knocked into theC/ebpα gene locus of C/EBPα-null mice restores liver function and, consequently, their viability.19 These mutant mice, C/ebpαβ/β, lack C/EBPα but have a concomitant gain of C/EBPβ in tissues. In the current study, we evaluate the ability of C/EBPβ functionally to replace C/EBPα in the hematopoietic system of C/ebpαβ/βmice. We find that bone marrow and peripheral blood cells from C/EBPβ knockin mice are indistinguishable from those of their wild-type littermates, indicating that hematopoiesis occurs normally in these animals. We confirm this finding on a molecular level by analyzing the expression of 21 myeloid-specific genes, including markers for distinct stages of granulocytic differentiation. Our results reveal no significant differences in the levels of expression of these genes in bone marrow of C/EBPβ knockin and wild-type mice, thus implying that C/EBPβ can substitute for C/EBPα during hematopoiesis when expressed from the C/ebpα gene locus.
Materials and methods
Mice
C/ebpαβ/β mice were generated by using gene-targeting technology and the Cre/loxP DNA recombination system as reported previously.19 Mice were maintained in a specific pathogen-free animal facility at the Institute of Molecular Biology, Academia Sinica, Taipei. Five- to 6-week-old littermates from the heterozygote interbreedings were used in this study.
Bone marrow morphologic analysis
Bone marrow was collected from the femoral bone of wild-type and C/EBPβ knockin mice. Smears were prepared and stained with Wright-Giemsa. Bone marrow smears were analyzed by light microscopy.
Differential cell counts
Differential white cell counts for peripheral blood and bone marrow were determined manually by 2 independent investigators. Percentages were calculated according to the cell morphology of a total of 600 cells per peripheral blood smear and 700 cells per bone marrow smear. Values are given as mean ± SD from 3 independent animals.
RNA isolation and reverse transcription–polymerase chain reaction
Total RNA was isolated from mononuclear bone marrow cells of 4 wild-type and 4 C/EBPβ knockin mice by using TRIzol (Life Technologies, Grand Islands, NY). Two micrograms of DNAse I-treated RNA was reverse transcribed by using Moloney murine leukemia virus reverse transcriptase (Life Technologies), and 50 ng of the resulting complementary DNAs (cDNAs) was used as templates for polymerase chain reaction (PCR). Amplification was carried out by using HotStarTaq DNA polymerase (Qiagen, Valencia, CA) under the following conditions: an initial denaturation step at 95°C for 15 minutes followed by 35 cycles of 95°C for 30 seconds, 60°C for 30 seconds, and 72°C for 1 minute. The specificity of primer pairs used for amplification was confirmed by Southern blot by using internal oligonucleotides as probes. Reaction products were visualized on ethidium bromide-stained agarose gels, and images of C/EBPα, C/EBPβ, and 18S DNAs were captured by using AlphaImager 2000 Gel Documentation software. Reverse transcriptase(RT)–PCR results were confirmed by varying input cDNA concentration and cycle number or by real-time PCR. For the latter, RT-PCR reactions were carried out by using HotStarTaq DNA polymerase (Qiagen), 50 ng cDNA for myeloid-specific genes (500-5 ng in serial dilutions for standard curves) or 1 pg for 18S (10-0.1 pg for standard curve), and SYBRGreenI nucleic acid gel staining solution in a 1:60 000 dilution. PCR conditions were as follows: a 95°C initial activation for 15 minutes followed by 45 cycles of 95°C for 15 seconds, 60°C for 15 seconds, and 72°C for 30 seconds, and fluorescence determination at the melting temperature of the product for 20 seconds on an ICycler detection system (BioRad, Hercules, CA).
Results
Construction of a C/EBPβ knockin targeting vector and generation of homozygous C/ebpαβ/β mice were described previously.19 These mice carry a mutantC/ebpα allele in which the protein-coding region ofC/ebpα was deleted and replaced with that ofC/ebpβ. C/ebpαβ/β mice are viable, fertile, and grossly normal and exhibit growth rates that are identical to their wild-type littermates. Furthermore, they show none of the liver abnormalities found in the C/EBPα-null mice, implying that C/EBPβ can functionally replace C/EBPα in the liver when expressed from the C/ebpα gene locus.
The functional redundancy of C/EBPα and C/EBPβ in liver raised the question of their redundancy in hematopoietic cells. Previous studies10 suggest that C/EBPα is indispensable for differentiation along the granulocytic pathway because C/EBPα-deficient mice lack mature neutrophils and eosinophils. Therefore, we evaluated the ability of C/EBPβ to compensate for C/EBPα in the hematopoietic system ofC/ebpαβ/β mice. Analysis of peripheral blood from C/ebpαβ/β mice and their wild-type littermates (Table 1) showed similar differential cell counts for all hematopoietic lineages, including myeloid elements from myeloblasts to mature neutrophils. Additionally, no differences in morphology were observed in Wright-Giemsa–stained bone marrow cells from C/EBPβ knockin and wild-type mice (Figure 1). C/EBPβ knockin and wild-type murine bone marrow also stained equally for the myeloid-specific, azurophilic protein myeloperoxidase and for Sudan black, a marker of myeloid progenitors (data not shown). The white blood cell counts of the C/EBPβ knockin mice were in the normal range (mean, 4.4 × 109/μL), and for unclear reasons the wild-type mice had a slightly elevated white blood cell count (mean, 7.8 × 109/μL).20 The morphology and number of neutrophils in the peripheral blood (Table 1 and data not shown) of the C/EBPβ knockin animals were normal,20indicating that expression of C/EBPβ from the C/ebpα gene locus overcomes the block in granulocytic differentiation observed in the C/EBPα-null mice. To rule out the possibility that this rescue is due to in vivo compensatory mechanisms other than the expression of C/EBPβ, bone marrow cells from wild-type and C/EBPβ knockin mice were used for in vitro colony assays in the presence of granulocyte colony-stimulating factor. The number of granulocyte colonies formed from bone marrow cells from knockin mice was not different from that seen when using bone marrow cells from wild-type animals (data not shown). The granulocyte colonies contained similar percentages of neutrophils in assays of bone marrow cells from knockin and wild-type mice (85.7 ± 3.1 and 82.3 ± 3.5, respectively). These data imply that differentiation along the granulocytic pathway is due to functional replacement of C/EBPα with C/EBPβ and is not likely to be attributable to C/EBPα-independent pathways.
Analysis of peripheral blood and bone marrow from wild-type and C/EBPβ knockin mice
| . | % ± SD . | |
|---|---|---|
| C/ebpα+/+ . | C/ebpαβ/β . | |
| Differential cell counts | ||
| Peripheral blood | ||
| Lymphocytes | 77.7 ± 3.1 | 81.7 ± 6.5 |
| Neutrophils | 18.3 ± 2.1 | 17.3 ± 7.5 |
| Monocytes | 4.0 ± 3.6 | 1.0 ± 1.0 |
| Bone marrow | ||
| Myeloblasts & promyelocytes | 4.3 ± 0.6 | 4.7 ± 2.1 |
| Myelocytes & metamyelocytes | 8.0 ± 1.0 | 6.7 ± 2.3 |
| Neutrophils | 47.0 ± 3.5 | 51.0 ± 2.6 |
| Nucleated red blood cells | 23.0 ± 2.6 | 20.3 ± 4.5 |
| Lymphocytes | 17.3 ± 2.3 | 17.5 ± 2.4 |
| White blood cell count, 109/μL* | 7.8 | 4.4 |
| Red blood cell count, 1012/L | 9.0 | 8.8 |
| Platelet count, 109/μL* | 771.3 | 691.0 |
| . | % ± SD . | |
|---|---|---|
| C/ebpα+/+ . | C/ebpαβ/β . | |
| Differential cell counts | ||
| Peripheral blood | ||
| Lymphocytes | 77.7 ± 3.1 | 81.7 ± 6.5 |
| Neutrophils | 18.3 ± 2.1 | 17.3 ± 7.5 |
| Monocytes | 4.0 ± 3.6 | 1.0 ± 1.0 |
| Bone marrow | ||
| Myeloblasts & promyelocytes | 4.3 ± 0.6 | 4.7 ± 2.1 |
| Myelocytes & metamyelocytes | 8.0 ± 1.0 | 6.7 ± 2.3 |
| Neutrophils | 47.0 ± 3.5 | 51.0 ± 2.6 |
| Nucleated red blood cells | 23.0 ± 2.6 | 20.3 ± 4.5 |
| Lymphocytes | 17.3 ± 2.3 | 17.5 ± 2.4 |
| White blood cell count, 109/μL* | 7.8 | 4.4 |
| Red blood cell count, 1012/L | 9.0 | 8.8 |
| Platelet count, 109/μL* | 771.3 | 691.0 |
For differential counts, percentages were based on cell morphology of a total of 600 cells per genotype in peripheral blood and 700 cells per genotype in bone marrow. Each value represents the mean ± SD from 3 independent animals. Student t test showed a P value > .1 for all numbers. Total white cell, red cell, and platelet counts were determined from 2 independentC/ebpαβ/β andC/ebpα+/+ mice.
Normal reported counts21: white blood cells, 4.0-5.6 × 109/μL; red blood cells, 9.0-9.9 × 1012/L; platelets, 800-1100 × 109/μL.
Wright-Giemsa–stained bone marrow cells from wild-type and C/EBPβ knockin mice.
Bone marrow smears from wild-type (A) andC/ebpαβ/β (B) mice show maturation of the myeloid lineage to mature granulocytes (indicated by G). Magnification × 100.
Wright-Giemsa–stained bone marrow cells from wild-type and C/EBPβ knockin mice.
Bone marrow smears from wild-type (A) andC/ebpαβ/β (B) mice show maturation of the myeloid lineage to mature granulocytes (indicated by G). Magnification × 100.
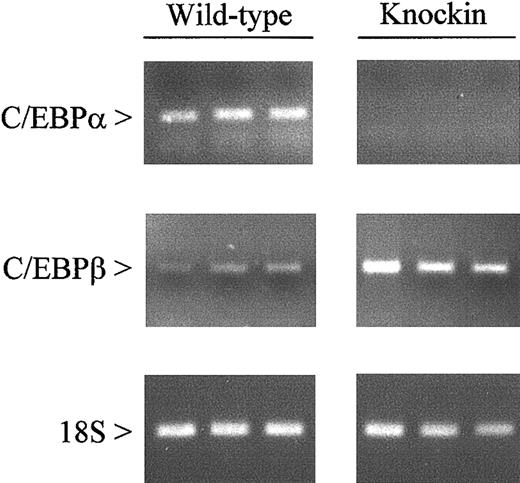
The presence of a mature granulocytic population inC/ebpαβ/β mice implies that genes necessary for differentiation are appropriately expressed. To verify this at the molecular level, we evaluated the expression of 21 myeloid-specific genes in bone marrow cells from C/EBPβ knockin mice and their wild-type littermates. We first confirmed the lack of C/EBPα expression in the bone marrow ofC/ebpαβ/β mice by RT-PCR (Figure2). The knockin mice showed increased expression of C/EBPβ compared with C/ebpα+/+mice, representing transcripts from both the C/ebpα andC/ebpβ gene loci.
Expression of C/EBPα and C/EBPβ messenger RNAs in wild-type and C/EBPβ knockin mice.
Gene expression was measured by RT-PCR using RNA from the bone marrow of wild-type and C/EBPβ knockin mice. After 35 (C/EBPα and C/EBPβ) and 15 (18S) cycles, amplification products were gel separated and stained with ethidium bromide.
Expression of C/EBPα and C/EBPβ messenger RNAs in wild-type and C/EBPβ knockin mice.
Gene expression was measured by RT-PCR using RNA from the bone marrow of wild-type and C/EBPβ knockin mice. After 35 (C/EBPα and C/EBPβ) and 15 (18S) cycles, amplification products were gel separated and stained with ethidium bromide.
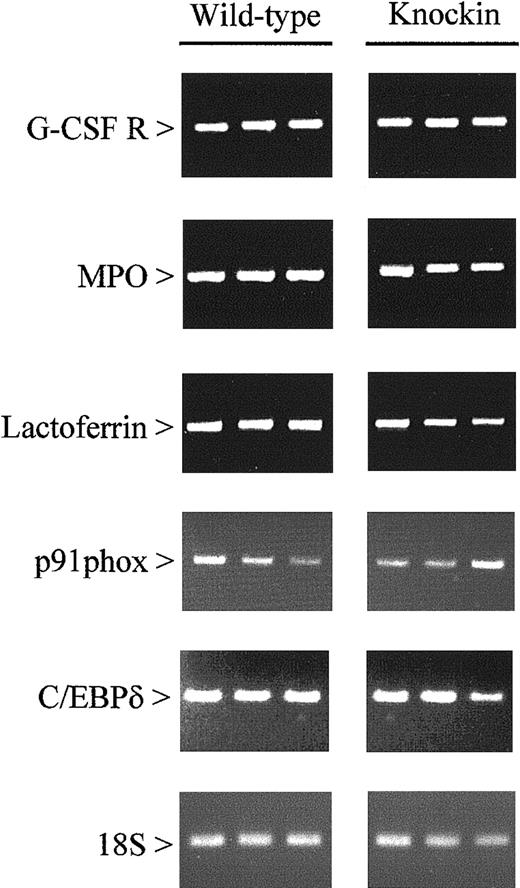
Other genes evaluated include (1) markers for different stages of maturation along the granulocytic pathway (primary and secondary granule proteins), (2) eosinophil-specific proteins, (3) colony-stimulating factor receptors and other cytokine-signaling proteins, and (4) proteins involved in phagocytosis (components of nicotinamide adenine dinucleotide phosphate oxidase and an antimicrobial protein). The individual genes analyzed are listed in Table 2. Our studies revealed no differences in the expression of these genes inC/ebpαβ/β andC/ebpα+/+ mice (Figure3). This finding is consistent with the lack of morphologic differences between the 2 genotypes.
Twenty-one myeloid-specific genes show no differences in expression in bone marrow cells ofC/ebpαβ/β as compared with wild-type mice
| Description . | Gene . |
|---|---|
| Myeloblastic & promyelocytic stages | Myeloperoxidase |
| Neutrophil elastase | |
| B9 | |
| Myelocytic stage | Lactoferrin |
| Neutrophil collagenase | |
| Neutrophil gelatinase | |
| CD11b (MAC-1) | |
| Eosinophil-specific markers | Eosinophil peroxidase |
| Major basic protein | |
| Cytokines | MIP2 |
| TNFα | |
| Cytokine receptors | G-CSF receptor |
| GM-CSF receptor | |
| M-CSF receptor | |
| IL-6 receptor | |
| IL-8 receptor | |
| Phagocytic proteins | p91phox |
| p47phox | |
| MCLP | |
| C/EBP family members | C/EBPδ |
| C/EBPε |
| Description . | Gene . |
|---|---|
| Myeloblastic & promyelocytic stages | Myeloperoxidase |
| Neutrophil elastase | |
| B9 | |
| Myelocytic stage | Lactoferrin |
| Neutrophil collagenase | |
| Neutrophil gelatinase | |
| CD11b (MAC-1) | |
| Eosinophil-specific markers | Eosinophil peroxidase |
| Major basic protein | |
| Cytokines | MIP2 |
| TNFα | |
| Cytokine receptors | G-CSF receptor |
| GM-CSF receptor | |
| M-CSF receptor | |
| IL-6 receptor | |
| IL-8 receptor | |
| Phagocytic proteins | p91phox |
| p47phox | |
| MCLP | |
| C/EBP family members | C/EBPδ |
| C/EBPε |
MIP2 indicates macrophage inflammatory protein-2; TNFα, tumor necrosis factor-α; G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; M-CSF, macrophage colony-stimulating factor; IL, interleukin; and MCLP, murine cathelinlike protein.
Expression of myeloid-specific genes in wild-type and C/EBPβ knockin mice.
Gene expression was measured by RT-PCR using RNA from the bone marrow of wild-type and C/EBPβ knockin mice. After 15 (18S) and 35 (all other genes) cycles, amplification products were gel separated and stained with ethidium bromide. G-CSF R indicates granulocyte-colony stimulating factor receptor; MPO, myeloperoxidase.
Expression of myeloid-specific genes in wild-type and C/EBPβ knockin mice.
Gene expression was measured by RT-PCR using RNA from the bone marrow of wild-type and C/EBPβ knockin mice. After 15 (18S) and 35 (all other genes) cycles, amplification products were gel separated and stained with ethidium bromide. G-CSF R indicates granulocyte-colony stimulating factor receptor; MPO, myeloperoxidase.
For 7 of these genes, expression levels were quantified by using quantitative RT-PCR. RNA from the bone marrow of 4 wild-type and 4 C/EBPβ knockin mice were reverse transcribed, and the resulting cDNAs were analyzed by real-time PCR. The values for individual mice within each group were averaged, and expression values in C/EBPβ knockin mice are given in Table 3 relative to expression in wild-type animals (arbitrarily set at 1.0). Again, our results indicate that the levels of expression of these genes in C/EBPβ knockin mice do not significantly differ from those observed for the wild-type animals, suggesting that gene expression is normal in the knockin mice.
Real-time PCR analysis of gene expression in wild-type and C/EBPβ knockin mice
| Gene . | WT ± SD . | KI ± SD . |
|---|---|---|
| G-CSF receptor | 1.0 ± 0.57 | 0.87 ± 0.89 |
| GM-CSF receptor | 1.0 ± 0.90 | 0.88 ± 0.97 |
| Myeloperoxidase | 1.0 ± 0.29 | 0.87 ± 0.29 |
| Neutrophil elastase | 1.0 ± 0.58 | 0.87 ± 0.72 |
| Neutrophil gelatinase | 1.0 ± 1.30 | 0.96 ± 0.58 |
| Eosinophil major basic protein | 1.0 ± 0.93 | 0.95 ± 1.35 |
| C/EBPε | 1.0 ± 0.72 | 0.71 ± 0.99 |
| Gene . | WT ± SD . | KI ± SD . |
|---|---|---|
| G-CSF receptor | 1.0 ± 0.57 | 0.87 ± 0.89 |
| GM-CSF receptor | 1.0 ± 0.90 | 0.88 ± 0.97 |
| Myeloperoxidase | 1.0 ± 0.29 | 0.87 ± 0.29 |
| Neutrophil elastase | 1.0 ± 0.58 | 0.87 ± 0.72 |
| Neutrophil gelatinase | 1.0 ± 1.30 | 0.96 ± 0.58 |
| Eosinophil major basic protein | 1.0 ± 0.93 | 0.95 ± 1.35 |
| C/EBPε | 1.0 ± 0.72 | 0.71 ± 0.99 |
Expression values were calculated as previously described.20 Mean ± SD for 4 wild-type (WT) and 4 C/EBPβ knockin (KI) mice are given. P values were > .1 in the Student t test for all genes tested. G-CSF indicates granulocyte colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor.
Discussion
Mice with targeted deletion of C/EBPα die soon after birth from hypoglycemia, and analysis of their peripheral blood and bone marrow reveal hematopoietic abnormalities, including the absence of granulocytic differentiation.10 Thus, normal expression of C/EBPβ does not compensate for the C/EBPα deficiency in the liver or hematopoietic tissue of C/EBPα-null mice. However, recent studies showed that expression of C/EBPβ from the C/ebpα gene locus, in addition to its expression from the C/ebpβ allele, restored liver function and overcomes the neonatal lethality of C/EBPα−/− mice.19 Similarly, in this report, we show that expression of C/EBPβ from theC/ebpα locus restores normal hematopoiesis by overcoming the selective block in granulocytic differentiation observed in the C/EBPα-null mice.
The ability of C/EBPβ to function for C/EBPα in hematopoietic cells of C/ebpαβ/β mice is likely related to at least 2 aspects of expression from the C/ebpα gene locus: (1) the level of transcriptional activity and (2) the timing of expression. In hepatic tissues ofC/ebpαβ/β mice, C/EBPβ messenger RNA expressed from the C/ebpα locus is significantly higher than expression of C/EBPβ from its endogenous allele.19Although our studies do not quantify the contribution of C/EBPβ messenger RNA from each locus in hematopoietic cells, we do observe higher expression of C/EBPβ in the knockin model, representing the combined expression from both loci. Radomska et al21suggested that high levels of C/EBPα at the stage of myeloid commitment is the molecular switch that directs myeloid precursors to the granulocytic pathway. Because myeloid progenitor cells express low levels of C/EBPβ, the amount of C/EBPβ in C/EBPα-null mice is possibly insufficient to transactivate genes whose expression is required for granulocytic differentiation.
Although it is low in early stages of myeloid differentiation, the level of C/EBPβ expression increases dramatically at later stages of differentiation especially in maturing macrophages.9C/EBPβ−/− mice have defects in macrophage function and develop lymphoproliferative disorders as a result, but deletion of C/EBPβ in mice does not adversely affect myeloid cell differentiation.13 Hence, its role in myelopoiesis is unclear. Expression of C/EBPβ from the C/ebpα locus places the protein at the scene of hematopoietic differentiation earlier and probably at higher levels than when expressed from theC/ebpβ gene locus. Therefore, the rescue of granulocytic differentiation in C/ebpαβ/β mice likely reflects changes in the temporal expression of C/EBPβ. Stages of differentiation in hematopoietic cells are driven not only by C/EBPs but also by other transcription factor groups including GATA-122,23 and GATA-2,24,25Myb,26-28 Ets,29-31 and AML1.32-34 Perhaps high expression of C/EBPβ early in myelopoiesis is sufficient to maintain the integrity of protein-protein interactions that direct myeloid progenitors toward mature granulocytes.
C/EBPs are highly homologous in their C-terminal dimerization and DNA-binding domains and are believed to bind the same recognition sites on DNA.35 Therefore, C/EBPβ conceivably can interact with dimerization partners of C/EBPα and bind C/EBPα-targeted promoters such as the receptors for granulocyte colony-stimulating factor36 and interleukin 6,37myeloperoxidase,38 and neutrophil elastase.39However, the N-terminal regions of C/EBPs are more diverse and mediate their transactivation functions. Given the complexity of transcriptional activation complexes, we are somewhat surprised that the transactivation domain of C/EBPβ can recruit necessary cofactors to activate promoters normally directed by the transactivation domain of C/EBPα. Interestingly, C/EBPβ does not substitute for C/EBPα in the expression of genes encoding adipocyte-specific factors adipsin and leptin.19 Abnormalities in fat storage but normal liver development and hematopoiesis suggest that the redundancy of C/EBPα and C/EBPβ may be tissue and gene specific. Previous studies implicate C/EBPα as a critical factor for granulocytic commitment of myeloid progenitor cells. Strain differences between mice may reconcile our findings with the strict requirement for C/EBPα in granulocytic differentiation reported previously. However, it is more likely that C/EBPα itself is less important than the timing and level of its expression.
Supported by grants from the National Institutes of Health (H.P.K.), the C. and H. Koeffler Fund, Parker Hughes Trust, Brian Harvey Fund, Frederick P. Begell Foundation, and the Joseph Troy Leukemia Foundation. L.C.J. is supported by grants from the American Cancer Society (PF-99-127-01-CNE) and the National Institutes of Health (T32 CA-75956). W.K.H. is a recipient of a scholarship from the Deutsche Forschungsgemeinschaft (HO2207/1-1). H.P.K. is a member of the Jonsson Comprehensive Cancer Center and holds the endowed Mark Goodson Chair of Oncology Research at Cedars-Sinai Medical Center/UCLA School of Medicine.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Letetia C. Jones, Division of Hematology and Oncology, Cedars-Sinai Medical Center, UCLA School of Medicine, 8700 Beverly Blvd, Suite BM-1, Rm 109, Los Angeles, CA 90048; e-mail:Letetia.Jones@cshs.org.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal