The tyrosine kinase inhibitor STI571 is a promising agent for the treatment of advanced Philadelphia chromosome positive (Ph+) acute lymphoblastic leukemia (ALL), but resistance develops rapidly in most patients after an initial response. To identify mechanisms of resistance to STI571, 30 complementary DNAs (including 9 matched samples) obtained from the bone marrow of individuals with Ph+ ALL were analyzed by direct sequencing of a 714–base pair region of ABL encoding for the adenosine triphosphate (ATP)–binding site and the kinase activation loop. A single point mutation was found at nucleotide 1127 (GI6382056) resulting in Glu255Lys. This mutation occurred in 6 of 9 patients (67%) following their treatment with STI571 but not in the samples from patients before beginning treatment with STI571. Glu255Lys is within the motif important for forming the pocket of the ATP-binding site in ABL and it is highly conserved across species. In conclusion, Ph+ ALL samples resistant to STI571 have a unique mutation Glu255Lys of BCR-ABL.
Introduction
The ABL-selective tyrosine kinase inhibitor STI571 (Glivec) is a major therapeutic advance for the management of chronic myelogenous leukemia (CML) in the chronic phase.1 It also has substantial activity in patients with advanced CML and Ph+ acute lymphoblastic leukemia (ALL), although responses are frequently not sustained.2,3 These leukemias result from the fusion gene p210BCR-ABL orp190BCR-ABL causing aberrantly high expression and constitutive activation of the ABL tyrosine kinase.4,5 The rationale for treatment of Ph+ALL with STI571 is based on the fact that the compound binds competitively to the adenosine triphosphate (ATP)–binding site of the ABL tyrosine kinase, thereby preventing the signaling by this oncogenic protein and consequently inhibiting growth of the affected leukemic cells.6 7
The binding of ATP to the catalytic domain of ABL kinase is required for autophosphorylation of BCR-ABL. Increased phosphotyrosine residues on BCR-ABL itself allow it to interact with and phosphorylate effector molecules responsible for activating downstream signaling pathways. The activation loop of ABL kinase contains a highly conserved Asp-Phe-Gly motif and is also critical for controlling the catalytic activity of the protein.8 To determine whether resistance to STI571 is associated with mutations in either of these critical regions, we analyzed the nucleotide sequence of the ABL kinase domain encoding the ATP-binding site and the kinase activation loop in matched bone marrow samples from patients with Ph+ ALL before and after undergoing treatment with STI571. During the course of our analysis, Gorre et al showed in CML that resistance to STI571 is associated with a single amino acid substitution (Thr315Ile) in the ABL kinase domain previously shown to be important for STI571 binding.9 We find a different site (Glu255Lys), which is frequently mutated in Ph+ ALL samples from patients only after they received therapy with STI571.
Study design
Patient samples
Thirty bone marrow samples from 21 patients with Ph+ALL enrolled into consecutive “Phase II study to determine the safety and antileukemic effect of STI571 in adult patients with Ph+ acute leukemias” were analyzed. Approval was obtained from the institutional review board at University Hospital Frankfurt/Main, Germany for these studies and informed consent was provided according to the Declaration of Helsinki. According to the study protocol, these patients had relapsed ALL or were refractory after at least 2 cycles of standard chemotherapy. Samples were obtained from all of the patients before treatment with STI571; 13 of these samples were from individuals who later were classified as good responders to STI571 (nos. 1-13, sensitive, S) including 12 patients with hematologic complete remission (CR) and 1 patient with partial remission (PR) but complete peripheral hematologic recovery (no. 1). Eight samples were collected from individuals who were resistant to STI571 (nos. 14-21, primarily resistant, R) including 6 patients without any hematologic response, 1 with cytoreduction in the bone marrow but persistent peripheral leukemic cells (no. 20), and another with PR but incomplete peripheral hematologic recovery (no. 16). Matched bone marrow samples from 9 patients (nos. 1-5 and 14-17) were also obtained while they were receiving treatment with STI571. Mononuclear cells were separated by density gradient centrifugation through Ficoll-Hypaque (Biochrom, Berlin, Germany). Total RNA was extracted using the acid guanidinium/phenol/chloroform method with minor modifications.10 Only samples with leukemic blast cell infiltration of more than 80% were included in the analysis.
Reverse transcription–polymerase chain reaction and sequencing analysis
Total RNA (1 μg) was used for reverse transcription by Superscript II RT (Life Technologies, Grand Island, NY) according to standard protocols. Primers specific for the ATP binding site of ABL including the “loop“ were: ATP-F 5′-GCG CAA CAA GCC CAC TGT CT-3′; ATP-R 5′-GCA CTC CCT CAG GTA GTC CA-3′; LOOP-F 5′-TGG ACT ACC TGA GGG AGT GC-3′; and LOOP-R 5′-CGG TAG TCC TTC TCT AGC AGC-3′. Oligonucleotides were synthesized by Life Technologies. Polymerase chain reaction (PCR) was performed as described previously11 using an annealing temperature of 58°C. PCR products were separated on a 2% agarose gel containing 0.3 mg/mL ethidium bromide and purified using the QIAquick purification system (Qiagen, Valencia, CA) according to the manufacturer's protocol. Purified amplification products were sequenced in both directions by the ABI PRISM dye terminator cycle sequencing reaction (Perkin-Elmer, Foster, CA).
Results and discussion
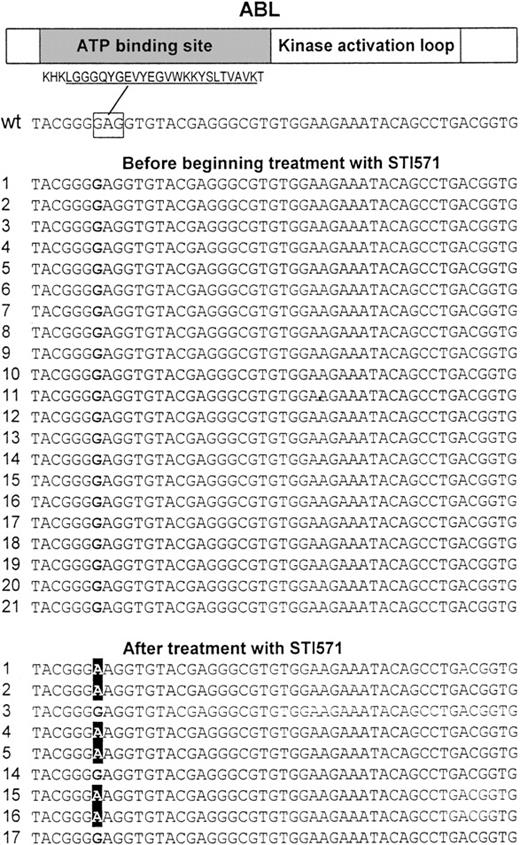
We analyzed 30 complementary DNAs (including 9 matched samples) obtained from the bone marrow of individuals with Ph+ ALL by direct sequencing of a 714–base pair (bp) region of ABL encoding for the ATP-binding site and the kinase activation loop. Analysis of the sequences of the ATP-binding site revealed a single point mutation at nucleotide 1127 (1127G > A, Figure 1) resulting in a substitution at codon 255 of lysine (mutant) for a glutamic acid (wild-type). This mutation was present in 6 patients (nos. 1, 2, 4, 5, 15, 16) following treatment with STI571, but it was absent in other samples including the matched samples from the patients before beginning treatment with STI571 (Table1 and Figure 1). The change was verified by sequencing from both the sense and antisense directions. In addition, one sample (no. 17) from a patient with an aberrant common ALL (cALL) had a single point mutation at nucleotide 1308 (1308C > T) resulting in a substitution at codon 315 of isoleucine (mutant) for a threonine (wild-type). This sample was unusual because the cells also expressed CD33, a cell surface protein expressed on myeloid cells. Interestingly, this type of mutation was described previously in CML samples.9 No mutations were found in the region encoding the kinase activation loop.
Point mutations at the ATP-binding site of the ABL kinase.
A 714-bp region of ABL that codes for the ATP-binding site and the kinase activation loop was PCR amplified using 2 primer pairs and directly sequenced in 30 complementary DNAs (including 9 paired samples) obtained from the bone marrow of individuals with Ph+ ALL. Sequencing revealed a single point mutation at nucleotide 1127 (GI6382056) changing a G to an A resulting in Glu255Lys. This mutation was found in 6 samples from patients who where treated with STI571 (nos. 1, 2, 4, 5, 15, 16) but not in any other sample including paired samples from the same individuals before treatment with STI571. (Top) Schematic protein structure of ABL including the amino acid sequence for the highly conserved ATP-binding site motif (underlined). The nucleotide sequence corresponding to Glu255, which is affected by the mutation is marked by a rectangle. The left hand column contains the patient number; wt indicates wild-type ABL.
Point mutations at the ATP-binding site of the ABL kinase.
A 714-bp region of ABL that codes for the ATP-binding site and the kinase activation loop was PCR amplified using 2 primer pairs and directly sequenced in 30 complementary DNAs (including 9 paired samples) obtained from the bone marrow of individuals with Ph+ ALL. Sequencing revealed a single point mutation at nucleotide 1127 (GI6382056) changing a G to an A resulting in Glu255Lys. This mutation was found in 6 samples from patients who where treated with STI571 (nos. 1, 2, 4, 5, 15, 16) but not in any other sample including paired samples from the same individuals before treatment with STI571. (Top) Schematic protein structure of ABL including the amino acid sequence for the highly conserved ATP-binding site motif (underlined). The nucleotide sequence corresponding to Glu255, which is affected by the mutation is marked by a rectangle. The left hand column contains the patient number; wt indicates wild-type ABL.
Matched bone marrow samples: development of mutations in the region coding for the ATP binding site of ABL during treatment of Ph+ ALL with STI571
| Patient no. . | Diagnosis . | BCR-ABL mutational status prior to treatment with STI571 . | Response to STI571 . | BCR-ABL mutational status after treatment with STI571 . |
|---|---|---|---|---|
| 1 | Ph+cALL | Wild-type | PR | Glu255Lys |
| 2 | Ph+ cALL | Wild-type | CR | Glu255Lys |
| 3 | Ph+ cALL | Wild-type | CR | Wild type |
| 4 | Ph+ cALL | Wild-type | CR | Glu255Lys |
| 5 | Ph+ cALL | Wild-type | CR | Glu255Lys |
| 14 | Ph+ cALL | Wild-type | No | Wild type |
| 15 | Ph+ cALL | Wild-type | No | Glu255Lys |
| 16 | Ph+ pre B-ALL | Wild-type | PR | Glu255Lys |
| 17 | Ph+, cALL, CD33+ | Wild-type | No | Thr315Ile |
| Patient no. . | Diagnosis . | BCR-ABL mutational status prior to treatment with STI571 . | Response to STI571 . | BCR-ABL mutational status after treatment with STI571 . |
|---|---|---|---|---|
| 1 | Ph+cALL | Wild-type | PR | Glu255Lys |
| 2 | Ph+ cALL | Wild-type | CR | Glu255Lys |
| 3 | Ph+ cALL | Wild-type | CR | Wild type |
| 4 | Ph+ cALL | Wild-type | CR | Glu255Lys |
| 5 | Ph+ cALL | Wild-type | CR | Glu255Lys |
| 14 | Ph+ cALL | Wild-type | No | Wild type |
| 15 | Ph+ cALL | Wild-type | No | Glu255Lys |
| 16 | Ph+ pre B-ALL | Wild-type | PR | Glu255Lys |
| 17 | Ph+, cALL, CD33+ | Wild-type | No | Thr315Ile |
Ph+ cALL indicates Philadelphia chromosome–positive common ALL (CD10+); PR, partial remission; CR, complete remission.
In vitro data suggest that reactivation of the ABL kinase activity12-15 may be an important mechanism to overcome the growth inhibitory effect of STI571. Mutations within the kinase activation loop or in the ATP-binding site, the target of STI571, may explain the development of resistance to STI571. A recent study showed that 6 of 9 samples from patients with CML resistant to STI571 had a Thr315Ile mutation.9 This mutation does not affect the site for ATP binding (Tyr272 and Tyr276). However, it changes a threonine residue that forms a critical hydrogen bond with STI571 resulting in the loss of binding of STI571 to BCR-ABL.7 In vitro results from the same investigators clearly demonstrate that STI571 does not inhibit phosphorylation of BCR-ABL in 293T cells, which were transfected with mutant BCR-ABLThr315Ile.
In bone marrow samples from individuals with Ph+ ALL, we found a mutation distinct from Thr315Ile in the ATP-binding site of BCR-ABL. Motif search (http://www.motif.genome.ad.jp/) revealed that Glu255, in contrast to Thr315, falls within a region that is highly conserved among Caenorhabditis elegans,16Drosophila melanogaster,17 and Rattus norvegicus.18 It is important for forming the pocket of the ATP-binding site in ABL. Substitution of glutamic acid (polar, negatively charged) with lysine (polar, positively charged) may lead to a conformational change that inhibits binding activity of STI571. On the other hand, mutation Glu255Lys does not affect either Tyr272 or Tyr276, both of which are key residues for binding ATP.7 This may provide the selected cells that have a mutated BCR-ABL with a growth advantage during the treatment with STI571. No mutation was present in samples from patients who had primary resistance to the drug, emphasizing that the selection of the resistant clone requires direct interaction of the leukemic cell with STI571. Our analysis of matched samples indicate that those from untreated patients did not contain this mutation. In contrast, 6 of 9 samples (67%) from these patients undergoing treatment with STI571 had this substitution at Glu255.
While our article was being reviewed, 3 commentaries to the original article by Gorre et al9 appeared online (Science online,www.scienceexpress.org). Barthe et al19 and Hochhaus et al20 studied 12 and 32 samples, respectively, of CML from individuals who had a relapse or were unresponsive to STI571. They found 2 BCR-ABL mutations, both at amino acid 255 (Glu255Lys and Glu255Val). In an additional commentary, Gorre's group updated their results21 and stated that Thr315Ile mutations occurred in 9 of 29 patients (31%) and Glu255Lys mutations were found in 4 of 29 patients (14%). Taken together with our results, both the Glu255Lys and the Thr315Ile mutation of the BCR-ABL gene are important in the development of STI571 resistance. Analyses of additional patients as well as in vitro studies are required to determine if selective mutations are associated with particular disease subtypes.
Supported by National Institutes of Health grants (H.P.K.), the C. and H. Koeffler Fund, Parker Hughes Trust, Brian Harvey Fund, Begell Foundation, the Joseph Troy Leukemia Foundation, the BMBF Competence Network Leukemias (01GI9971) and the German Genom Research Network. W.K.H. is a recipient of a scholarship from the Deutsche Forschungsgemeinschaft (HO2207/1-1). L.C.J. is a fellow of the American Cancer Society. S.d.V. was supported by the UCLA STAR Program as a Hematology/Oncology fellow and as an Advanced Research fellow. H.P.K. is a member of the Jonsson Comprehensive Cancer Center and holds the endowed Mark Goodson Chair of Oncology Research at Cedars Sinai Medical Center/UCLA School of Medicine.
W.K.H. and L.C.J. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Wolf-K. Hofmann, Division of Hematology and Oncology, Cedars Sinai Medical Center, UCLA School of Medicine, 8700 Beverly Blvd, Suite BM-1, Rm 109, Los Angeles, CA 90048; e-mail:w.k.hofmann@em.uni-frankfurt.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal