Abstract
The recombination activating gene-1 (RAG-1) andRAG-2 are expressed specifically in immature lymphoid cells undergoing the recombination of antigen receptor genes. The regulation of murine RAG-2 promoter was studied and it was revealed that the −41/−17 RAG-2 promoter region, which is conserved between humans and mice, was indispensable for theRAG-2 promoter activity in B-cell lines. The region contained 2 cis elements that bound c-Myb and Pax-5. Mutation in the c-Myb–binding site in the promoter reduced the promoter activity in B-cell lines. Cooperative activation of theRAG-2 promoter was seen by a combination of c-Myb and Pax-5 in a human embryonic kidney cell line (293T), via their synergistic DNA-binding. Deletion experiments showed that the C-terminus of c-Myb was responsible for their interaction. Furthermore, the dominant-negative c-Myb mutant suppressed the activation of theRAG-2 promoter in a pre–B-cell line as well as in 293T cells. These results suggest that cooperative binding of c-Myb and Pax-5 to the RAG-2 promoter is one of the mechanisms to direct the restricted expression of the RAG-2 in immature B cells.
Introduction
Immunoglobulin (Ig) and T-cell receptor(TCR) variable genes consist of germline variable (V), diversity (D), and joining (J) segments and are assembled during lymphocyte development by V(D)J recombination. The recombination activating gene-1 (RAG-1) and RAG-2 encode the essential and lymphocyte-specific components of V(D)J recombination machinery. Their products are sufficient for the recognition and initial cleavage of DNA containing recombination signal sequences that flank each coding segment.1,2 During lymphocyte development, expression of RAG-1 and RAG-2 is tightly regulated. RAG genes are expressed in immature B- or T-lineage cells undergoing Ig or TCR gene rearrangements.3-6 The failure of functional expression ofRAG causes a defect in the formation of the functional antigen receptor of lymphocytes, and hence causes the block of lymphocyte development in mice and humans.7-10
The transcription of RAG is regulated at different levels. At the chromatin level, Fuller and Storb11 and Kitagawa et al12 have demonstrated that alteration of chromatin structure detected by DNase I hypersensitivity was noted in the promoter region of mouse and human RAG-1 only inRAG-expressing lymphocytes, indicating that chromatin remodeling is one of the mechanisms for regulating RAGexpression. At the cis-element level, Yu et al have demonstrated that about 10 kb 5′ upstream region of RAG-2 is necessary for the expression of RAG in B-lineage cells and in CD4−CD8− thymocytes, and that further upstream region is required for the expression of RAG in CD4+CD8+ thymocytes.13 Monroe et al have demonstrated that about 10 kb 5′ upstream region ofRAG-2 is enough for rescuing B- and T-cell development inRAG2−/− mice by usingRAG-2−/− blastocyst complementation.14 These results suggest that expression of RAG is regulated by the cis-element, such as enhancer. At the promoter level, it was reported that the humanRAG-1 promoter region does not confer the lymphocyte-specific expression ofRAG-1.12,15-17 Regarding the promoter ofRAG-2, about 300 bp 5′ upstream region from the major transcription initiation site of mouse RAG-2 is conserved between mice and humans,17-19 indicating that this region is important for the promoter activity of RAG-2. It was also demonstrated that the human RAG-2 promoter is activated not only in lymphoid cells but also in nonlymphoid cells.17Concerning the mouse RAG-2 promoter, Schlissel and his colleagues and Kishi et al have demonstrated that the core promoter of mouse RAG-2 confers lymphoid specificity and may be regulated with distinct transcription factors: Pax-518,19in B cells and GATA318 or c-Myb20 in T cells.
c-Myb protein is a transcription factor that is predominantly expressed in immature hematopoietic cells of all lineages and regulates cell proliferation and differentiation.21 c-Myb antisense oligonucleotide inhibited hematopoietic colony formation22and mice deficient for c-Myb showed defective hematopoiesis in fetus.23 To activate lineage-specific genes in different hematopoietic precursor cells, c-Myb functioned in a combinational manner with other transcription factors.24Some transcription factors, such as core-binding factor/PEBP2,25 C/EBPα, or PU.1,26 have been shown to cooperate with c-Myb to activate enhancer or promoter without specifically binding to c-Myb. The other transcription factors, such as C/EBPε27 have been reported to directly bind to c-Myb and cooperatively activate the transcription. It was also reported that c-Myb interacted with coactivators, such as CBP, to activate transcription.28 Here we extended the study on transcriptional regulation of mouse RAG-2 in B-lineage cells and demonstrated that c-Myb interacts with Pax-5 and activates the mouse RAG-2 promoter in B cells.
Materials and methods
Cells and cell culture
The 18.8.1 pre-B cell line18 and the BAL17 B cell line,18 both of which express endogenous murineRAG-1 and RAG-2, were cultured in RPMI 1640 supplemented with 10% fetal calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in 5% CO2 atmosphere. 293T, a human embryonic kidney cell line, was grown in Dulbecco modified Eagle medium containing 10% fetal calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C and 5% CO2.
Plasmid constructs
Luciferase constructs used for promoter assay were prepared using PicaGene basic vector 2 (Nippon Gene, Tokyo, Japan) as described previously.18 Luciferase constructs containing the mutation in putative c-Myb binding sites (c-Myb1 and c-Myb2 in Figure 2A) were prepared by producing the promoter fragment by a sequential polymerase chain reaction (PCR). First, a 5′ fragment and a 3′ fragment containing the mutated sequence were prepared by PCR using oligonucleotide 1 (5′-TTCTGTCTCCCTCAACCATC-3′, −85/−66 ofRAG-2) and either oligonucleotide 2 (5′-ACGGGAGTAAATCACTGTGACCT-3′, for c-Myb1m, −19/−41 ofRAG-2) or oligonucleotide 3 (5′-TGCTGGTGTAAAGCGAGTAACTGA-3′, for c-Myb2m, −9/−32 of RAG-2), and by PCR using either oligonucleotide 4 (5′-AGGTCACAGTGATTTACTCCCGT-3′, for c-Myb1m, −41/−19 of RAG-2) or oligonucleotide 5 (5′-TCAGTTACTCGCTTTACACCAGCA-3′, for c-Myb2m, −32/−9 ofRAG-2) and oligonucleotide 6 (5′-GGGGTACCATGGCCAGAGGGGCTGCTTATC-3′, +147/+118 of RAG-2), respectively. Then the full length −86/+147 fragments containing mutated sequences were prepared by PCR using oligonucleotides 1 and 6 and cloned into PicaGene basic vector 2. Mouse βTCR 3′ enhancer was cloned into the luciferase constructs as described previously18 and used for transfection into B-cell lines. Luciferase constructs without enhancer were used for transfection into 293T cells. The mouse c-Myb expression vector (pAct–c-Myb) and its mutants29 were kindly provided by Dr Ishii at Riken, Tsukuba, Japan. Pax-5 expression vector (pEFBOS–Pax-5) was generated by preparing the Pax-5 cDNA by reverse transcriptase (RT)–PCR and cloning it to the pEF-BOS vector.30
Luciferase reporter assay
For transfection into B-cell lines, luciferase constructs were transfected using diethylaminoethyl (DEAE) dextran method as described previously.18 pSRα-LacZ gene was used as an internal control. When the expression vector encoding dominant-negative mutant c-Myb (pAct-ΔTA) was cotransfected with different doses, the total amount of DNA was adjusted by adding pAct vector DNA. Twenty-four hours after transfection, cells were harvested, and luciferase activity and β-galactosidase activity were measured as described previously.18 For transfection into 293T cells, the calcium/phosphate method was used as described previously.18 Forty-eight hours after transfection, cells were harvested, and luciferase activity and β-galactosidase activity were measured. When c-Myb or Pax-5 expression vectors were transfected with different doses, the total amount of DNA was adjusted by adding either pAct vector DNA or pEF-BOS vector DNA.
Electrophoresis mobility shift assay
Nuclear extracts were prepared.18Electrophoresis mobility shift assay (EMSA) was performed by incubating nuclear extracts with radiolabeled oligonucleotides and then subjecting them to electrophoresis as described previously.18 For supershift assay, 0.5 μg polyclonal Pax-5 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or control IgG was added to the extract prior to oligonucleotide addition. Where indicated, the radioactivity of the DNA/protein complex was measured by BAS2000 (Fuji Film, Tokyo, Japan). Oligonucleotides containing the consensus binding site for c-Myb, mutated c-Myb binding site, Pax-5 binding site, mutated Pax-5 binding site, and the GATA binding site used for competition were 5′-TTACAGGCATAACGGTTCCGTAGTGA-3′, 5′-TACAGGCATATCGGTTCCGTAGTGA-3′, 5′-TACCCTTGATCAAAGCAGTGTGACGGTAGC-3′, 5′-GACCCTTGATCAAAGCAGTATGATGGTAGC-3′, and 5′-CACTTGATAACAGAAAGTGATAACTCT-3′, respectively.
Precipitation of c-Myb and Pax-5 by DNA-sepharose
To prepare DNA-sepharose beads, oligonucleotides containing −41/−17 sequences of mouse RAG-2 promoter, 5′-ATGCATGCATGAGGTCACAGTCAGTTACTCCCGTT -3′ and 5′-ATGCATGCATAACGGGAGTAACTGACTGTGACCTC-3′, were annealed and coupled to cyanogen bromide– activated sepharose 4B (Amersham Pharmacia Biotech, Uppsala, Sweden) as previously described.31 As a control, oligonucleotides containing the STAT3 binding site (5′-TCGACTCGTTCCCAGCAGCAC-3′) were coupled to the beads. A quantity of 100 μL nuclear extracts was added to 100 μL of 1x binding buffer (4% ficoll, 20 mM HEPES, pH 7.9, 50 mM KCl, 1 mM ethylenediaminetetraacetic acid [EDTA], 1 mM dithiothreitol [DTT], 0.25 mg/mL bovine serum albumin [BSA]) and precleared with 10 μL of sepharose 4B beads at 4°C for 1 hour. Then 50 μL extracts was diluted to 90 μL in 1x binding buffer containing 0.13 μg/μL poly(dI-dC) and incubated with 10 μL DNA-sepharose beads at 4°C for 3 hours. The beads were washed with 1x binding buffer 5 times. Bound proteins were eluted by boiling in sodium dodecyl sulfate (SDS)–sample buffer, resolved in SDS–polyacrylamide gel electrophoresis (PAGE), and transferred to PolyScreen polyvinylidenefluoride (PVDF) membrane (NEN Life Science Products, Boston, MA). The membrane was incubated with c-Myb monoclonal antibody (clone 1.1; Upstate Biotechnology, Lake Placid, NY) or polyclonal Pax-5 antibody described above, followed by peroxidase-conjugated anti–mouse IgG or anti–goat IgG, and finally developed using the ECL system (Amersham Pharmacia Biotech).
Results
The −41/−17 region is essential for the murineRAG-2 promoter
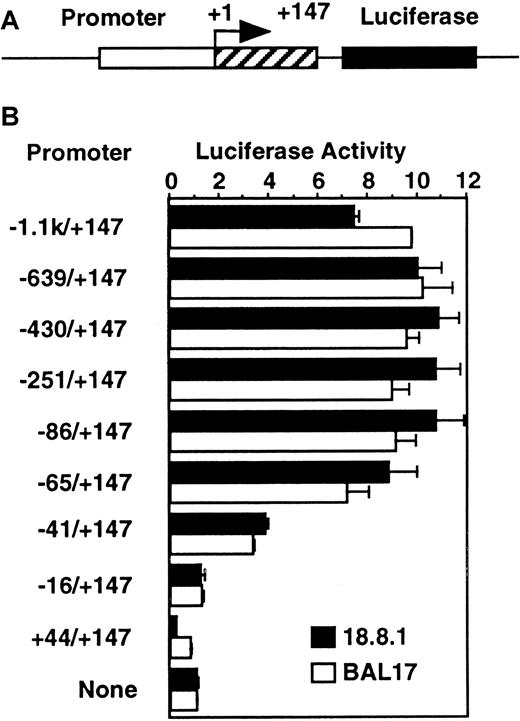
To determine the minimal region for activation of the murineRAG-2 promoter, the luciferase reporter gene connected to the −1.1 kb to +147 region of the murine RAG-2 or its serial deletion was transfected into the 18.8.1 pre–B-cell line or the BAL17 B-cell line (Figure 1). Luciferase constructs linked to −1.1 kb/+147 showed the luciferase activity about 8-fold to 10-fold higher than that of the promoterless construct. Deletion of the RAG-2 promoter from −1.1 kb to −65 did not affect the promoter activity in both cell lines. Deletion to −41 reduced the promoter activity to about half, and deletion to −16 completely abolished the promoter activity, showing that the −41 to −16 region was essential for murine RAG-2 promoter activity.
The −41/−17 region is essential for the murine
RAG-2 promoter in B-cell lines. (A) Diagram of promoter/luciferase construct. Mouse RAG-2 gene spanning −1.1 kb to +147 bp or its serial deletion (open box for −1.1 kb to −1 and shaded box for +1 to +147) was linked to 5′ of luciferase reporter gene (closed box). +1 indicates major transcription initiation site. (B) Promoter activity in B-cell lines with serially deletedRAG-2 promoter region. A quantity of 10 μg luciferase constructs linked to the serially deleted mouse RAG-2promoter region (indicated on left) was transfected into 18.8.1 cells or BAL17 cells. pSRα-LacZ was included as an internal control. Twenty-four hours later, luciferase and β-galactosidase assays were performed on cell extracts. The activity of the luciferase construct without promoter in each cell is set to 1. Error bars indicate deviation of 3 experiments.
The −41/−17 region is essential for the murine
RAG-2 promoter in B-cell lines. (A) Diagram of promoter/luciferase construct. Mouse RAG-2 gene spanning −1.1 kb to +147 bp or its serial deletion (open box for −1.1 kb to −1 and shaded box for +1 to +147) was linked to 5′ of luciferase reporter gene (closed box). +1 indicates major transcription initiation site. (B) Promoter activity in B-cell lines with serially deletedRAG-2 promoter region. A quantity of 10 μg luciferase constructs linked to the serially deleted mouse RAG-2promoter region (indicated on left) was transfected into 18.8.1 cells or BAL17 cells. pSRα-LacZ was included as an internal control. Twenty-four hours later, luciferase and β-galactosidase assays were performed on cell extracts. The activity of the luciferase construct without promoter in each cell is set to 1. Error bars indicate deviation of 3 experiments.
c-Myb and Pax-5 bind the −41/−17 region of theRAG-2 promoter
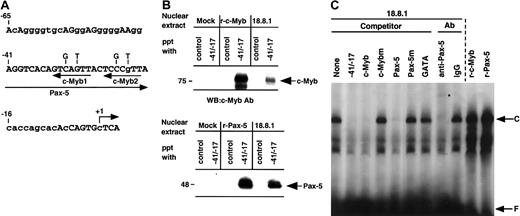
We and others have previously shown that the −41/−17 region of the RAG-2 promoter is highly conserved between humans and mice (Figure 2A and see Kishi et al18 and Lauring and Schlissel19) and that Pax-5 binds the −41/−17 region.17-19 A database search for putative binding sites of transcription factors in the murineRAG-2 promoter showed the putative c-Myb–binding elements (5′-AACKGNC-3′) at −27 (c-Myb1) and at −18 (c-Myb2). In order to examine whether c-Myb as well as Pax-5 bind the −41/−17RAG-2 promoter, the DNA-sepharose precipitation assay was performed. Nuclear extracts prepared from 18.8.1 cells or 293T transfectant which had been transfected with mock, pAct–c-Myb, or pEFBOS–Pax-5 were incubated with the sepharose beads conjugated with the −41/−17 fragment, and the binding of c-Myb or Pax-5 to the fragment was assessed by Western blot analysis using an antibody against either c-Myb or Pax-5. As a control, sepharose beads conjugated with oligonucleotide containing STAT3 binding site were used. As shown in Figure 2B, the −41/−17 fragment precipitated c-Myb and Pax-5 from nuclear extracts of 18.8.1 cells as well as from c-Myb or Pax-5 transfectant, but not from Mock transfectants. On the contrary, the control fragment did not precipitate either c-Myb or Pax-5.
c-Myb with Pax-5 binds −41/−17 region of the
RAG-2 promoter. (A) DNA sequence of the mouseRAG-2 promoter region of −65 to +4. Putative binding sites for Pax-5 and c-Myb (c-Myb1 and c-Myb2) are shown by arrows under the sequence. Nucleotides used for c-Myb binding site mutation are indicated above the sequence. Nucleotides conserved between humans and mice are shown by capital letters. +1 indicates major transcription initiation site. (B) DNA-sepharose precipitation assay. Nuclear extracts were prepared from 293T cells transfected with pAct vector (Mock) or pAct–c-Myb (r-c-Myb), or from 18.8.1 cells (top panel) or 293T cells transfected with pEF-BOS vector (Mock) or pEFBOS–Pax-5 (r-Pax-5), or 18.8.1 cells (bottom panel). Extracts were incubated with sepharose beads conjugated with either oligonucleotides containing the STAT3 binding site (control) or −41/−17. Bound proteins were eluted and resolved in SDS-PAGE and detected by immunoblotting using c-Myb or Pax-5 antibody. (C) Binding of c-Myb and Pax-5 in nuclear extracts from 18.8.1 cells to the −41/−17 region. Nuclear extracts were prepared from 18.8.1 cells or 293T cells transfected with either pAct–c-Myb (r-c-Myb) or pEFBOS–Pax-5 (r-Pax-5) and subjected to EMSA with a radiolabeled −41/−17 oligonucleotide. Oligonucleotide competition was carried out with 200-fold excess of unlabeled −42/−17 oligonucleotide, the oligonucleotide containing the consensus c-Myb binding sequence (c-Myb) or its mutant binding sequence (c-Mybm), the oligonucleotide containing the consensus Pax-5 binding sequence (Pax-5) or its mutant binding sequence (Pax-5m), or the oligonucleotide containing the consensus GATA binding sequence (GATA). For supershift assay, EMSA was performed in the presence of 0.5 μg polyclonal Pax-5 antibody or control IgG. Specific protein/DNA complex is indicated as C and free probe as F. Supershifted band is indicated by an asterisk.
c-Myb with Pax-5 binds −41/−17 region of the
RAG-2 promoter. (A) DNA sequence of the mouseRAG-2 promoter region of −65 to +4. Putative binding sites for Pax-5 and c-Myb (c-Myb1 and c-Myb2) are shown by arrows under the sequence. Nucleotides used for c-Myb binding site mutation are indicated above the sequence. Nucleotides conserved between humans and mice are shown by capital letters. +1 indicates major transcription initiation site. (B) DNA-sepharose precipitation assay. Nuclear extracts were prepared from 293T cells transfected with pAct vector (Mock) or pAct–c-Myb (r-c-Myb), or from 18.8.1 cells (top panel) or 293T cells transfected with pEF-BOS vector (Mock) or pEFBOS–Pax-5 (r-Pax-5), or 18.8.1 cells (bottom panel). Extracts were incubated with sepharose beads conjugated with either oligonucleotides containing the STAT3 binding site (control) or −41/−17. Bound proteins were eluted and resolved in SDS-PAGE and detected by immunoblotting using c-Myb or Pax-5 antibody. (C) Binding of c-Myb and Pax-5 in nuclear extracts from 18.8.1 cells to the −41/−17 region. Nuclear extracts were prepared from 18.8.1 cells or 293T cells transfected with either pAct–c-Myb (r-c-Myb) or pEFBOS–Pax-5 (r-Pax-5) and subjected to EMSA with a radiolabeled −41/−17 oligonucleotide. Oligonucleotide competition was carried out with 200-fold excess of unlabeled −42/−17 oligonucleotide, the oligonucleotide containing the consensus c-Myb binding sequence (c-Myb) or its mutant binding sequence (c-Mybm), the oligonucleotide containing the consensus Pax-5 binding sequence (Pax-5) or its mutant binding sequence (Pax-5m), or the oligonucleotide containing the consensus GATA binding sequence (GATA). For supershift assay, EMSA was performed in the presence of 0.5 μg polyclonal Pax-5 antibody or control IgG. Specific protein/DNA complex is indicated as C and free probe as F. Supershifted band is indicated by an asterisk.
In order to verify the binding of c-Myb and Pax-5 in the nuclear extracts of B-cell lines to the −41/−17 fragment, EMSA was performed using nuclear extract prepared from 18.8.1 cells (Figure 2C) or BAL17 cells. A specific complex (C) was detected in the gel and the complex formation was completely inhibited by an excess of the oligonucleotide containing consensus binding site for c-Myb or Pax-5, but not by an excess of oligonucleotides containing the mutant c-Myb, the mutant Pax-5, or unrelated GATA binding sites. The addition of Pax-5 antibody shifted the majority of this complex higher in the gel, whereas control IgG had no effect, suggesting that Pax-5 is present in this complex. We could not detect a supershift of the complex using several commercially available antibodies against c-Myb, probably because these antibodies were unsuitable for the supershift assays (data not shown). Nevertheless, the results indicate that c-Myb and Pax-5 in nuclear extracts of the B-cell line concurrently bind the −41/−17 RAG-2 promoter region. The mobility of this complex was almost similar to that of the complex formed with recombinant Pax-5 or recombinant c-Myb produced in 293T cells (Figure2C). This may be due to the similar electrostatic properties of the DNA/protein complexes formed with Pax-5 alone, c-Myb alone, and their combination. The similar observation was reported in glucocorticoid-responsive element (GRE) that the mobility of the complex of GRE and glucocorticoid receptor (GR) alone was the same as that of the tertiary complex of GRE/GR/STAT3.32
c-Myb activates the RAG-2 promoter by binding to the c-Myb1 site in the promoter region
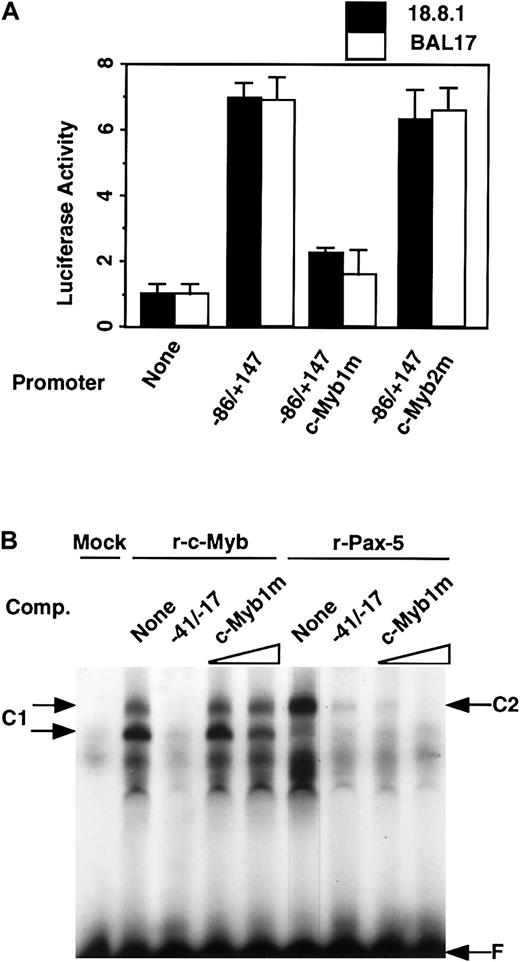
To determine which putative c-Myb elements are involved in the activation of the RAG-2 promoter in B-cell lines, luciferase construct with the wild-type −86/+147 region or its mutation at the c-Myb1 or c-Myb2 site (Figure 2A) was transfected into 18.8.1 or BAL17 cells (Figure 3A). The mutation at the c-Myb1 site markedly reduced the promoter activity of the −86/+147 fragment, although mutation of the c-Myb2 site did not affect its promoter activity. To verify whether c-Myb binds the c-Myb1 element at −27 of the murine RAG-2 promoter, a radiolabeled −41/−17 fragment was incubated with nuclear extracts prepared from 293T cells which had been transfected with pAct–c-Myb (Figure 3B). The complex formation (C1) was noted in the gel, which was inhibited by an excess of unlabeled wild-type oligonucleotides, but not mutant oligonucleotides for c-Myb1. This result indicates that c-Myb binds the c-Myb1 site in the −41/−17 region. Recombinant Pax-5 also bound the −41/−17 fragment and formed the complex (C2), which was inhibited by the unlabeled probe oligonucleotide (Figure 3B). This complex formation was inhibited by oligonucleotide containing consensus Pax-5 binding site, but not by oligonucleotide containing mutated Pax-5 binding site as previously reported.18 Importantly, the binding of recombinant Pax-5 was completely inhibited by an excess of unlabeled −41/−17 oligonucleotide containing mutant c-Myb1 site (Figure 3B), indicating that mutation of c-Myb1 site did not affect the binding of Pax-5 to the mouse RAG-2promoter.
c-Myb activates the RAG-2 promoter by binding the c-Myb1 site in the promoter region.
(A) Promoter activity of wild-type versus c-Myb binding site–mutatedRAG-2 promoter. A quantity of 10 μg luciferase constructs linked to the −86/+147 promoter fragment or that with mutation in c-Myb binding sites (c-Myb1m or c-Myb2m) together with pSRα-LacZ was transfected into 18.8.1 cells or BAL17 cells, and luciferase activity was analyzed as in Figure 1. Error bars indicate deviation of 3 experiments. (B) Effect of mutation in c-Myb1 site on binding of recombinant c-Myb and Pax-5 to the −41/−17 region. Nuclear extracts were prepared 48 hours after transient transfection of 293T cells with pAct vector (Mock), pAct–c-Myb (r-c-Myb), or pEFBOS–Pax-5 (r-Pax-5) DNA and subjected to EMSA as in Figure 2C. To verify specific binding, 50-fold excess of unlabeled −41/−17 oligonucleotide (WT), or 50-fold or 200-fold excess of a similar oligonucleotide containing mutations at the c-Myb1 site (c-Myb1m) was included in EMSA. Specific DNA/protein complex is indicated as C1 (for c-Myb) and C2 (for Pax-5), and free probe as F. Two bands are detected for c-myb (C1), probably reflecting the monomer (lower band) and dimer (upper band) formation of c-Myb protein(s) in the gel Pax-5.29
c-Myb activates the RAG-2 promoter by binding the c-Myb1 site in the promoter region.
(A) Promoter activity of wild-type versus c-Myb binding site–mutatedRAG-2 promoter. A quantity of 10 μg luciferase constructs linked to the −86/+147 promoter fragment or that with mutation in c-Myb binding sites (c-Myb1m or c-Myb2m) together with pSRα-LacZ was transfected into 18.8.1 cells or BAL17 cells, and luciferase activity was analyzed as in Figure 1. Error bars indicate deviation of 3 experiments. (B) Effect of mutation in c-Myb1 site on binding of recombinant c-Myb and Pax-5 to the −41/−17 region. Nuclear extracts were prepared 48 hours after transient transfection of 293T cells with pAct vector (Mock), pAct–c-Myb (r-c-Myb), or pEFBOS–Pax-5 (r-Pax-5) DNA and subjected to EMSA as in Figure 2C. To verify specific binding, 50-fold excess of unlabeled −41/−17 oligonucleotide (WT), or 50-fold or 200-fold excess of a similar oligonucleotide containing mutations at the c-Myb1 site (c-Myb1m) was included in EMSA. Specific DNA/protein complex is indicated as C1 (for c-Myb) and C2 (for Pax-5), and free probe as F. Two bands are detected for c-myb (C1), probably reflecting the monomer (lower band) and dimer (upper band) formation of c-Myb protein(s) in the gel Pax-5.29
c-Myb and Pax-5 cooperatively activate the RAG-2promoter
To examine the effect of c-Myb and Pax-5 on the activation of theRAG-2 promoter, 293T cells were transfected with luciferase construct with −86/+147 RAG-2 promoter or its mutants alone, or together with either pAct–c-Myb or pEFBOS–Pax-5, or both, and luciferase activity was determined (Figure4). Each Pax-5 or c-Myb maximally activated the RAG-2 promoter about 3-fold when compared with the promoter activity without Pax-5 and c-Myb, and their combination maximally activated the promoter about 9-fold (Figure 4A). This cooperation between c-Myb and Pax-5 was found by the wild-type promoter (WT) and c-Myb2 site-mutant promoter (c-Myb2m). However, the cooperation was significantly reduced when the c-Myb1 site-mutant promoter (c-Myb1m) was used (Figure 4B). These results show that c-Myb and Pax-5 cooperatively activate the RAG-2 promoter and that the c-Myb1 site in the promoter is important for the cooperation.
c-Myb and Pax-5 cooperatively activate the
RAG-2 promoter in 293T cells. (A)RAG-2 promoter activity in 293T cells with expression vector for Pax-5, c-Myb, or both. A quantity of 5 μg luciferase construct linked to the −86/+147 promoter region was transfected into 293T cells in the presence of 0.04 μg, 0.2 μg, 1 μg, or 5 μg of pEFBOS–Pax-5 alone or with 0.2 μg of pAct–c-Myb (indicated as +), or 0.008 μg, 0.04 μg, 0.2 μg, or 1 μg of pAct–c-Myb alone or together with 1 μg of pEFBOS–Pax-5 (indicated as +). pSRα-LacZ was included as an internal control. Total amount of DNA was adjusted with either pAct vector or pEF-BOS vector. Luciferase and β-galactosidase activities were assayed 48 hours later. The luciferase activity without Pax-5 and c-Myb was set to 1. Error bars indicate deviation of 3 experiments. (B) Promoter activity of wild-type versus c-Myb binding site–mutated RAG-2 promoter in the presence of Pax-5, c-Myb, or both. Luciferase construct linked to −86/+147 promoter region containing wild-type c-Myb binding site (WT) or its mutation (c-Myb1m or c-Myb2m) was transfected into 293T cells in the absence or presence of Pax-5, c-Myb, or both Pax-5 and c-Myb expression vectors, and the promoter activities were assessed as above. Error bars indicate deviation of 3 experiments.
c-Myb and Pax-5 cooperatively activate the
RAG-2 promoter in 293T cells. (A)RAG-2 promoter activity in 293T cells with expression vector for Pax-5, c-Myb, or both. A quantity of 5 μg luciferase construct linked to the −86/+147 promoter region was transfected into 293T cells in the presence of 0.04 μg, 0.2 μg, 1 μg, or 5 μg of pEFBOS–Pax-5 alone or with 0.2 μg of pAct–c-Myb (indicated as +), or 0.008 μg, 0.04 μg, 0.2 μg, or 1 μg of pAct–c-Myb alone or together with 1 μg of pEFBOS–Pax-5 (indicated as +). pSRα-LacZ was included as an internal control. Total amount of DNA was adjusted with either pAct vector or pEF-BOS vector. Luciferase and β-galactosidase activities were assayed 48 hours later. The luciferase activity without Pax-5 and c-Myb was set to 1. Error bars indicate deviation of 3 experiments. (B) Promoter activity of wild-type versus c-Myb binding site–mutated RAG-2 promoter in the presence of Pax-5, c-Myb, or both. Luciferase construct linked to −86/+147 promoter region containing wild-type c-Myb binding site (WT) or its mutation (c-Myb1m or c-Myb2m) was transfected into 293T cells in the absence or presence of Pax-5, c-Myb, or both Pax-5 and c-Myb expression vectors, and the promoter activities were assessed as above. Error bars indicate deviation of 3 experiments.
c-Myb and Pax-5 synergistically bind the RAG-2promoter
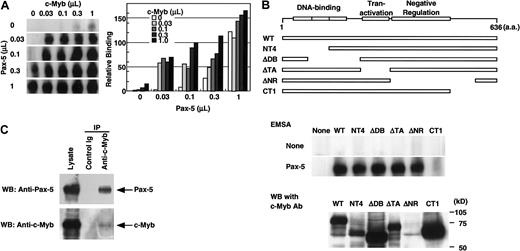
To test the possibility that c-Myb and Pax-5 synergistically bind the RAG-2 promoter, a radiolabeled −41/−17 fragment was incubated with various amounts of nuclear extracts prepared from 293T cells transfected with pAct–c-Myb or pEFBOS–Pax-5, or with the combination of these extracts (Figure5A). Although c-Myb alone or low doses of Pax-5 alone produced the faint bands of the complex, their combination markedly augmented the complex formation. This complex formation by recombinant c-Myb and Pax-5 was specifically blocked by the oligonucleotide containing either the consensus c-Myb binding site or the Pax-5 binding site but not by the oligonucleptide containing the GATA binding site (data not shown). The mobility of the complex formed with the combination of recombinant c-Myb and recombinant Pax-5 was the same as that of the complex formed with each recombinant protein (Figure 5A) as shown in Figure 3C.
c-Myb and Pax-5 synergistically bind the
RAG-2 promoter. (A) Binding of c-Myb and Pax-5 to the −41/−17 region. A radiolabeled −41/−17 oligonucleotide was incubated with various doses of nuclear extract containing either Pax-5, c-Myb, or their combinations, and subjected to EMSA as in Figure 2. Radioactivity of complexes seen in the left gel were measured by BAS2000 and are shown on the right panel. (B) Effect of various c-Myb mutants on the synergistic binding to −41/−17 region with Pax-5. Expression vectors for various c-Myb mutants shown on the top were transfected into 293T cells, and nuclear extracts were prepared 48 hours later. A quantity of 0.1 μL of the nuclear extract containing recombinant c-Myb was incubated with a radiolabeled −41/−17 oligonucleotide in the absence or presence of 0.01 μL 293T cell nuclear extract containing Pax-5, and subjected to EMSA as in Figure 5A (middle panel). Protein concentrations of the nuclear extracts containing c-Myb, c-Myb mutants, or Pax-5 corresponded to those used above. Expression of wild-type or mutant c-Myb in nuclear extracts was verified by Western blot analysis using monoclonal antibody (clone 1.1), which binds to the negative regulation domain of c-Myb (lower panel). ΔNR could not be detected by this antibody. (C) Direct binding of Pax-5 with c-Myb. Expression vectors for both c-Myb and Pax-5 were transfected into 293T cells. After 48 hours, the nuclear extract was prepared and incubated with anti–c-Myb antibody or control antibody. Immunoprecipitates were resolved in SDS-PAGE, transferred to nylon membrane, and probed with anti–Pax-5 antibody or anti–c-Myb antibody. Lysate was applied to SDS-PAGE as a positive control for Western blotting.
c-Myb and Pax-5 synergistically bind the
RAG-2 promoter. (A) Binding of c-Myb and Pax-5 to the −41/−17 region. A radiolabeled −41/−17 oligonucleotide was incubated with various doses of nuclear extract containing either Pax-5, c-Myb, or their combinations, and subjected to EMSA as in Figure 2. Radioactivity of complexes seen in the left gel were measured by BAS2000 and are shown on the right panel. (B) Effect of various c-Myb mutants on the synergistic binding to −41/−17 region with Pax-5. Expression vectors for various c-Myb mutants shown on the top were transfected into 293T cells, and nuclear extracts were prepared 48 hours later. A quantity of 0.1 μL of the nuclear extract containing recombinant c-Myb was incubated with a radiolabeled −41/−17 oligonucleotide in the absence or presence of 0.01 μL 293T cell nuclear extract containing Pax-5, and subjected to EMSA as in Figure 5A (middle panel). Protein concentrations of the nuclear extracts containing c-Myb, c-Myb mutants, or Pax-5 corresponded to those used above. Expression of wild-type or mutant c-Myb in nuclear extracts was verified by Western blot analysis using monoclonal antibody (clone 1.1), which binds to the negative regulation domain of c-Myb (lower panel). ΔNR could not be detected by this antibody. (C) Direct binding of Pax-5 with c-Myb. Expression vectors for both c-Myb and Pax-5 were transfected into 293T cells. After 48 hours, the nuclear extract was prepared and incubated with anti–c-Myb antibody or control antibody. Immunoprecipitates were resolved in SDS-PAGE, transferred to nylon membrane, and probed with anti–Pax-5 antibody or anti–c-Myb antibody. Lysate was applied to SDS-PAGE as a positive control for Western blotting.
To determine the functional domain in c-Myb necessary for the interaction with Pax-5, a radiolabeled −41/−17 fragment was incubated with nuclear extracts prepared from 293T cells transfected with expression vectors for wild-type c-Myb or its deletion mutants in the absence or presence of that of Pax-5 (Figure 5B). Deletion of the C-terminus of c-Myb (CT1) abolished the cooperation of c-Myb and Pax-5 for DNA binding (Figure 5B, middle panel), although the amount of CT1 in the nuclear extract was comparable to that in the other mutants or wild type (Figure 5B, lower panel). Deletion of the DNA-binding domain (NT4, ΔDB), the transactivation domain (ΔTA), or the negative regulation domain (ΔNR) of c-Myb did not affect the cooperative binding of c-Myb mutant and Pax-5 to the −41/−17 fragment. The result shows that the C-terminal of c-Myb is involved in the interaction with Pax-5.
To explore whether c-Myb and Pax-5 physically interact, nuclear extracts prepared from 293T cells transfected with expression vectors for both c-Myb and Pax-5 were incubated with either control antibody or anti–c-Myb antibody, then precipitated with protein G beads. Immunoprecipitates were resolved by SDS-PAGE, transferred to nylon membrane, and probed with anti–Pax-5 antibody. Anti–c-Myb antibody, but not control antibody, coprecipitated Pax-5 together with c-Myb (Figure 5C). The result demonstrates that c-Myb and Pax-5 directly bind each other.
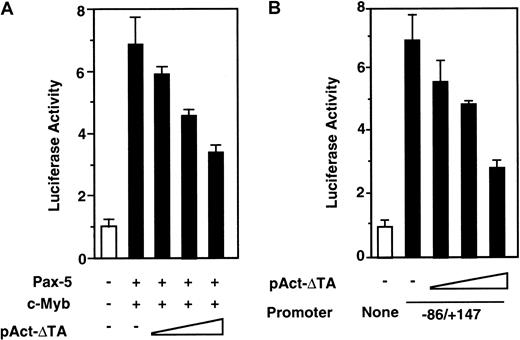
Dominant-negative c-Myb suppresses RAG-2 promoter activity
In order to examine the role of c-Myb in the activation of theRAG-2 promoter in vivo, 293T cells were transfected with luciferase construct with −86/+147 RAG-2 promoter alone, or together with pEFBOS–Pax-5 plus pAct–c-Myb and various amounts of pAct-ΔTA, which encodes transactivation domain-deleted c-Myb mutant (ΔTA) (Figure 5B). After transfection, luciferase activity was determined. As shown in Figure 6A, ΔTA suppressed c-Myb– and Pax-5–dependent RAG-2 promoter activation in a dose-dependent manner. To verify the role of c-Myb in the activation of the RAG-2 promoter in B cells, 18.8.1 cells were transfected with luciferase construct with −86/+147RAG-2 promoter alone, or together with pAct–c-Myb and various amounts of pAct-ΔTA, and the luciferase activity was assessed. ΔTA dose-dependently suppressed the RAG-2promoter activation in the B-cell line (Figure 6B). The result indicates the involvement of endogenous c-Myb in the activation of theRAG-2 promoter in a B-cell line.
Dominant-negative c-Myb suppresses
RAG-2 promoter activity. (A) Effect of dominant-negative c-Myb on RAG-2 promoter activity in 293T cells. A quantity of 5 μg luciferase construct linked to the −86/+147 promoter region was transfected into 293T cells with or without 1 μg Pax-5 expression vector and 0.2 μg c-Myb expression vector, along with various amounts (0.013 μg, 0.32 μg, 8 μg) of pAct-ΔTA. Forty-eight hours later, luciferase activities were determined as in Figure 4. Error bars indicate deviation of 3 experiments. (B) Effect of dominant-negative c-Myb on RAG-2promoter activity in 18.8.1 cells. A quantity of 10 μg luciferase construct linked to −86/+147 promoter region was transfected into 18.8.1 cells together with various amounts (0.013 μg, 0.32 μg, and 8 μg) of pAct-ΔTA. Luciferase construct without promoter (None) was transfected as a control. Forty-eight hours later, luciferase activities were assayed as in Figure 1. Error bars indicate deviation of 3 experiments.
Dominant-negative c-Myb suppresses
RAG-2 promoter activity. (A) Effect of dominant-negative c-Myb on RAG-2 promoter activity in 293T cells. A quantity of 5 μg luciferase construct linked to the −86/+147 promoter region was transfected into 293T cells with or without 1 μg Pax-5 expression vector and 0.2 μg c-Myb expression vector, along with various amounts (0.013 μg, 0.32 μg, 8 μg) of pAct-ΔTA. Forty-eight hours later, luciferase activities were determined as in Figure 4. Error bars indicate deviation of 3 experiments. (B) Effect of dominant-negative c-Myb on RAG-2promoter activity in 18.8.1 cells. A quantity of 10 μg luciferase construct linked to −86/+147 promoter region was transfected into 18.8.1 cells together with various amounts (0.013 μg, 0.32 μg, and 8 μg) of pAct-ΔTA. Luciferase construct without promoter (None) was transfected as a control. Forty-eight hours later, luciferase activities were assayed as in Figure 1. Error bars indicate deviation of 3 experiments.
Discussion
We and others have shown that the mouse RAG-2 promoter region confers lymphocyte-specificity.18,19 In B-lineage cells, it was demonstrated that Pax-5 plays an important role for activation of mouse RAG-2 promoter.18,19 In T-lineage cells, we indicated the role of GATA-3 in the activation of the promoter,18 and recently Wang et al have demonstrated the possible involvement of c-Myb in the promoter activation.20 The present study demonstrates that cooperative binding of c-Myb and Pax-5 to the RAG-2 promoter directed expression of the RAG-2 in immature B cells.
The sequences of the −41 to −16 region of mouse RAG-2 and that of human RAG-2 are almost identical and confer core promoter activity in B-cell lines (Figure 1 and Figure 2A). c-Myb and Pax-5 in the nuclear extract of the pre–B-cell line bound the −41/−17 fragment, and the formation of DNA/protein complex was completely inhibited by either oligonucleotide containing consensus c-Myb or Pax-5 binding site (Figure 2B-C). However, the formation of the complex by a recombinant Pax-5 alone was not inhibited by oligonucleotide containing consensus c-Myb binding site (data not shown). The results suggested that the binding of Pax-5 in B-cell nuclear extracts to the −41/−17 fragment is dependent on c-Myb in the extracts. This notion was confirmed by the following findings: (1) ectopic expression of c-Myb and Pax-5 cooperatively activated the promoter in 293T cells (Figure 4), and (2) recombinant c-Myb and Pax-5 synergistically bound the −41/−17 sequences (Figure 5). These results indicate that c-Myb and Pax-5 cooperatively bind and activate theRAG-2 core promoter in B-lineage cells. As to the −65/−41 of RAG-2 5′ upstream region, the promoter activity became about half when the −65/−42 region was deleted (Figure 1). The −65/−42 region was not conserved between mice and humans as previously shown.18,19 When we compared this region between mice and humans in more detail, guanine in −53 to −42 in the mouse RAG2 promoter region was substituted to adenine in the human RAG2 promoter (Figure 2A and Kishi et al18 and Lauring and Schlissel19). This indicates that some transcription factor(s) might bind this site, although it is only speculation.
It was demonstrated that c-Myb was expressed in the cortex of thymus, comprised of immature T-cells, but not in the medulla.33Expression of the dominant-negative c-Myb mutant in the T-lineage in mice partially blocked T-cell differentiation in thymus,34and in homozygous null c-Myb/RAG1 chimeric mice, T-cell development was blocked at the CD4−CD8−stage.35 The results indicate the inevitable role of c-Myb in T-cell development. In accordance with these results, Wang et al indicated that c-Myb is essential for RAG-2 promoter activity in T-lineage cells.20 c-Myb expression during B-cell development remains controversial. Bender and Kuehl have demonstrated that c-Myb is expressed in pre–B-cell lines at high levels but its expression is significantly low or undetectable in mature B-cell lines or plasmacytoma cell lines.36Recently, Akashi et al have reported that c-Myb is not expressed in freshly isolated pro-B or pre-B populations.37 However, Allen et al have shown that in homozygous null c-Myb/RAG1chimeric mice, B-cell development is blocked at the pre–B-cell stage in the bone marrow.35 The result verifies an important role of c-Myb in early B-cell development and is consistent with our present results.
Regarding the expression of Pax-5, it initiated from the early B-lineage precursor cells and persisted to mature B-cells. It was down-regulated in terminally differentiated plasma cells.38,39 In Pax-5 mutant mice, B-cell development was completely arrested at the pro-B stage.40 Precursor cells in the bone marrow of Pax-5 mutant mice could give rise to pre-B cells, in which the DH-to-JH rearrangement occurred at normal frequency, but the frequency of the VH-to-DJH rearrangement reduced about 50-fold,41 showing that Pax-5 is dispensable for DH-to-JH recombination. Concerning the role of Pax-5 for RAG expression, Lauring and Schlissel19 and Kishi et al18 have demonstrated that Pax-5 plays an important role for activation of mouseRAG-2 promoter in B-lineage cells. In this study, we have demonstrated that not only Pax-5 but also c-Myb is involved in the promoter activation in B-cell lines. It is conceivable that the activation of the mouse RAG-2 promoter is regulated with cooperation of multiple transcription factors including c-Myb and Pax-5. It is worthy to note that in the 5′ upstream region of the mouse and human RAG-2 promoter, up to 300 bp from the major transcription initiation site are conserved.17-19 These promoter sequences must have been conserved because of their inevitable roles in RAG-2 expression, although the in vitro luciferase system so far failed to demonstrate its essential roles (Figure 1). With this regard, some regulatory regions such as the local control region (LCR)42 may exist which control chromatin remodeling as well as transcriptional activity in vivo, but show very low transactivation activity in vitro as employed in this study.
In the present study, we showed that c-Myb and Pax-5 cooperatively bind to synergistically activate the mouse RAG-2 core promoter in B-cell lines (Figure 2-Figure 5). Concerning the interaction of c-Myb and Pax-5, we observed the synergistic activation, albeit reduced, of the RAG-2 promoter with the mutated c-Myb binding site (c-Myb1 site) by recombinant Pax-5 and c-Myb (Figure 4B). We also found that the c-Myb mutant without its DNA-binding domain (NT4 and ΔDB) could augment the binding of Pax-5 to the promoter (Figure 5B). Finally, we demonstrated that c-Myb and Pax-5 were coprecipitated by an antibody to c-Myb in the absence of target DNA (Figure 5C). These observations strongly suggest the direct protein-protein interaction between c-Myb and Pax-5. With respect to the interaction of Pax-5 and c-Myb, we demonstrated that interaction between c-Myb and Pax-5 required the C-terminal region of c-Myb (Figure 5B). Although the DNA-binding domain and transactivation domain of c-Myb have been reported to be necessary for the interaction of c-Myb with other transcription factors,24 the role of the C-terminal region has not been described. Thus, this study demonstrates for the first time the functional role of the C-terminal of c-Myb.
Regarding the transcriptional role of c-Myb functional domains (Figure5),24,29 it has been demonstrated that the transactivation domain of c-Myb directly or by binding to a coactivator, such as CBP or p300,24 activates transcription. In the present study, we showed that the c-Myb mutant of the transactivation domain (ΔTA) suppressed RAG-2 promoter activation in B-cell lines (Figure6). The result indicates that ΔTA functioned as a dominant-negative c-Myb mutant and implies that the transactivation domain of c-Myb may play a role in activation of the RAG-2 promoter in B cells. Alternative interpretation of the result would be that ΔTA bound the promoter and inhibited the binding of Pax-5 to the promoter. This is unlikely, because the ΔTA mutant did not block but rather augmented the binding of Pax-5 to the promoter (Figure 5B). The DNA-binding domain of c-Myb also plays a role in the promoter activation, because mutation of c-Myb binding sites in the promoter reduced the promoter activity in B cells (Figure 3A). However, as shown in Figure 5B, the c-Myb deletion mutant of the DNA-binding domain, NT4, could also bind the promoter by cooperating with Pax-5, indicating that the DNA-binding domain may play a role other than DNA-binding in RAG-2promoter activation. In this context, it is noteworthy that NT4 inhibited the RAG-2 promoter activation in B cells (data not shown). The DNA-binding domain of c-Myb not only binds DNA but also interacts with other transcription factors or coactivators, such as C/EBPβ or p100.24 Thus, the DNA-binding domain of c-Myb may play a role not only in DNA-binding but also in interacting with other transcription factors to activate the RAG-2 promoter. Taken together, the hematopoietic but not lineage-specific transcription factor, c-Myb, cooperates with the B-cell–specific transcription factor, Pax-5,38 39 and activates theRAG-2 transcription in B-lineage cells. Further study is necessary to clarify whether other transcription factors are involved in RAG-2 promoter activation in B-lineage cells.
We thank S. Ishii for c-Myb and its mutant expression vectors.
Supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science, Tokyo, Japan.
H.K. and Z.-X.J. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Atsushi Muraguchi, Department of Immunology, Faculty of Medicine, Toyama Medical and Pharmaceutical University, 2630, Sugitani, Toyama, 930-0194 Japan; e-mail:gucci@ms.toyama-mpu.ac.jp.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal