External stimuli act in concert with intracellular signals to regulate a cell's genetic program, activating genes important in granulocytic lineage commitment, proliferation, and maturation. Signal transducer and activator of transcription 3 (STAT3), a transcription factor, has been implicated in mediating granulocytic differentiation. We have examined the role of STAT3 as a physiologic mediator of granulocytic kinetics. Distinct isoforms—the long form STAT3α, the truncated forms STAT3β and STAT3γ, and a putative novel form STAT3δ—were expressed and activated in a maturation stage–specific manner. With the progression of differentiation, the ratio of isoforms shifted from predominantly STAT3α to STAT3β. The kinetics of STAT3γ, generated through proteolytic cleavage of STAT3α, coincided with but were inverse to those of STAT3α. STAT3δ was expressed at low levels and decreased with differentiation but was preferentially phosphorylated during an intermediate stage of maturation. Under different culture conditions (pH, O2 tension [pO2], IL-3), we found that the expression and phosphorylation status of the different STAT3 isoforms displayed unique kinetic patterns that correlated with the effects on granulocyte differentiation. The evidence suggests that signals triggered by pH, pO2, and IL-3 each converge on STAT3 through independent mechanisms, exploiting the flexibility granted by the diversity in expression and phosphorylation of the different STAT3 isoforms, to regulate distinct granulocytic cell responses. The selective expression of STAT3 isoforms and their activation is a major determinant of granulocytic cell development and provides a molecular basis for evaluating the effects of various environmental factors on the STAT3-mediated signaling pathway.
Introduction
Hematopoietic lineage determination by transcriptional collaboration with cytokines is well established.1 In particular, members of the STAT (signal transducer and activator of transcription) transcription factor family mediate many cytokine-induced responses in hematopoietic cells, including proliferation, differentiation, and survival.2Upon ligand binding, STATs are recruited to activated cell-surface receptors and become phosphorylated on a specific residue either directly or through association with a Janus kinase. Subsequently, the phosphorylated STATs homodimerize or heterodimerize and translocate to the nucleus, where they regulate transcription by binding to specific DNA promoter elements.2,3 The STAT family consists of 7 members, most of which are ubiquitously expressed. However, individual STAT proteins may be differentially activated depending on the cell type or tissue.2 4-6
During myeloid differentiation, various isoforms of STAT3 and STAT5 are activated in a cell type– and maturation state–dependent manner. While STAT5 appears to be important for proliferative responses to interleukin-3 (IL-3), granulocyte-macrophage colony-stimulating factor (GM-CSF), and G-CSF,7,8 STAT3 is activated by G-CSF and appears to be the major STAT protein driving G-CSF–mediated granulocytic differentiation.9-16 Expression of dominant negative STAT39 or G-CSF receptor (G-CSFR) mutants lacking regions in the membrane distal domain, which contains recruitment sites for STAT3,16-20 blocks granulocytic differentiation, demonstrating a requirement for STAT3 activation in growth arrest and morphologic granulocytic differentiation.
Three distinct isoforms of STAT3, all derived from a single gene, have been identified11,21: STAT3α (p92), the full-length isoform expressed in most cells; STAT3β (p83), an alternatively spliced RNA form of STAT3α in which 55 amino acids of the C-terminal transactivation domain are replaced by 7 unique residues22; and STAT3γ (p72), also a C-terminal truncated form of STAT3α but derived posttranslationally through limited proteolysis.23Experimental evidence indicates that the α and β isoforms are functionally distinct,10,22,24-26 although all of these studies used cell lines or uncultured leukemic patient samples whose signaling processes may have been altered by cellular transformation and may lack critical components of normal differentiation. Two reports, using uncultured normal human cells—bone marrow10or mobilized peripheral blood CD34+ cells, bone marrow metamyelocytes, and mature peripheral blood neutrophils27—do indeed suggest that the balance between α and β isoforms may alter a cell's capacity to differentiate, with a predominance of STAT3β activation favoring differentiation. The physiologic function of STAT3γ, which has been shown to be primarily activated in terminally differentiated neutrophils,23 is less clear. Diversity not only in the expression of different STAT3 isoforms, but also in their phosphorylation status, may increase the specificity of the cellular response. Tyrosine phosphorylation of STATs is considered essential for activation,3,12,18,28,29 while serine phosphorylation may also be important, albeit controversial, in a cell type–dependent manner.30-34
Analyzing STAT3 signaling in primary cells is key to understanding its physiologically relevant function. However, as mentioned above, in primary cells STAT3 has previously been examined only in uncultured human myeloid cells isolated at 3 different stages of differentiation.10,27 We designed an ex vivo granulopoietic system that expands granulocytic cells from CD34+ cells in sufficient numbers and purity (> 90%) to examine STAT3 isoform expression and phosphorylation in coordination with granulocytic proliferation and differentiation. Little is known about the production of the different STAT3 isoforms—more specifically, about the enzymes involved in the proteolytic cleavage of STAT3α to STAT3γ or in the splicing events leading to the generation of STAT3β. This lack of mechanistic details limits the range of experimental strategies that can link one particular isoform with a specific biologic effect. Thus, we devised a practical scheme to establish the potential relationship between STAT3 isoforms and granulocytic development. Previously, we have shown that selected environmental factors (pH, O2 tension [pO2], and IL-3) have clearly distinct effects on the dynamics of granulocytic proliferation, differentiation, and survival.35,36 While pO2 has no impact on differentiation, pH affects both the differentiation and proliferation processes throughout the entire granulocytic pathway. IL-3 also influences differentiation but, unlike the effects of pH, those of IL-3 are isolated to a specific stage of maturation. These findings provide a gateway for evaluating the mechanistic role of STAT3 as the physiologic mediator of distinct granulocytic responses. In addition, the convergence of 3 independent factors on a cell-specific, STAT-mediated mechanism may help explain how distinct hematopoietic environments, created by the highly organized arrangement of cells in vivo, guide granulocytic development. This is the first report that explores involvement of such a mechanism as a possible explanation for recent evidence showing that exposure of cells to low pH and pO2, conditions that reflect the topology of granulopoiesis deep within the bone marrow,37-40 favors production of granulocytic cells.35
Material and methods
Cells and cell culture
Fresh G-CSF–mobilized peripheral blood CD34+ cells from normal donors were obtained from AllCells (Foster City, CA) within 24 hours after collection. The experimental design and methods have been described previously.35 Briefly, CD34+cells were seeded at 2 × 104/mL and cultured for 15 days in serum-containing, modified McCoy 5A medium (Sigma, St Louis, MO)35 supplemented with 50 ng/mL stem cell factor (SCF) (Amgen, Thousand Oaks, CA), 10 ng/mL IL-6 (Peprotech, Rocky Hill, NJ), and 10 ng/mL G-CSF (Amgen) in the presence or absence of 10 ng/mL IL-3 (R&D Systems, Minneapolis, MN). An additional 10 ng/mL G-CSF was added to the cultures every 2 days to compensate for its depletion due to degradation at 37°C. Beginning on day 3, cultures were assessed every 2 days for total cell concentration using a Coulter Multisizer (Coulter Corporation, Miami, FL), cell viability using 0.25% trypan blue dye exclusion, cellular phenotype and cell-surface receptor expression using flow cytometry, and expression of STAT3 and its phosphorylated forms via Western blot and immunofluorescence microscopy. Cultures were diluted as necessary to maintain cell concentrations between 7.5 × 104 and 2.5 × 105 cells per milliliter. Cells were examined under 4 different conditions: 5% O2, pH 7.25, +IL-3 (presence of IL-3); 20% O2, pH 7.25, +IL-3; 5% O2, pH 7.4, +IL-3; and 5% O2, pH 7.25, −IL-3 (absence of IL-3). Granulocytic cell production was greatest at 5% O2, pH 7.25, +IL-3, which was designated as the “control” condition. Cultures from 3 independent donor samples were evaluated, except for studies on G-CSFR expression (n = 6). The CD34+ cell purity and viability were more than 97%, with viabilities remaining more than 95% throughout culture.
Flow cytometric analysis
Cultured cells were evaluated for expression of the granulocyte lineage–associated antigens CD15 and CD11b (clones MMA and D12, respectively; Becton Dickinson, San Jose, CA) using a FACScan flow cytometer (Becton Dickinson) as described.35 G-CSFR surface content was assessed using a biotinylated anti–G-CSFR monoclonal antibody (clone LMM741) and phycoerythrin-streptavidin system (Pharmingen, San Diego, CA).
Western blot analysis
Cells numbering 6 × 106 to 12 × 106 (day 0) or 1.5 × 106 to 3.0 × 106 (at later times) were mixed with an equal volume of ice-cold phosphate-buffered saline (PBS) supplemented with 1 mM Na3VO4 and 5 mM NaF and immediately spun down (4°C, 300g, 10 minutes). Cells were rinsed with 1 mL PBS solution and pelleted again at 4°C. Whole-cell extracts were prepared by adding 60 μL of modified radioimmunoprecipitation (RIPA) buffer per 1.0 × 106 cells. Preparation of RIPA buffer and cell lysate was carried out according to the manufacturer (Upstate Biotechnology, Lake Placid, NY)41 with the following modifications to the RIPA solution: 2 μg/mL each of aprotinin, leupeptin, and pepstatin and 20 mM NaF. Protein was quantified in duplicate with the BCA kit (Pierce, Rockford, IL) using bovine serum albumin (BSA) as standard.
From each sample, 20 μg total protein was separated by 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (Biorad, Hercules, CA) and transferred to a polyvinylidene fluoride membrane (0.45 μm Immobilon-P; Millipore, Bedford, MA). Membranes were incubated with either a mouse monoclonal anti-STAT3 (1:2000; clone 84; Transduction Laboratories, Lexington, KY), rabbit polyclonal anti-STAT3 pTyr 705 (1:1000; Cell Signaling Technology, Beverly, MA), or rabbit polyclonal anti-STAT3 pSer 727 (1:500; Upstate Biotechnology) antibody, followed by horseradish peroxidase–conjugated secondary antibody, either sheep anti–mouse (1:5000; Amersham Pharmacia Biotech, Piscataway, NJ) or donkey anti–rabbit (1:2500; Amersham Pharmacia Biotech), as appropriate. Immunoreactive bands were visualized by using the enhanced chemiluminescence (ECL) Plus detection kit (Amersham Pharmacia Biotech) and directly scanned using the Molecular Dynamics Storm imaging system (Sunnyvale, CA). Signals were quantified using ImageQuant software (Molecular Dynamics).
Immunofluorescence microscopy
A total of 2 × 104 cells were washed in PBS supplemented with 2% BSA, cytocentrifuged (Cytospin 3, Shandon, Pittsburgh, PA) onto glass microscope slides, fixed in 4% paraformaldehyde for 10 minutes, washed in PBS/0.1 M glycine 5 times for 3 minutes each, and permeabilized in 0.3% Triton X-100 in PBS for 5 minutes. After blocking nonspecific binding sites for 1 hour with 10% normal goat serum (Vector Laboratories, Burlingame, CA) in PBS/2% BSA, cells were incubated with a polyclonal rabbit anti-STAT3 or anti-STAT3 pSer 727 (1:300; Cell Signaling Technology) antibody simultaneously with monoclonal mouse anti-CD15 antibody (1:100; clone 80H5; immunoglobulin M [IgM]; Immunotech, France) in PBS/2% BSA overnight at 4°C. Cells were washed in PBS/0.1 M glycine 5 times for 3 minutes each, incubated with both fluorescein isothiocyanate (FITC)–conjugated goat anti–rabbit IgG (1:200; Vector Laboratories) and Texas Red–conjugated goat anti–mouse IgM μ-chain–specific (1:100; Jackson Immunoresearch Laboratories, West Grove, PA) secondary antibodies in PBS/2% BSA containing 2% goat serum for 2 hours in the dark, and washed again. Slides were mounted in Vectashield Mounting Media (Vector Laboratories) containing DAPI. Each fluorochrome was analyzed individually: DAPI fluorescence (white pixels) to distinguish the nuclei, FITC fluorescence (green pixels) to assess STAT3 expression, and Texas Red fluorescence (red pixels) to identify CD15+ cells. No significant background or overlap between fluorescence signals was detected. All images were captured with a Zeiss Axiophot fluorescence microscope using a 40× or 100× oil immersion objective and processed by IPlab software (VayTek, Farfield, IA).
Statistical analysis
Statistical comparisons of the values representing production of total cells and CD15dim/CD11b−, CD15bright/CD11b−, and CD15bright/CD11b+ granulocyte subpopulations and receptor expression levels under different culture conditions were performed using a 2-tailed paired Student t test. Data are reported as the mean ± SEM. Immunofluorescence microscopy and Western analysis were carried out on 3 sets of experiments that examined the same 4 culture conditions with different donor CD34+ cells. The results were consistent across experiments, and thus one representative data set is shown. Immunofluorescence images are representative of 2 to 3 randomly selected fields per culture condition for each of the 3 different sets of experiments.
To control for possible error introduced during protein quantitation and gel loading, each Western blot was probed for β-actin protein. However, β-actin levels were found to change with the different experimental conditions and with differentiation, with an increasing proportion of total protein expressed as β-actin as the cells matured (data not shown). It has been reported that some commonly used “housekeeping” genes, including GAPDH, β-actin, and cyclophilin, are susceptible to regulation by culture conditions42-45—in particular, to O2tension.45 Thus, to determine the cumulative error in quantifying STAT3 protein by Western analysis, 4 separate lysates were collected simultaneously from an individual culture, run in parallel on a single gel, and quantified. Signal intensities varied on average by 6% when comparing individual bands across lanes. Lane-to-lane variability was not a factor in comparing relative band intensities within a single lane; the error was less than 2% when comparing ratios of bands within a lane to the corresponding ratios in the other lysates. Furthermore, to minimize the error in determining STAT3 isoform percentages on a given day, duplicate blots were run on protein samples from days 5, 7, 9, and 11 within a single experiment, and the values were averaged.
Results
Manipulation of the experimental conditions under which the CD34+ cells were cultured elicited a variety of distinct cellular responses. Granulocytic differentiation and proliferation were clearly altered in unique ways depending upon exposure to a different pH, pO2, or IL-3 environment. The ability of these 3 factors to independently induce effects on a specific cellular process or stage of maturation allowed potential functional links with STAT3 isoform expression and/or phosphorylation to be explored. As detailed below, this experimental approach entailed a thorough kinetic characterization of each culture as it progressed through different stages of granulocytic development, ie, detecting phenotypic and morphologic changes in conjunction with changes in STAT3 isoform expression and phosphorylation.
Characterization of ex vivo granulocytic differentiation of CD34+ cells
To assess STAT3 kinetics throughout the granulocytic pathway, we used flow cytometry and fluorescence microscopy to clearly define the differentiation status of our cultures. By monitoring the differential expression of CD15 and CD11b, in combination with nuclear morphology, the neutrophil lineage can be classified into discrete maturational stages. As cells progressed from CD15−/CD11b−to CD15dim/CD11b− (myeloblasts), CD15bright/CD11b− (promyelocytes/early myelocytes), and CD15bright/CD11b+ (early myelocytes/myelocytes/metamyelocytes/bands),35 their morphology and size also changed dramatically from the initial small size of the CD15−/CD11b− (CD34+) cells. The nuclei increased in size to days 5 to 7 (Figure1) and contained multiple nucleoli that were readily visible from the lack of STAT3 within their interior (Figure 2). As the cells matured into myelocytes, the nucleoli disappeared and the nuclei decreased in size, becoming more rounded so that the nuclear-cytoplasmic ratio decreased (Figure 2). With further maturation, cells decreased in size while the nuclei underwent drastic changes that allowed for the more mature CD15bright/CD11b+ population to be further compartmentalized into distinct stages (Figures 1 and 2)—going from the indented, kidney-shaped appearance of the metamyelocyte to the more elongated, horseshoelike appearance of the band form, which showed the initial signs of nuclear cleavage that characterizes segmented neutrophils.
Localization and expression of STAT3 and CD15 proteins during ex vivo granulocytic differentiation.
CD34+ cells from mobilized peripheral blood were directed along the granulocyte lineage with SCF, IL-6, G-CSF, with or without IL-3, and harvested for analysis by dual-color immunofluorescence microscopy. Detection of STAT3 and CD15 in cultures at (A) 5% O2, pH 7.25, +IL-3 (control condition); (B) 5% O2, pH 7.4, +IL-3 (high pH); and (C) 5% O2, pH 7.25, −IL-3. Negative controls (background) for each day of analysis using FITC- and Texas Red–conjugated secondary antibodies in the absence of the relevant primary antibodies are shown in the first row. The same field of cells is shown in the upper and lower panels of panels A-C. Upper panels of panels A-C: DAPI staining of nucleic acid (white). Lower panels of panels A-C: codetection of STAT3 (green) and CD15 (red) using polyclonal rabbit anti-STAT3 and monoclonal mouse anti-CD15 primary antibodies, followed by FITC-conjugated goat antirabbit IgG and Texas Red–conjugated goat antimouse IgM μ-chain–specific secondary antibodies. Images were captured using a × 40 oil immersion objective and are representative of 3 randomly selected fields per culture condition for each of 3 different sets of experiments. No appreciable differences in STAT3 and CD15 staining were visible in cells cultured at 20% O2 versus the control condition (data not shown). A 10-μm scale bar is shown in the day 0 panels.
Localization and expression of STAT3 and CD15 proteins during ex vivo granulocytic differentiation.
CD34+ cells from mobilized peripheral blood were directed along the granulocyte lineage with SCF, IL-6, G-CSF, with or without IL-3, and harvested for analysis by dual-color immunofluorescence microscopy. Detection of STAT3 and CD15 in cultures at (A) 5% O2, pH 7.25, +IL-3 (control condition); (B) 5% O2, pH 7.4, +IL-3 (high pH); and (C) 5% O2, pH 7.25, −IL-3. Negative controls (background) for each day of analysis using FITC- and Texas Red–conjugated secondary antibodies in the absence of the relevant primary antibodies are shown in the first row. The same field of cells is shown in the upper and lower panels of panels A-C. Upper panels of panels A-C: DAPI staining of nucleic acid (white). Lower panels of panels A-C: codetection of STAT3 (green) and CD15 (red) using polyclonal rabbit anti-STAT3 and monoclonal mouse anti-CD15 primary antibodies, followed by FITC-conjugated goat antirabbit IgG and Texas Red–conjugated goat antimouse IgM μ-chain–specific secondary antibodies. Images were captured using a × 40 oil immersion objective and are representative of 3 randomly selected fields per culture condition for each of 3 different sets of experiments. No appreciable differences in STAT3 and CD15 staining were visible in cells cultured at 20% O2 versus the control condition (data not shown). A 10-μm scale bar is shown in the day 0 panels.
IL-3 and pH modulate STAT3 and CD15 expression as well as changes in nuclear morphology during granulocytic differentiation.
Similar to Figure 1, except that immunofluorescence was evaluated on days 3, 7, and 15 of culture for images captured using a × 100 oil immersion objective. Images are representative of 2 randomly selected fields per culture condition for each of 3 different sets of experiments. The same field of cells is shown in the upper and lower panels of each day for each culture condition. Left panels: negative control (background) using FITC- and Texas Red–conjugated secondary antibodies in the absence of the relevant primary antibodies. Upper panels of each day: DAPI staining of nucleic acid (white). Lower panels of each day: codetection of STAT3 (green) and CD15 (red) using polyclonal rabbit anti-STAT3 and monoclonal mouse anti-CD15 primary antibodies, followed by FITC-conjugated goat anti–rabbit IgG and Texas Red–conjugated goat anti–mouse IgM μ-chain–specific secondary antibodies. DAPI staining indicates that cells sequentially exhibit features characteristic of typical granulocytic maturation—from the enlarged nucleus of the immature myeloblast to the lobulated nucleus of the mature neutrophil—with their progression dependent on the culture conditions. Arrows in day 3 panels indicate nucleoli. Arrows in day 7 panels indicate the decreased nuclear-cytoplasmic ratio of the myelocyte stage. Arrows in day 15 panels indicate typical nuclear morphology of metamyeloctyes (M), bands (B), and segmented neutrophils (SN). Advancement through the morphologic transitions did not appear different at 20% O2 compared with control cultures (data not shown). Ten-micrometer scale bars are shown in the negative control panels.
IL-3 and pH modulate STAT3 and CD15 expression as well as changes in nuclear morphology during granulocytic differentiation.
Similar to Figure 1, except that immunofluorescence was evaluated on days 3, 7, and 15 of culture for images captured using a × 100 oil immersion objective. Images are representative of 2 randomly selected fields per culture condition for each of 3 different sets of experiments. The same field of cells is shown in the upper and lower panels of each day for each culture condition. Left panels: negative control (background) using FITC- and Texas Red–conjugated secondary antibodies in the absence of the relevant primary antibodies. Upper panels of each day: DAPI staining of nucleic acid (white). Lower panels of each day: codetection of STAT3 (green) and CD15 (red) using polyclonal rabbit anti-STAT3 and monoclonal mouse anti-CD15 primary antibodies, followed by FITC-conjugated goat anti–rabbit IgG and Texas Red–conjugated goat anti–mouse IgM μ-chain–specific secondary antibodies. DAPI staining indicates that cells sequentially exhibit features characteristic of typical granulocytic maturation—from the enlarged nucleus of the immature myeloblast to the lobulated nucleus of the mature neutrophil—with their progression dependent on the culture conditions. Arrows in day 3 panels indicate nucleoli. Arrows in day 7 panels indicate the decreased nuclear-cytoplasmic ratio of the myelocyte stage. Arrows in day 15 panels indicate typical nuclear morphology of metamyeloctyes (M), bands (B), and segmented neutrophils (SN). Advancement through the morphologic transitions did not appear different at 20% O2 compared with control cultures (data not shown). Ten-micrometer scale bars are shown in the negative control panels.
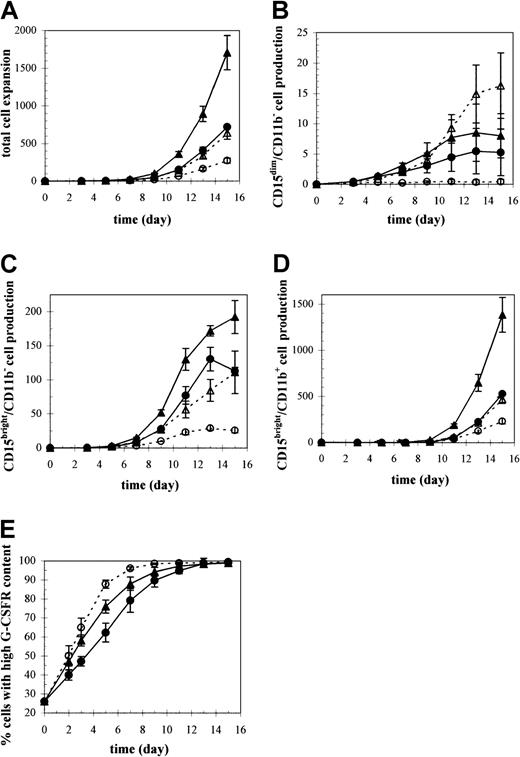
Cultures under all conditions were predominantly granulocytic, containing at least 90% CD15bright cells by day 15. However, the rate at which the cells matured as well as the expansion of each granulocytic subpopulation (Figure3B-D, Table1) varied under each culture condition.35,36 Total cell expansion was dramatically favored in cultures supplemented with IL-3 at low pH (pH 7.25) and low pO2 (5% O2), with a 2.4- or 2.7-fold enhancement in cell numbers by day 15 over cultures either at high pH (pH 7.4) or under high pO2 (20% O2), respectively (Figure 3A, Table 1). A pH of 7.4 delayed maturation throughout the entire differentiation pathway compared with control cultures.35,36 For example, by day 15, cultures at high pH consisted mostly of metamyelocytes that were just beginning to take on the appearance of bands and showed very few signs of nuclear cleavage or lobulations (Figure 2), whereas at low pH, cultures contained more mature cells, with nuclei segmented into 2 to 5 lobes. In contrast to pH, pO2 did not affect differentiation based on CD15/CD11b35 36 or morphologic characterization (data not shown) during the first week of culture and had only minor effects on differentiation at later time points.
Ex vivo granulocytic proliferation and differentiation under different culture conditions including pH, pO2, and IL-3.
CD34+ cells from mobilized peripheral blood were directed along the granulocyte lineage with SCF, IL-6, G-CSF, with or without IL-3, and harvested for total cell counts and characterized by flow cytometry for both cellular phenotype and receptor expression. Production of (A) total cells and the granulocytic subpopulation (B) CD15dim/CD11b− cells, (C) CD15bright/CD11b− cells, and (D) CD15bright/CD11b+ cells. Results are expressed as mean values ± SEM (n = 3) relative to the number of CD34+ cells used to initiate the cultures. (E) Kinetics of G-CSFR expression presented as mean ± SEM (n = 6) percentage of cells expressing high levels of G-CSFR at different culture times. Significant differences between cultures under the different conditions for each parameter examined are indicated in Table 1. ▴ indicates 5% O2, pH 7.25, +IL-3; ▵ indicates 20% O2, pH 7.25, +IL-3; ● indicates 5% O2, pH 7.4, +IL-3; ○ indicates 5% O2, pH 7.25, −IL-3.
Ex vivo granulocytic proliferation and differentiation under different culture conditions including pH, pO2, and IL-3.
CD34+ cells from mobilized peripheral blood were directed along the granulocyte lineage with SCF, IL-6, G-CSF, with or without IL-3, and harvested for total cell counts and characterized by flow cytometry for both cellular phenotype and receptor expression. Production of (A) total cells and the granulocytic subpopulation (B) CD15dim/CD11b− cells, (C) CD15bright/CD11b− cells, and (D) CD15bright/CD11b+ cells. Results are expressed as mean values ± SEM (n = 3) relative to the number of CD34+ cells used to initiate the cultures. (E) Kinetics of G-CSFR expression presented as mean ± SEM (n = 6) percentage of cells expressing high levels of G-CSFR at different culture times. Significant differences between cultures under the different conditions for each parameter examined are indicated in Table 1. ▴ indicates 5% O2, pH 7.25, +IL-3; ▵ indicates 20% O2, pH 7.25, +IL-3; ● indicates 5% O2, pH 7.4, +IL-3; ○ indicates 5% O2, pH 7.25, −IL-3.
Significant differences (P < .05) between culture conditions for the experiments shown in Figure 3
| Parameter . | Days with significant differences . | ||
|---|---|---|---|
| pH 7.25 vs pH 7.4 at 5% O2, +IL-3 . | 5% O2vs 20% O2 at pH 7.25, +IL-3 . | +IL-3 vs −IL-3 at 5% O2, pH 7.25 . | |
| Total cells | 3-15 | 3-15 | 3-15 |
| CD15dim/CD11b− cells | 7, 11, 15 | 5, 7 | 3, 5, 7 |
| CD15bright/CD11b−cells | 5-13 | 3-15 | 7-15 |
| CD15bright/CD11b+cells | 3-15 | 7-15 | 5-15 |
| G-CSFR content | 3-11 | * | 3-7 |
| Parameter . | Days with significant differences . | ||
|---|---|---|---|
| pH 7.25 vs pH 7.4 at 5% O2, +IL-3 . | 5% O2vs 20% O2 at pH 7.25, +IL-3 . | +IL-3 vs −IL-3 at 5% O2, pH 7.25 . | |
| Total cells | 3-15 | 3-15 | 3-15 |
| CD15dim/CD11b− cells | 7, 11, 15 | 5, 7 | 3, 5, 7 |
| CD15bright/CD11b−cells | 5-13 | 3-15 | 7-15 |
| CD15bright/CD11b+cells | 3-15 | 7-15 | 5-15 |
| G-CSFR content | 3-11 | * | 3-7 |
Control condition: 5% O2, pH 7.25, +IL-3.
Not evaluated at 20% O2, pH 7.25, +IL-3.
Total cell expansion was significantly reduced by 6.3-fold in cultures without IL-3 (Figure 3A, Table 1). However, the rate of differentiation was accelerated compared with that in the presence of IL-3.36 The kinetic effects of IL-3 on granulocytic maturation were distinct from the pH effects in that they did not persist through the entire granulocytic differentiation pathway but, rather, were isolated to the CD15dim/CD11b−myeloblast stage. The segmented nuclei of cells cultured without IL-3 indicated the prevalence of fully differentiated neutrophils by day 15 (Figure 2). The adverse effects of high pH, high pO2, or the absence of IL-3 were evidenced by the lower production of each granulocytic subtype compared with control conditions (Figure 3B-D).
G-CSFR up-regulation that occurs with granulocytic maturation was retarded at high pH and accelerated in the absence of IL-3 compared with control cultures (Figure 3E, Table 1). Approximately 90% of the cells acquired a high content of G-CSFR after 9 days at high pH, whereas control cultures approached this level on day 7 and as early as day 5 in cultures without IL-3. An independent set of experiments showed that high pO2 (20% O2) had no effect on G-CSFR up-regulation.35
Localization and expression of STAT3 during granulocytic differentiation
Cells were stained with antibodies specific for STAT3 and the granulocytic differentiation marker CD15 to assess protein expression and subcellular distribution. STAT3 was present early in hematopoiesis, staining intensely in day 0 CD34+ cells (Figure 1). On day 3, STAT3 protein appeared in both the nucleus and cytoplasm but not in the nucleoli (Figure 2). Subsequently, STAT3 continued to stain intensely but with an increased preference for the nucleus. Coexpression of STAT3 and CD15 clearly revealed their selective partitioning—STAT3 (green) in the nucleus and CD15 (red) in an extranuclear location, such as on the cell membrane—with minimal colocalization (yellow). STAT3 expression appeared strongest in CD15− cells and gradually decreased upon granulocytic differentiation (Figures 1 and 2). The most remarkable decrease in STAT3 expression was evidenced in day 15 CD15+ segmented neutrophils (Figure 2).
Expression of distinct STAT3 isoforms during granulocytic differentiation
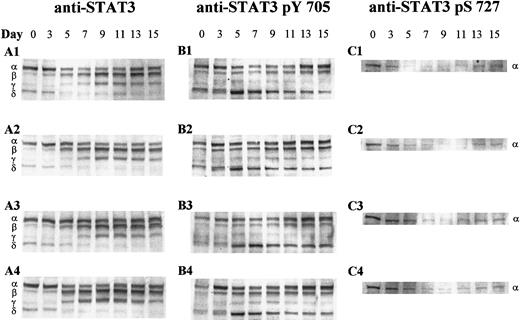
Western blot analysis was used to discriminate between the different STAT3 proteins present during granulocytic development. A widely used N-terminal anti-STAT3 antibody, generated against amino acids 1 to 178, recognized at least 3 previously characterized STAT3 isoforms with different electrophoretic mobilities: full-length STAT3α (92 kd) and the carboxyl-truncated forms, STAT3β (83 kd) and STAT3γ (72 kd) (Figure 4A). An additional faster-migrating protein of approximately 64 kd, which we designated as STAT3δ, was detected, likely representing a new putative truncated isoform. Protein levels of the different STAT3 species were assessed by quantitating the densities of the respective immunoreactive bands. Unlike the microscopic images, which depict STAT3 expression on a per-cell basis, the Western analysis of Figure 4reflects changes in STAT3 expression relative to equivalent amounts of total protein loaded on each day. Total protein per cell decreased as the cells matured, making it necessary to lyse a greater number of cells on later days to yield an equivalent amount of protein as on earlier days of analysis.
Western analysis of STAT3 isoform expression and phosphorylation during granulocytic differentiation.
Whole cell extracts (20 μg) of CD34+-derived cells undergoing granulocytic differentiation were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, blotted onto polyvinylidene fluoride membranes, and probed with antibodies against (A) STAT3, (B) STAT3 pTyr 705, and (C) STAT3 pSer 727. The 4 culture conditions evaluated are designated as 1-4 and are as follows: 1 indicates 5% O2, pH 7.25, +IL-3; 2 indicates 20% O2, pH 7.25, +IL-3; 3 indicates 5% O2, pH 7.4, +IL-3; and 4 indicates 5% O2, pH 7.25, −IL-3. The relative positions of STAT3α, β, γ, and δ are indicated on the left. The results shown are representative of 3 sets of experiments from different mobilized peripheral blood samples.
Western analysis of STAT3 isoform expression and phosphorylation during granulocytic differentiation.
Whole cell extracts (20 μg) of CD34+-derived cells undergoing granulocytic differentiation were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, blotted onto polyvinylidene fluoride membranes, and probed with antibodies against (A) STAT3, (B) STAT3 pTyr 705, and (C) STAT3 pSer 727. The 4 culture conditions evaluated are designated as 1-4 and are as follows: 1 indicates 5% O2, pH 7.25, +IL-3; 2 indicates 20% O2, pH 7.25, +IL-3; 3 indicates 5% O2, pH 7.4, +IL-3; and 4 indicates 5% O2, pH 7.25, −IL-3. The relative positions of STAT3α, β, γ, and δ are indicated on the left. The results shown are representative of 3 sets of experiments from different mobilized peripheral blood samples.
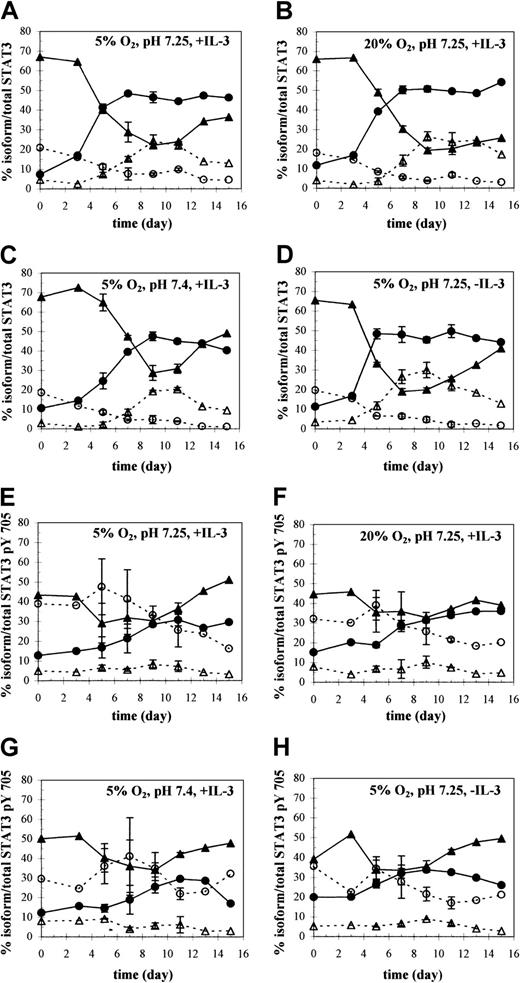
The ratio of isoforms shifted markedly during differentiation, creating a distinct pattern of expression at different stages of maturation (Figure 4A, culture condition 1). CD34+ cells expressed more than 65% of STAT3 as STAT3α, about 20% as the novel short form STAT3δ, about 10% as STAT3β, and the remainder as STAT3γ (Figure5A). Expression of the different isoforms changed appreciably between days 3 and 9 as indicated by the pronounced shift in expression of STAT3α to STAT3β (Figures 4A and 5A). Concomitant with STAT3α down-regulation during this time, expression of STAT3γ, a proteolytic fragment of STAT3α, was up-regulated. Enhancement of STAT3β expression plateaued under control conditions on day 7 at 50% (Figure 5A), where it was maintained throughout the culture period. A decrease in the STAT3β/STAT3α ratio after day 9 was indicative of an increase in STAT3α levels, not of down-regulated STAT3β expression. In summary, the ratio of the 4 STAT3 isoforms was dependent on the stage of differentiation, with STAT3α prevailing during early stages of granulocytic differentiation and STAT3β later during maturation.
Western analysis of STAT3 isoform expression and tyrosine phosphorylation kinetics throughout granulocytic differentiation.
(A-D) The changing distribution of STAT3α (▴), β (●), γ (▵), and δ (○) with time, expressed as a percentage of total STAT3, under the indicated culture conditions. (E-H) The corresponding changes in the distribution of tyrosine-phosphorylated STAT3α (▴), β (●), γ (▵), and δ (○) with time, expressed as percentages of total STAT3 pTyr 705. Percentages were calculated by quantifying protein levels of the immunoblots shown in Figure 4A (culture conditions 1-4) and 4B (culture conditions 1-4), respectively. Values on days 5 to 11 represent averages ± SD of 2 separate immunoblots from samples collected during the same experiment.
Western analysis of STAT3 isoform expression and tyrosine phosphorylation kinetics throughout granulocytic differentiation.
(A-D) The changing distribution of STAT3α (▴), β (●), γ (▵), and δ (○) with time, expressed as a percentage of total STAT3, under the indicated culture conditions. (E-H) The corresponding changes in the distribution of tyrosine-phosphorylated STAT3α (▴), β (●), γ (▵), and δ (○) with time, expressed as percentages of total STAT3 pTyr 705. Percentages were calculated by quantifying protein levels of the immunoblots shown in Figure 4A (culture conditions 1-4) and 4B (culture conditions 1-4), respectively. Values on days 5 to 11 represent averages ± SD of 2 separate immunoblots from samples collected during the same experiment.
STAT3 expression under different pH, pO2, and IL-3 conditions
Total STAT3 expression per cell (Figures 1 and 2) as well as the kinetics and magnitude of the shift between isoforms (Figure 4A, culture conditions 1-4) varied with culture conditions (pH, pO2, IL-3) and was consistent with differences in culture differentiation. At pH 7.4, STAT3 levels per cell were sustained for a longer period than at pH 7.25. For example, the less differentiated cells at high pH demonstrated a higher intensity of STAT3 in their nuclear portion at days 13 and 15 (Figures 1 and 2). The primary effect on isoform patterns was the maintenance of higher STAT3α levels throughout culture (Figures 4A, culture condition 3, and 5C). At its lowest level on day 9, STAT3α at pH 7.4 was still 50% higher than at pH 7.25. This, together with lower levels of STAT3β, resulted in a lower STAT3β/STAT3α ratio at pH 7.4 versus 7.25 throughout culture.
In contrast, in the absence of IL-3, total STAT3 levels per cell decreased more rapidly than in control cultures, along with a faster rise in the fraction of CD15+ cells and earlier signs of nuclear cleavage (Figures 1 and 2). The shift from predominantly STAT3α to predominantly STAT3β was also accelerated (Figure 4A, culture condition 4). By day 5, 50% of total STAT3 was expressed as the β isoform in −IL-3 cultures compared with day 7 in +IL-3 cultures (Figure 5D vs 5A). The accelerated decrease in STAT3α expression occurred concomitantly with an accelerated increase in STAT3γ expression in −IL-3 cultures (Figures 4A, culture condition 4, and 5D). The effects of O2 tension on STAT3 isoform expression were less remarkable, showing little difference in protein levels, particularly during the first week of culture (Figures 4A, culture condition 2, and 5B). However, during the second week, lower levels of STAT3α and higher levels of STAT3γ persisted for longer times under 20% O2.
STAT3 tyrosine phosphorylation
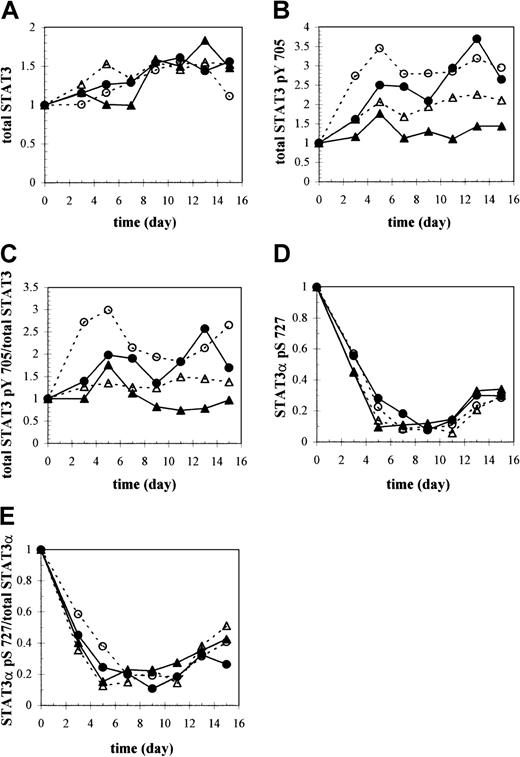
A strong correlation is known to exist between tyrosine phosphorylation specifically on residue 705 and rapid nuclear translocation of STAT3 and the formation of STAT3-STAT3-DNA complexes.26 30 We used an antibody against tyrosine-phosphorylated STAT3 (anti-STAT3 pTyr 705) to assess activation of total STAT3 and of the individual isoforms under different culture conditions (Figure 4B, culture conditions 1-4) and thereby determine whether selective phosphorylation of one isoform versus another occurred, leading to its preferential translocation to the nucleus. Although overall levels of STAT3 did not vary much between culture conditions when comparing equal amounts of total protein (Figure 6A), the extent of tyrosine phosphorylation did. Overall levels of tyrosine-phosphorylated STAT3 remained relatively constant under control conditions but were about 2.5 to 3 times greater in the absence of IL-3 (Figure 6B). STAT3 tyrosine phosphorylation at high pH (7.4) also reached levels of 2.5 to 3 times that at the lower pH (7.25) of control conditions but increased more gradually until about day 13 (Figure 6B,C). Under 20% O2, STAT3 tyrosine phosphorylation also appeared to increase steadily, although to a lesser extent (Figure 6B,C).
Western analysis of STAT3, tyrosine-phosphorylated STAT3, and serine-phosphorylated STAT3.
Total (A) STAT3, (B) tyrosine-phosphorylated STAT3 (STAT3 pTyr 705), and (D) serine-phosphorylated STAT3 (STAT3α pSer 727) protein as a function of time and culture condition were quantified based on equivalent amounts of total protein by analyzing the immunoblots shown in Figure 4A-C (culture conditions 1-4), respectively, with ImageQuant software. The signal intensities for each isoform within a lane were added to yield the total amount of STAT3, STAT3 pTyr 705, or STAT3 pSer 727 protein and expressed relative to the day 0 value. Anti-STAT3 pSer 727 antibody specifically recognized the phosphorylated serine 727 site found only in the STAT3α isoform. (C) STAT3 pTyr 705 was normalized to total STAT3 levels, or (E) STAT3 pSer 727 protein was normalized to total STAT3α levels at each time point to show changes in the fraction of STAT3 that is either tyrosine- or serine-phosphorylated, respectively, during differentiation. ▴ indicates 5% O2, pH 7.25, +IL-3; ▵ indicates 20% O2, pH 7.25, +IL-3; ● indicates 5% O2, pH 7.4, +IL-3; and ● indicates 5% O2, pH 7.25, −IL-3.
Western analysis of STAT3, tyrosine-phosphorylated STAT3, and serine-phosphorylated STAT3.
Total (A) STAT3, (B) tyrosine-phosphorylated STAT3 (STAT3 pTyr 705), and (D) serine-phosphorylated STAT3 (STAT3α pSer 727) protein as a function of time and culture condition were quantified based on equivalent amounts of total protein by analyzing the immunoblots shown in Figure 4A-C (culture conditions 1-4), respectively, with ImageQuant software. The signal intensities for each isoform within a lane were added to yield the total amount of STAT3, STAT3 pTyr 705, or STAT3 pSer 727 protein and expressed relative to the day 0 value. Anti-STAT3 pSer 727 antibody specifically recognized the phosphorylated serine 727 site found only in the STAT3α isoform. (C) STAT3 pTyr 705 was normalized to total STAT3 levels, or (E) STAT3 pSer 727 protein was normalized to total STAT3α levels at each time point to show changes in the fraction of STAT3 that is either tyrosine- or serine-phosphorylated, respectively, during differentiation. ▴ indicates 5% O2, pH 7.25, +IL-3; ▵ indicates 20% O2, pH 7.25, +IL-3; ● indicates 5% O2, pH 7.4, +IL-3; and ● indicates 5% O2, pH 7.25, −IL-3.
The most notable difference between trends in expression of the individual STAT3 isoforms and their respective tyrosine phosphorylation kinetics was the preferential activation of STAT3δ over the other isoforms (Figure 4B, culture condition 1). Under control conditions, at day 0 STAT3δ made up about 20% of total STAT3 protein and diminished thereafter (Figure 5A); however, STAT3δ pTyr 705 made up 30% to 40% of total STAT3 pTyr 705 at day 0 and increased to a maximum value of about 50% at day 5 before slowly decreasing (Figure5E). Consistent with greater total STAT3 phosphorylation at high pH and in −IL-3 cultures (Figure 6B), the absolute level of STAT3δ pTyr 705 was greater under both conditions compared with control and peaked later on day 7 at high pH (data not shown).
The kinetics of STAT3γ tyrosine phosphorylation were also in clear contrast to the kinetics of total STAT3γ protein. Whereas total STAT3γ protein levels increased to a maximum between days 9 and 11 and reached as high as 30% of the total STAT3 species (Figure 5A), its tyrosine phosphorylation was minimal throughout culture (Figure 4B, culture condition 1) at levels averaging 5% of total tyrosine-phosphorylated STAT3 in every culture condition (Figure 5E-H).
STAT3α and STAT3β were also strongly phosphorylated on tyrosine 705. However, the pronounced shift from predominantly STAT3α to predominantly STAT3β that was detected in their overall expression was not observed with their activated forms. Phosphorylated STAT3β levels approached but never surpassed those of STAT3α (Figures 4B and5E). Activation of STAT3α was enhanced at high pH, but this increase was commensurate with the increase in total STAT3α levels at pH 7.4 compared with those at pH 7.25 (data not shown). Therefore, the fraction of STAT3α that was tyrosine-phosphorylated relative to total STAT3α was not dependent on pH during the first 9 days. Similar to high pH cultures, STAT3α was also more strongly phosphorylated in −IL-3 cultures (Figure 4B, culture condition 4) compared with control. However, unlike cultures at high pH, lower total STAT3α levels in −IL-3 cultures (data not shown) compared with control indicate that the absence of IL-3 enhanced the fraction of STAT3α that was phosphorylated. This effect served to minimize the difference in the STAT3β/STAT3α ratio of tyrosine-phosphorylated forms between cultures with and without IL-3.
STAT3 serine phosphorylation
Immunoreactivity for STAT3 serine phosphorylation was detected only with the STAT3α isoform, reflecting the absence of serine residue 727 in the carboxyl-truncated isoforms, STAT3β and STAT3γ, and in STAT3δ (Figure 4C, culture condition 1). Therefore, unlike all STAT3 isoforms that are potential targets for tyrosine phosphorylation of residue 705, only STAT3α is subject to serine phosphorylation on residue 727. Strongest reactivity of anti-STAT3 pSer 727 antibody was observed on day 0, after which there was a dramatic drop in expression levels (Figure 6D).
The effects of different culture conditions on serine phosphorylation of STAT3α were similar to the effects on its tyrosine phosphorylation: pH elicited differences that were reflected in total STAT3α levels, while IL-3 altered the fraction of total STAT3α that was serine-phosphorylated. Specifically, at high pH, an increased presence of STAT3α pSer 727 compared with control (Figures 4C, culture condition 3, and 7B) paralleled higher levels of total STAT3α under this condition, suggesting that the higher STAT3α pSer 727 levels observed (Figure 6D) resulted from greater total STAT3α levels. In the absence of IL-3, despite lower levels of total STAT3α than control, relative levels of STAT3α pSer 727 were enhanced, particularly on days 3 and 5 (Figure 6D). Furthermore, STAT3 pSer 727 immunofluorescence was noticeably more intense in the nuclei of −IL-3 cultures on day 5 (Figure 7C). No differences in STAT3 pSer 727 were apparent under different O2 tensions (Figure 6D,E or as observed microscopically [data not shown]).
Localization and expression of serine-phosphorylated STAT3 and CD15 proteins during granulocytic differentiation.
Detection of STAT3 pSer 727 and CD15 in CD34+ cell-derived granulocytic cultures at (A) 5% O2, pH 7.25, +IL-3 (control condition); (B) 5% O2, pH 7.4, +IL-3 (high pH); and (C) 5% O2, pH 7.25, −IL-3 using dual-color immunofluorescence microscopy. Negative controls (background) for each day of analysis using FITC- and Texas Red–conjugated secondary antibodies in the absence of the relevant primary antibodies are shown in the first row. The same field of cells is shown in the upper and lower panels of panels A-C. Upper panels of panels A-C: DAPI staining of nucleic acid (white). Lower panels of panels A-C: codetection of STAT3 pSer 727 (green) and CD15 (red) using polyclonal rabbit anti-STAT3 pSer 727 and monoclonal mouse anti-CD15 primary antibodies, followed by FITC-conjugated goat antirabbit IgG and Texas Red–conjugated goat antimouse IgM μ-chain–specific secondary antibodies. Images were captured using a × 100 oil immersion objective and are representative of 2 randomly selected fields per culture condition for each of 3 different sets of experiments. No appreciable differences in STAT3 pSer 727 and CD15 staining were visible in cells cultured at 20% O2 versus the control condition (data not shown).
Localization and expression of serine-phosphorylated STAT3 and CD15 proteins during granulocytic differentiation.
Detection of STAT3 pSer 727 and CD15 in CD34+ cell-derived granulocytic cultures at (A) 5% O2, pH 7.25, +IL-3 (control condition); (B) 5% O2, pH 7.4, +IL-3 (high pH); and (C) 5% O2, pH 7.25, −IL-3 using dual-color immunofluorescence microscopy. Negative controls (background) for each day of analysis using FITC- and Texas Red–conjugated secondary antibodies in the absence of the relevant primary antibodies are shown in the first row. The same field of cells is shown in the upper and lower panels of panels A-C. Upper panels of panels A-C: DAPI staining of nucleic acid (white). Lower panels of panels A-C: codetection of STAT3 pSer 727 (green) and CD15 (red) using polyclonal rabbit anti-STAT3 pSer 727 and monoclonal mouse anti-CD15 primary antibodies, followed by FITC-conjugated goat antirabbit IgG and Texas Red–conjugated goat antimouse IgM μ-chain–specific secondary antibodies. Images were captured using a × 100 oil immersion objective and are representative of 2 randomly selected fields per culture condition for each of 3 different sets of experiments. No appreciable differences in STAT3 pSer 727 and CD15 staining were visible in cells cultured at 20% O2 versus the control condition (data not shown).
Discussion
STAT3 kinetics were maturation stage–dependent and varied with pH, pO2, and IL-3 in a manner consistent with the marked differences induced by each condition on granulocytic growth and differentiation. Overall levels of STAT3 per cell were found to decrease with granulocytic differentiation (Figures 1 and 2). This phenomenon, and the disappearance of nucleoli past the myelocyte stage, may reflect the end of mitosis and the need for new protein synthesis, while the lack of STAT3 expression in segmented neutrophils could be representative of their maturation status as the terminal differentiation stage. These cell-to-cell differences in overall STAT3 protein production were less obvious by Western analysis, especially because more cells were used on later days to account for decreasing levels of total protein with differentiation. Western analysis, however, was essential in revealing that different STAT3 isoforms—STAT3α, STAT3β, STAT3γ, and the putative STAT3δ—were regulated during granulocytic differentiation in a maturation stage–specific manner.
The most pronounced change in the relative concentrations of the different STAT3 isoforms occurred with the shift in expression from predominantly STAT3α to predominantly STAT3β between days 3 and 9, which coincided with the rapid increase in the number of cells expressing high levels of G-CSFR and CD15 antigen. These results are consistent with those of Biethahn et al,27 who showed that while STAT3α levels are much higher than STAT3β in CD34+ cells, the reverse is true in bone marrow granulocytes consisting mostly of metamyelocytes. These findings and others that support a role for STAT3α in proliferation11,24,46 47—together with our results—provide compelling evidence to link differential expression of the STAT3α and β isoforms to the maturation process, with a shift away from STAT3α expression toward STAT3β favoring differentiation.
Variations in STAT3α protein levels can be attributed in large part to proteolytic cleavage of STAT3α to STAT3γ. The apparent impaired ability of STAT3γ to become tyrosine-phosphorylated indicates that it has less transactivating potential than other STAT3 isoforms and suggests that proteolytic activity could be an important event in normal granulocytic development.
A fourth putative STAT3 isoform, STAT3δ, which was recognized by both anti-STAT3 and anti-STAT3 pTyr 705 antibodies, was also regulated in a developmental fashion. STAT3δ levels were higher in the early stages of differentiation and were subsequently down-regulated. Although STAT3δ was not a major presence in our cultures, it was preferentially phosphorylated. This suggests that it could be an important determinant of STAT3 activity by participating in the formation of active STAT3 dimers.
The balance among the various STAT3 isoforms does indeed appear to be functionally relevant, given the coordination between differential expression and/or activation of the different STAT3 isoforms and distinct granulocytic cell responses induced by pH, pO2, and IL-3. At high pH, differentiation and proliferation were slower throughout the granulocytic pathway in parallel with slower G-CSFR up-regulation. This correlates with a slower shift from STAT3α to STAT3β and with lower STAT3β:STAT3α ratios throughout differentiation. Higher levels of tyrosine and serine phosphorylated species, particularly of STAT3α, reflect greater total expression levels rather than preferential phosphorylation. Thus, slower differentiation at high pH through all stages of granulocytic maturation appears to be due primarily to altered protein levels of the different isoforms rather than to altered isoform phosphorylation.
In contrast to slower overall differentiation at high pH, the absence of IL-3 accelerated differentiation and G-CSFR up-regulation. These data are suggestive of an antagonistic action of IL-3 on G-CSF–dependent differentiation, which is consistent with earlier cell line–based data.48-50 It seems likely, therefore, that the enhanced differentiation rate in −IL-3 cultures is due to an uninhibited signal that may involve STAT3 proteins. This is supported by the more rapid shift from STAT3α to STAT3β that we observed between days 3 and 9 in −IL-3 cultures. Although the proportion of each tyrosine-phosphorylated isoform was similar between cultures with and without IL-3, we detected increased absolute levels of tyrosine-phosphorylated STAT3 species in the absence of IL-3. Because of G-CSF's well-known ability to induce tyrosine phosphorylation of STAT proteins,51 it is possible that the accelerated G-CSFR up-regulation that we observed under this condition triggered enhanced G-CSF–mediated STAT3 activation. This is consistent with the finding by Steinman and Iro,52 who used a cell line model to show that the presence of IL-3 inhibits G-CSF–mediated STAT3 activation because of decreased tyrosine phosphorylation.
Exposure of cells to 20% O2 was also found to increase STAT3 tyrosine phosphorylation, particularly during the second week of culture. Given that we found no link between pO2 and G-CSFR expression, a mechanism distinct from the proposed G-CSFR–mediated pathway leading to enhanced STAT3 tyrosine phosphorylation in −IL-3 cultures may be involved in the pO2 effects. Carballo et al53 provided evidence that H2O2and other oxidants directly enhance STAT3 tyrosine phosphorylation in human lymphocytes and lead to STAT3 translocation to the nucleus by inhibiting tyrosine phosphatases. It is likely that production of reactive oxygen species, which has been shown to occur in hematopoietic cultures under 20% O2,54-57 resulted in the increase of STAT3 tyrosine phosphorylation that we observed.
In contrast to tyrosine phosphorylation, which is obligatory for transcriptional activity of STAT3, the role of serine phosphorylation has been controversial; it can enhance,30-32inhibit,34,58 or have no effect6,59 60 on tyrosine phosphorylation, DNA binding, and transcriptional activity. Serine phosphorylation in STAT3 occurs primarily on a single residue, serine 727, and in our cultures levels were greatest on early days, when cells occupied relatively immature stages of granulopoiesis and were highly proliferative, but decreased rapidly with differentiation. This suggests that serine phosphorylation may be a more influential determinant of the expansion of primitive cell types (myeloid stem and progenitor cells) than in the differentiation of granulocytic postprogenitors.
These data collectively reveal the complexity of STAT regulation and show that STAT3 can integrate diverse signals, ranging from cytokines to pH and pO2, so as to coordinate proliferation and differentiation. The evidence strongly indicates that the multiple STAT3 isoforms, each possessing distinct physiologic functions, potentiate the range of granulocytic responses observed under the different experimental conditions. Selective expression and/or phosphorylation of the different STAT3 isoforms provides a molecular basis for these responses to the culture environment, thereby emphasizing the importance of cellular context in determining granulocytic kinetics. These results are particularly relevant in the physiologic setting where cells are exposed to numerous stimuli simultaneously. Other potential mechanisms that were not investigated here, such as the activation of other STAT family members like STAT5 or functional interactions with other transcriptional coactivators, may also contribute to the balance between proliferation and differentiation and will be important in further defining a molecular model of granulopoiesis.
We thank Amgen for donation of SCF. We are grateful to Dr B. He for numerous valuable discussions on quantitative Western analysis. We also thank Dr M. Swartz, Dr R. Holmgren, and Yu Kuang for technical assistance.
Supported by National Institutes of Health grant R01 HL48276.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Eleftherios T. Papoutsakis, Northwestern University, Dept of Chemical Engineering, 2145 Sheridan Rd, Evanston, IL 60208-3120; e-mail: e-paps@northwestern.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal