Glucose-6-phosphate dehydrogenase (G6PD) A-is a common G6PD variant among Africans that may cause acute hemolysis triggered by infections and certain drugs, as well as by fava beans. This class-3 phenotype can be caused by a combination of the common 376A>G (Asn126Asp) mutation and either of 3 additional mutations: 202G>A (Val68Met), 680G>T (Arg227Leu), or 968T>C (Leu323Pro).1,2 The structure and function relationship of the most common type with 376A>G + 202G>A has been studied in detail.3,4 The missense mutation 376A>G by itself causes an asymptomatic class-4 variant G6PD A with normal enzyme activity,5 whereas the other mutation 202G>A has never been found in humans by itself. Some investigators insisted that both mutations in G6PD A- are necessary to produce the G6PD-deficient phenotype.6 Here we report a symptomatic G6PD deficiency case associated with the missense mutation 202G>A but not the 376A>G.
A 3-year-old Japanese boy was noted to have jaundice and anemia, and was referred to the Department of Pediatrics, Asahi General Hospital (K.K. and A.Ho.), for evaluation. At the time of admission, he showed clinical and laboratory findings compatible with acute hemolysis, including hemoglobin concentration, 9.6 g/dL; reticulocyte count, 2.9%; and serum total bilirubin, 4.7 mg/dL (83% of which was indirect reacting). His anemia and jaundice improved spontaneously within 5 days. At the time of follow-up visits, no signs of chronic hemolysis were observed. He had a history of prolonged neonatal jaundice.
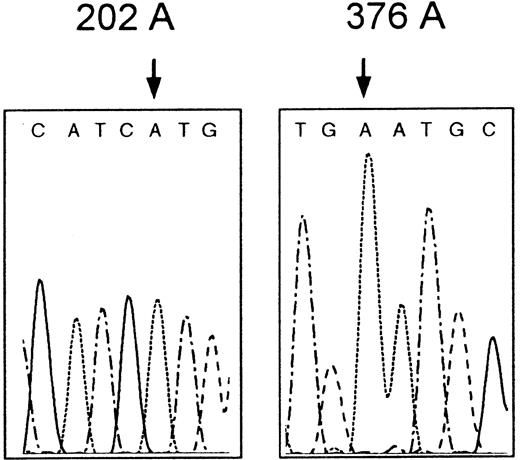
The patient's red cell G6PD activity was decreased (2.9 IU/gHb; referential range (mean ± SD): 7.12 ± 0.79 IU/gHb). The enzyme activity of the mother was also decreased (2.8 IU/gHb). Using PCR-SSCP (polymerase chain reaction–single strand conformation polymorphism) analysis combined with direct sequencing,7 we identified a missense mutation 202G>A, which predicts the amino acid substitution Val68Met in the proband'sG6PD gene, but surprisingly failed to find the counterpart 376A>G (Figure 1). Because it was quite unusual for 202G>A to be found alone, we further sequenced the whole coding exons and flanking regions of adjacent introns to search the second mutation. But we did not find any additional mutation except for a single base deletion in intron 5 (636delT or 637delT), which has been identified in various Mongoloid populations, including Japanese.7 Thus we concluded that the patient's reduced enzyme activity was caused entirely by the 202G>A mutation alone. Produced unique variant designated G6PD Asahi falls into class 3 in the World Health Organization criteria.8 The mother was found to be heterozygous for this mutation.
Partial nucleotide sequence of G6PD Asahi.
The proband's G6PD gene has the mutation 202G>A (left) but not the 376A>G (right).
Partial nucleotide sequence of G6PD Asahi.
The proband's G6PD gene has the mutation 202G>A (left) but not the 376A>G (right).
It is evident that the 202G>A mutation found in our patient has arisen separately from those common in Africans, because the patient had none of the silent mutations closely linked with the African mutation,9 while he had an intronic single base deletion common in Mongoloid.7
An in vitro study6 using recombinant human G6PDmutants expressed in Escherichia coli suggested that 202G>A, as well as 376A>G, does not cause enzyme deficiency by itself, and the synergistic action of these 2 mutations is necessary to produce the class-3 phenotype of G6PD A-. This synergistic interaction was also supported by the fact that Val68 and Asn126 are closely located in a 3-dimensional model of human G6PD.3 Our results are inconsistent with those previous observations. Although there still remains a rare possibility that we have overlooked a second mutation in introns or in 5′- and 3′-noncoding regions, the inconsistency suggests that the function and the fate of a mutant enzyme in vivo might be very different from those deduced from experiments using the recombinant enzyme.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal