Constitutive activation of the FLT3 receptor tyrosine kinase, either by internal tandem duplication (ITD) of the juxtamembrane region or by point mutations in the second tyrosine kinase domain (TKD), has been described in patients with acute myelogenous leukemia (AML). We analyzed the prevalence and the potential prognostic impact of FLT3 mutations in 979 AML patients. Results were correlated with cytogenetic data and the clinical response. FLT3-ITD mutations were found in 20.4% and FLT3-TKD mutations in 7.7% of the patients. Each mutation was associated with similar clinical characteristics and was more prevalent in patients with normal karyotype. Significantly more FLT3 aberrations were found in patients with FAB M5, and fewer were found in patients with FAB M2 and M6. Although less frequent in patients with cytogenetic aberrations, FLT3-ITDs were found in 13 of 42 patients with t(15;17) and in 9 of 10 patients with t(6;9). The prevalence of the ITD allele on the DNA level was heterogeneous, ranging from faint mutant bands in some patients to predominant mutant bands in others. Based on quantitative analysis, the mutant–wild-type (wt) ratio ranged from 0.03 to 32.56 (median, 0.78). Patients with a high mutant/wt ratio (ie, greater than 0.78) had significantly shorter overall and disease-free survival, whereas survival in patients with ratios below 0.78 did not differ from those without FLT3 aberrations. Multivariate analysis confirmed a high mutant/wt ratio to be a strong independent prognostic factor. Taken together, these data confirm that FLT mutations represent a common alteration in adult AML. Constitutive activation may be associated with monocytoid differentiation. A high mutant/wt ratio in ITD-positive patients appears to have a major impact on the prognostic relevance.
Introduction
Acute myelogenous leukemia (AML) describes a group of hematopoietic stem cell malignancies, affecting approximately 2 to 3 adults per 100 000 each year in Western countries. Cytogenetic aberrations represent one of the most important independent prognostic factors. Several studies have shown that good-risk cytogenetics, such as t(8;21), are associated with significantly better survival than poor-risk chromosomal aberrations, such as monosomy 7 or monosomy 5 (reviewed in Lowenberg et al1). Novel treatment strategies take into account these prognostic factors to develop risk-stratified treatment options. The ultimate goal is to offer more intensive treatment options—autologous or allogeneic blood stem cell transplantation—to patients at high risk, thus sparing patients with favorable prognosis from toxicity. However, 40% to 50% of the patients have a normal karyotype according to standard cytogenetics. This patient group has intermediate disease-free survival (DFS) and overall survival (OS),2 but individual courses may differ substantially. Little is known about the genetic factors responsible for the development of the disease in these patients. Identification of prognostic factors suitable to further subdivide this group may help to optimize their treatment.
The growth and differentiation of hematopoietic cells is governed by the concerted action of growth factors and their receptors. One of these receptors, FMS-like tyrosine kinase 3 (FLT3),3 also called stem cell kinase 1 (STK1)4 or fetal liver kinase 2 (flk2),5 belongs to the group of class 3 receptor tyrosine kinases, together with other growth factor receptors such as c-Kit, PDGF-R, and c-fms (for review, see Shurin et al6). The FLT3 protein is expressed on early hematopoietic and lymphoid progenitors4,7 and seems to play an important role in early stem cell survival and myeloid differentiation.6 The protein is highly expressed in most patients with AML and in up to 50% of leukemic blasts in patients with acute lymphoblastic leukemia (ALL).7-11
Recently, internal tandem duplication (ITD) mutations of theFLT3 gene have been described in approximately 20% of patients with adult AML12-16; a lower incidence (5%-16.5%) has been reported in childhood leukemia.17-19These mutations cluster in exons 11 and 12 of the human FLT3gene on chromosome 13q12, a part that codes for the juxtamembrane domain of the FLT3 protein. Mutations typically consist of an in-frame inserted sequence corresponding to the region between amino acids (AA) 575 and 613 of the FLT3 protein.20 This alteration induces constitutive activation of the protein20 and leads to activation of downstream signal molecules, including STAT5, Ras, and MAP kinase, when transfected into 32D or BA/F3 cells.21,22In AML patients, the FLT3-ITD mutations were found to be associated with increased leukocyte counts and were frequent in patients lacking other cytogenetic aberrations. The presence of FLT3 aberrations appears to be associated with an unfavorable clinical response.14-16,23 However, most studies published so far are limited in patient number, and patients were not treated according to a single protocol, which renders an assessment of the potential prognostic impact of this mutation difficult. More recently, point mutations in codon 835 of the FLT3 gene have been described in approximately 7% of patients with AML.24 These mutations are located in the activation loop of the second tyrosine kinase domain (TKD) of FLT3 and constitutively activate the protein.24 The prognostic relevance of this alteration is not yet defined. The aims of the present study were to characterize the incidence of FLT3-ITDs and FLT3-TKD mutations in a large (n = 979), unselected, and well-characterized cohort of patients with AML and to study the potential prognostic impact of these alterations in patients treated on a single protocol. Finally, we addressed the origin and the prognostic relevance of differences in the prevalence of mutant and wild-type (wt) FLT3-ITD alleles.
Patients, materials, and methods
Patients
Nine hundred seventy-nine patients with AML (818 with de novo AML, 123 with AML and history of myelodysplastic syndrome [MDS], and 38 with therapy-related AML) and 34 patients with RAEB-t were examined for the presence of FLT3 mutations. Most (n = 816) of these patients were treated according to the AML-96 multicenter protocol of the German Süddeutsche Hämoblastose Group (SHG). A list of the participating study centers is given in the . A detailed description of this study has been published previously.25In this protocol, postinduction therapy was stratified according to cytogenetic risk groups for patients 60 years of age and younger. These risk groups were defined according to results of standard cytogenetic analysis as follows: high risk, −5/del(5q),−7/del(7q), hypodiploid karyotypes (besides 45, X,−Y, or −X), inv(3q), abn 12p, abn 11q, +11, +13, +21, +22, t(6;9); t(9;22); t(9;11); t(3;3), multiple aberrations (= 3 independent aberrations); intermediate risk, patients without low-risk or high-risk constellation;low risk, t(8;21) and t(8;21) combined with other aberrations. Patients with AML FAB-M3 were included in the European APL-93 trial when possible (Fenaux et al26).
In patients younger than 60, first induction therapy consisted of triple therapy with 10 mg/m2 mitoxantrone (days 4-8), 100 mg/m2 cytosine arabinoside (ara-C) (days 1-8), and 100 mg/m2 VP16 (days 4-8) (MAV). Second induction consisted of 2 × 1000 mg/m2 ara-C (days 1-5) and 100 mg/m2 m-AMSA (days 1-5) (MAMAC). Patients at intermediate cytogenetic risk were referred for allogeneic hematopoietic stem cell transplantation (HSCT) from an HLA-identical sibling donor. Patients at intermediate cytogenetic risk without a sibling donor and patients at low risk were randomized to receive intermediate dose ara-C (2 × 1000 mg/m2 every 12 hours days 1-6) (I-MAC) or high-dose ara-C (2 × 3000 mg/m2 every 12 hours days 1-6) (H-MAC) plus mitoxantrone (10 mg/m2 days 4-6), which was followed by autologous peripheral blood SCT (intermediate cytogenetic risk) or MAMAC (low cytogenetic risk). Patients at high cytogenetic risk were referred for allogeneic HSCT, including the option of unrelated HSCT. Patients without a donor were treated with either I-MAC or H-MAC and were referred for autologous peripheral blood SCT.
Patients older than 60 received 2 induction cycles containing 45 mg/m2 daunorubicin (days 3-5) and 100 mg/m2ara-C (days 1-7) (DA). Postremission therapy consisted of MAMAC.
Complete remission (CR) was defined as the presence of less than 5% blasts cells in a standardized bone marrow aspirate after the second course of induction. Only patients with a fully regenerated peripheral blood count were considered to be in CR.
This study was approved by the ethics board of the Technical University Dresden. Each patient gave written informed consent to participate in the study.
Patient samples
All samples investigated in this study were obtained at the time of diagnosis. Bone marrow samples were used whenever available. In all other cases, peripheral blood samples were examined. All samples were enriched for the blast fraction using density-gradient centrifugation (density, 1.077; BIOCOLL separating solution; Biochrom, Berlin, Germany). Genomic DNA was extracted from 5 × 106 cells using either phenol–chloroform extraction after proteinase K digestion27 or a silica-based procedure (Qiagen DNA Blood Kit; Qiagen, Hilden, Germany) according to the manufacturer's protocols. Total RNA was prepared using acid guanidinium thiocyanate–phenol–chloroform extraction28 (RNAzol B; Biozol, Munich, Germany).
Polymerase chain reaction for exons 11 and 12
Polymerase chain reaction (PCR) was performed on genomic DNA using published primer molecules.14 In brief, 1 μL DNA was amplified in a volume of 50 μL containing 50 mM KCl, 10 mM Tris-HCl, pH 8.3, 1.5 mM MgCl2, 0.001% (wt/vol) gelatin, 200 μM dNTPs, oligonucleotides (FLT3-11F and FLT3-12R; 0.5 μM each), and 1 U Taq DNA-polymerase (Perkin-Elmer, Norwalk, CT). The PCR consisted of an initial incubation step at 94°C for 150 seconds followed by 35 cycles at 94°C for 30 seconds, 57°C for 60 seconds, and 72°C for 120 seconds, and a final elongation step at 94°C for 30 seconds and 60°C for 10 minutes. PCR products were analyzed on standard 3% agarose gels. Reverse transcriptase (RT)–PCR was performed for those 38 samples for which no DNA was available. The RT reaction was performed as outlined recently.29 In brief, 1 μg total RNA was reverse transcribed using 200 U M-MLV RT (Superscript; Gibco BRL, Karlsruhe, Germany), in the presence of 40 U RNAse-inhibitor (RNAsin; Promega, Mannheim, Germany) in a total volume of 20 μL. One microliter of the RT reaction was used for PCR.
PCR for exon 17 and RFLP analysis
PCR for the detection of mutations in codon 835/836 was performed using published primer molecules.24 The conditions were identical to those mentioned above; however, hot start was performed using AmpliTaq Gold DNA polymerase (Perkin-Elmer). Five microliters PCR product was mixed with 5 μL restriction mix containing 5 U EcoRV (New England Biolabs, Frankfurt, Germany) in 1× Buffer 3 and was digested for 1 hour at 37°C. Agarose gel electrophoresis was performed as described above.
Genescan analysis of the mutant to wild-type FLT3-ITD ratio
For Genescan analysis, PCR primer FLT3 11F14 was labeled with 6-FAM (TIB MOLBIOL, Berlin, Germany). PCR setup was identical to the standard PCR; however, AmpliTaq Gold DNA-polymerase (1 U; Perkin-Elmer) was used. PCR conditions were modified as follows: preincubation at 95°C for 11 minutes, followed by 30 seconds at 94°C, 30 seconds at 57°C, and 60 seconds at 72°C. The total number of cycles was 27. Quantitative addition of +A overhangs of all PCR molecules30 was achieved using a final elongation step with 60°C for 45 minutes. The DNA concentration was determined and adjusted to 5 ng. Precautions taken to achieve reproducible results and the conditions of the Genescan analysis have been described recently.31 Several parameters were tested for their influence on the quantitative results. Among these, the DNA concentration was particularly important (see above). Experiments using dilutions of cloned ITD-fragments with different sizes (18-111 bp) showed no effect of the size of the ITD on the quantitative result (data not shown). We analyzed DNA from the peripheral blood of 50 healthy persons using this technique and did not find any evidence for the presence of additional FLT3 signals.
PCR assay for the detection of allelic loss at 13q12
The principal basis of this assay has been described.32,33 The relative amplification of the FLT3 exon 11/12 region was compared to the genomic amplification of the human growth hormone gene (HGH), located on chromosome 17q22-24.34 Primers for HGH were as described recently35; primer HGH-s was labeled with HEX. PCR conditions were identical to those described for the analysis of the mutant/wt ratio (see above). DNA from 22 healthy persons and 12 AML patients with an FLT3 mutant/wt allelic ratio greater than 2 was analyzed. To assess the ability of this assay to measure an increase in FLT3 genomic equivalents, BAC (1-3 pg) DNA (see below) was added to a DNA sample of a healthy person, showing a linear increase with the added amount of BAC 85P08.
Sequence analysis
For sequence analysis of the FLT3-ITD mutations, DNA showing additional bands was amplified in a second reaction. PCR products were separated on 3% agarose gels, and the mutant bands were isolated and cloned into pCR 2.1 TOPO vectors (Invitrogen, Leek, The Netherlands). After the preparation of plasmid DNA, samples were sequenced using Big Dye Terminator cycle sequencing chemistry (ABI). Sequences were compared to the wild-type sequence (accession no. E970630). For analysis of the FLT3-TKD mutations, digested PCR products were separated electrophoretically, and the undigested bands were isolated as described above. The resultant DNA was used for direct cycle sequencing. If this approach did not yield sufficient data,EcoRV digestion was performed, and the undigested fragments were cloned and sequenced as described above.
Fluorescence in situ hybridization
Fifty different bacterial artificial chromosome (BAC) clones (RPCI-11 human male BAC library; BACPAC Resources, Oakland, CA) containing DNA from the chromosomal region 13q12 were selected and screened for the presence of the FLT3 gene by PCR. BAC-DNA was isolated from clone 85P08 (Qiagen Large-Construct Kit; Qiagen) and was labeled by nick translation (Vysis, Downers Grove, IL) with fluorophore-labeled dUTP (SpectrumRed; Vysis). For each fluorescence in situ hybridization (FISH) experiment, 200 ng probe mixed with 1 μg COT-1 DNA (Boehringer Mannheim, Germany) and 2 μg herring sperm DNA (Boehringer Mannheim) were applied per target area. FISH was performed as outlined recently.36 A second probe (LSI 13, 13q14, SpectrumGreen, Vysis) was included in the analysis.
Statistical analysis
Using the Kaplan-Meier method, OS and DFS were calculated only for those patients who had been included in the SHG AML-96 study.37 Cox regression analysis was performed to calculate the relative risk of patients with high FLT3 ratios. Comparisons between different groups were made using 2-sided Fisher exact test (dichotomic variables) or the nonparametric Mann-WhitneyU test (continuous variables). P < .05 was considered significant. For multivariate analysis of prognostic factors, a proportional hazard regression model was used. Stepwise forward selection was performed. Variables were added atP < .01 and were deleted at P > .05. All calculations were performed using the SPSS software package, version 4.1 (SPSS, Chicago, IL).
Results
We have analyzed samples from 979 patients with newly diagnosed AML and 34 patients with transformed MDS (RAEB-t) for the presence of ITDs and for mutations in the second tyrosine kinase domain (TKD; codons 835/836) of the FLT3 gene. An ITD was found in 200 of 979 (20.4%) AML patients and in 3 of 34 (8.8%) patients with RAEB-t. A mutation in the TKD was present in 75 of 979 (7.7%) patients with AML and in 2 of 34 (5.9%) patients with MDS. Seventeen AML patients had an FLT3-ITD and a mutation in the TKD. Thus, 258 of 979 (26.4%) AML patients had activation of FLT3, and 5 of 34 (14.7%) with transformed MDS.
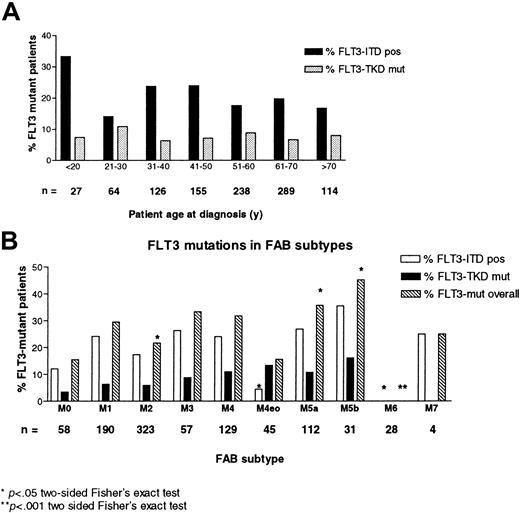
The presence of any type of FLT3 mutation was significantly associated with absolute leukocyte counts and bone marrow blast counts (Table1). No association was seen between patient age at diagnosis and the prevalence of FLT3 mutations (Figure1A). Compared with the overall incidence, the frequency of FLT3 mutations with respect to French-American-British (FAB) subtype was significantly lower in patients with M2 and M6 morphologies and was increased in patients with M5a and M5b (Figure1B).
Clinical features and FLT3 mutations in AML and RAEB-t patients at diagnosis
| . | ITD neg + TKD wt (median, range) . | ITD pos + TKD wt (median, range) . | ITD neg + TKD mut (median, range) . | ITD pos + TKD mut (median, range) . |
|---|---|---|---|---|
| Absolute leukocyte counts (× 109/L) | 10.9 (0.32-380.0) | 52.0* (0.4-465.9) | 40.1* (0.9-320.0) | 50.7† (3.6-183.0) |
| N = | 704 | 177 | 54 | 16 |
| Bone marrow blasts (%) | 57.0 (3.5-99.0)1-153 | 73.5* (20-99)1-153 | 68.0† (19.5-96.5)1-153 | 71.3 (42.0-83.0)1-153 |
| N = | 687 | 164 | 51 | 14 |
| CD34+ cells (%) | 30 (0-98) | 20‡ (0-96) | 22 (0-91) | 10 (3-73) |
| N = | 636 | 167 | 49 | 15 |
| . | ITD neg + TKD wt (median, range) . | ITD pos + TKD wt (median, range) . | ITD neg + TKD mut (median, range) . | ITD pos + TKD mut (median, range) . |
|---|---|---|---|---|
| Absolute leukocyte counts (× 109/L) | 10.9 (0.32-380.0) | 52.0* (0.4-465.9) | 40.1* (0.9-320.0) | 50.7† (3.6-183.0) |
| N = | 704 | 177 | 54 | 16 |
| Bone marrow blasts (%) | 57.0 (3.5-99.0)1-153 | 73.5* (20-99)1-153 | 68.0† (19.5-96.5)1-153 | 71.3 (42.0-83.0)1-153 |
| N = | 687 | 164 | 51 | 14 |
| CD34+ cells (%) | 30 (0-98) | 20‡ (0-96) | 22 (0-91) | 10 (3-73) |
| N = | 636 | 167 | 49 | 15 |
P < .001;
P < .01;
P < .05.
Some patients had FAB M6.
Prevalence of FLT3 mutations in different age groups and FAB-subtypes.
(A) Percentage of patients with FLT3 mutations according to age cohorts. The number below each column gives the total number of patients in the respective cohort. (B) Prevalence of FLT3-ITDs in different FAB subtypes, given as a percentage of the total number of patients (mentioned above each column).
Prevalence of FLT3 mutations in different age groups and FAB-subtypes.
(A) Percentage of patients with FLT3 mutations according to age cohorts. The number below each column gives the total number of patients in the respective cohort. (B) Prevalence of FLT3-ITDs in different FAB subtypes, given as a percentage of the total number of patients (mentioned above each column).
Correlation of cytogenetic data with the FLT3-ITD and TKD mutations is shown in Table 2. Most FLT3-ITD mutations were observed in patients with normal karyotype as assessed by standard cytogenetics (134 of 415 with 46,XX or 46,XY compared to 44 of 445 with cytogenetic aberrations; P < .0001). Likewise, FLT3-TKD mutations were also significantly more frequent in patients with normal karyotype (50 of 415) than in those with cytogenetic aberrations (23 of 445) (P = .0003). A significantly higher prevalence of FLT3-ITD mutations was found in patients with t(15;17) (31%;P < .0001), and in patients with TKD mutations there was a trend for a significantly higher prevalence (19%,P = .056). The highest proportion of FLT3-ITDs was observed in patients carrying the t(6;9)/DEK-CAN fusion—9 of 10 patients were positive for the alteration (P < .0001).
Comparison of cytogenetic aberrations and FLT3 mutations in patients with AML (n = 979)
| Karyotype characteristics . | ITD neg/TKD wt N (%) . | ITD pos N (%) . | TKD mut N (%) . | ITD pos + TKD mut N (%) . |
|---|---|---|---|---|
| All patients | 721 (73.6) | 183 (18.7) | 58 (5.9) | 17 (1.7) |
| Karyotype not available | 59 (8.2) | 22 (12.0) | 2 (3.4) | 0 (0) |
| Normal (XX,XY) | 282 (39.1) | 119 (65.0)‡ | 35 (60.3)2-153 | 15 (88.2)‡ |
| Aberrant | 380 (52.7) | 42 (23.0) | 21 (36.2) | 2 (11.8) |
| Individual aberrations | ||||
| t(8;21) | 38 (5.3) | 2 (1.1) | 1 (1.7) | 0 (0) |
| t(15;17) | 26 (3.6) | 13 (7.1)‡ | 4 (6.9) | 0 (0) |
| inv(16);t(16;16) | 36 (5) | 1 (0.6) | 5 (8.6) | 1 (4.8) |
| t(6;9)* | 1 (0.1) | 9 (4.9)‡ | 0 (0) | 0 (0) |
| t(3;3), inv(3q) | 10 (1.4) | 1 (0.6) | 0 (0) | 0 (0) |
| +8 | 69 (9.6) | 6 (3.3) | 6 (10.3) | 0 (0) |
| t(9;11);t(9;22) | 14 (1.9) | 0 (0) | 0 (0) | 0 (0) |
| +11/+13/+21/+22 | 75 (10.4) | 1 (0.6)2-153 | 4 (6.9) | 1 (4.8) |
| −5/5q− | 75 (10.4) | 2 (1.1)2-155 | 0 (0)2-155 | 0 (0) |
| −7/7q− | 86 (11.9) | 0 (0)‡ | 5 (8.6) | 0 (0) |
| Other monosomies | 94 (13.3) | 3 (1.6)2-155 | 1 (1.7)2-155 | 0 (0) |
| Multiple aberrations | 132 (18.3) | 3 (1.6)‡ | 5 (8.6) | 1 (4.8) |
| Karyotype characteristics . | ITD neg/TKD wt N (%) . | ITD pos N (%) . | TKD mut N (%) . | ITD pos + TKD mut N (%) . |
|---|---|---|---|---|
| All patients | 721 (73.6) | 183 (18.7) | 58 (5.9) | 17 (1.7) |
| Karyotype not available | 59 (8.2) | 22 (12.0) | 2 (3.4) | 0 (0) |
| Normal (XX,XY) | 282 (39.1) | 119 (65.0)‡ | 35 (60.3)2-153 | 15 (88.2)‡ |
| Aberrant | 380 (52.7) | 42 (23.0) | 21 (36.2) | 2 (11.8) |
| Individual aberrations | ||||
| t(8;21) | 38 (5.3) | 2 (1.1) | 1 (1.7) | 0 (0) |
| t(15;17) | 26 (3.6) | 13 (7.1)‡ | 4 (6.9) | 0 (0) |
| inv(16);t(16;16) | 36 (5) | 1 (0.6) | 5 (8.6) | 1 (4.8) |
| t(6;9)* | 1 (0.1) | 9 (4.9)‡ | 0 (0) | 0 (0) |
| t(3;3), inv(3q) | 10 (1.4) | 1 (0.6) | 0 (0) | 0 (0) |
| +8 | 69 (9.6) | 6 (3.3) | 6 (10.3) | 0 (0) |
| t(9;11);t(9;22) | 14 (1.9) | 0 (0) | 0 (0) | 0 (0) |
| +11/+13/+21/+22 | 75 (10.4) | 1 (0.6)2-153 | 4 (6.9) | 1 (4.8) |
| −5/5q− | 75 (10.4) | 2 (1.1)2-155 | 0 (0)2-155 | 0 (0) |
| −7/7q− | 86 (11.9) | 0 (0)‡ | 5 (8.6) | 0 (0) |
| Other monosomies | 94 (13.3) | 3 (1.6)2-155 | 1 (1.7)2-155 | 0 (0) |
| Multiple aberrations | 132 (18.3) | 3 (1.6)‡ | 5 (8.6) | 1 (4.8) |
Note that the number of patients with individual aberrations adds to a higher number than the absolute number of patients with aberrations because several patients had more than one aberration.
P values were calculated using Fisher exact test. For individual cytogenetic aberrations, the groups were compared with other patients with abnormal karyotypes.
Patients had t(6;9) as the sole abnormality (n = 9); one patient with a FLT3-ITD had t(13;13)(q11;q11).
Three or more structural or numerical chromosomal aberrations.
P < .001;
P< .01;
P < .05.
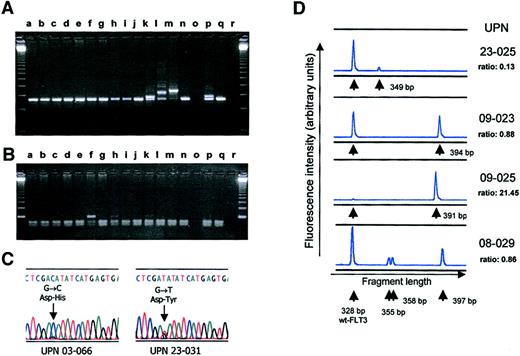
DNA sequence analysis of FLT3-ITDs was performed in 97 patients, in whom 101 sequences were identified. In 93 patients a single sequence was found, and 4 patients displayed 2 different mutant FLT3-sequences. All fragments contained in-frame rearrangements; the median length of the inserted sequence was 54 bases (range, 6-180 bases). The ITD sequences included sequences from AA573 to AA620 of the FLT3 protein, thus the vast majority of mutations contained sequences from exon 11. A major cluster was seen for the stretch between codons 591 and 601, which was present in 60% of the ITDs. In 3 patients, sequences derived from intron 11 were identified in the inserted segment. Additional bases not matching FLT3 sequences were found in 45 of 101 FLT3-ITDs (45%), and in 2 patients the lengthened FLT3-PCR products contained 18 and 24 bases not related to FLT3 sequences.
Examples of the FLT3 exon 17 mutations are shown in Figure 2C. Confirmation of mutations by sequence analysis was performed in all 77 patients who had been identified by RFLP-screening. Eighty-seven mutations were found; 8 samples contained 2 different mutations, and 1 sample contained 3 different mutations (Table3). Transversion mutations of the first position of codon 835, either G-T or G-C, leading to amino acid exchanges from aspartic acid to tyrosine or histidine respectively, were the most common alterations, accounting for 52 of 87 mutations (60%). In contrast to these single-base exchanges, in 13 patient samples a complete deletion of codon 836 was identified. Two patient samples contained insertion mutations: one 3 bases, coding for arginine, the other sample contained a complex change with the amino acids leucine and lysine inserted after codon 835 in combination with a point mutation, changing isoleucine to serine in codon 836. None of the changes found altered the reading frame. In 10 of the 17 patients with an FLT3-ITD mutation and an FLT3-TKD mutation, analysis could be performed on the distribution of both mutations between the 2 copies of the FLT3 gene using PCR for exons 11 to 17 on cDNA material. Six of these patients showed a TKD mutation of the wt allele, whereas in 4 patients the analysis indicated the presence of the TKD mutation on the allele carrying the ITD mutation (data not shown).
PCR analysis of FLT3-ITDs and TKD mutations.
(A) Agarose gel electrophoresis of PCR with (lanes k, l, m, p) and without FLT3-ITDs. Lane q contains DNA of a normal donor, and lane r contains the water control. (B) The same set of samples analyzed for the FLT3 TKD mutation. Lanes f, h, and k showed an undigested band, arguing for a mutation. All 3 mutations were confirmed by sequencing. (C) Results of direct sequencing of PCR products obtained from patient UPN 03-066, corresponding to lane f in panel B, and UPN 23-031, corresponding to lane h in panel B. Note the residual wt FLT3 sequence visible in both samples. (D) Genescan analysis of FLT3-ITDs using denaturing PAA-gel electrophoresis and fluorescence detection. The ratio given for each sample denotes the relative proportion of the AUC of mutant and wt FLT3 alleles (ie, AUC FLT3-ITD/AUC FLT3-wt). Sample UPN 08-029 shows 3 different FLT3-ITDs of 355, 358, and 397 bp.
PCR analysis of FLT3-ITDs and TKD mutations.
(A) Agarose gel electrophoresis of PCR with (lanes k, l, m, p) and without FLT3-ITDs. Lane q contains DNA of a normal donor, and lane r contains the water control. (B) The same set of samples analyzed for the FLT3 TKD mutation. Lanes f, h, and k showed an undigested band, arguing for a mutation. All 3 mutations were confirmed by sequencing. (C) Results of direct sequencing of PCR products obtained from patient UPN 03-066, corresponding to lane f in panel B, and UPN 23-031, corresponding to lane h in panel B. Note the residual wt FLT3 sequence visible in both samples. (D) Genescan analysis of FLT3-ITDs using denaturing PAA-gel electrophoresis and fluorescence detection. The ratio given for each sample denotes the relative proportion of the AUC of mutant and wt FLT3 alleles (ie, AUC FLT3-ITD/AUC FLT3-wt). Sample UPN 08-029 shows 3 different FLT3-ITDs of 355, 358, and 397 bp.
Types of FLT3 mutations found in codons 835 and 836 of the second TKD
| Mutation . | N . | Amino acid change . | Remarks . |
|---|---|---|---|
| 835 GAT → TAT | 34 | Asp → Tyr | — |
| 835 GAT → CAT | 18 | Asp → His | — |
| 835 GAT → GTT | 5 | Asp → Val | — |
| 835 GAT → GAG | 8 | Asp → Glu | — |
| 835 GAT → GAG | 2 | Asp → Asn | Combined with other mutations |
| 835 GAT → GGT | 4 | Asp → Gly | — |
| 835 GAT → multiple | 1 | Asp → ins. Leucin-Lysin | Combined with 836 isoleucin-serin |
| — | |||
| 836 ATC → Del | 13 | Ile → Del | — |
| 836 ATC → ATGCGC | 1 | Ile → MetArg | Insertion of arginin |
| 836 ATC → ACC | 1 | Ile → Thr | — |
| Mutation . | N . | Amino acid change . | Remarks . |
|---|---|---|---|
| 835 GAT → TAT | 34 | Asp → Tyr | — |
| 835 GAT → CAT | 18 | Asp → His | — |
| 835 GAT → GTT | 5 | Asp → Val | — |
| 835 GAT → GAG | 8 | Asp → Glu | — |
| 835 GAT → GAG | 2 | Asp → Asn | Combined with other mutations |
| 835 GAT → GGT | 4 | Asp → Gly | — |
| 835 GAT → multiple | 1 | Asp → ins. Leucin-Lysin | Combined with 836 isoleucin-serin |
| — | |||
| 836 ATC → Del | 13 | Ile → Del | — |
| 836 ATC → ATGCGC | 1 | Ile → MetArg | Insertion of arginin |
| 836 ATC → ACC | 1 | Ile → Thr | — |
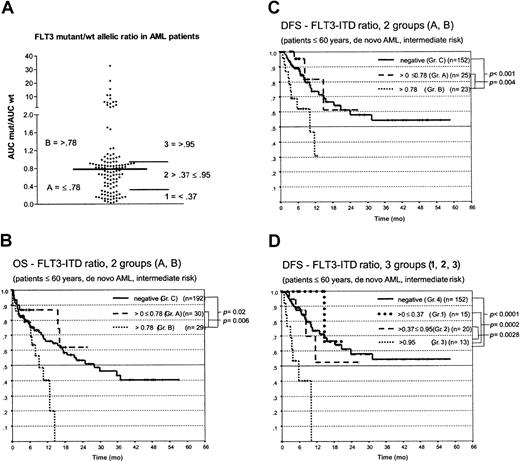
Missing or very weak amplification of the genomic FLT3 wt alleles was observed in a subgroup of patients with FLT3-ITDs on standard agarose gels (Figure 2A). This finding led us to study this phenomenon in more detail. First, a quantitative assay based on Genescan analysis was developed. This assay compares the relative abundance of wt and mutant FLT3 alleles. Genomic DNA from 121 patients with FLT3-ITD was analyzed. The ratio of mutant to wt FLT3 ranged from 0.03 to 32.56, with a median of 0.78. Repeated analysis in 42 patients gave almost identical results (median deviation, 0.04). Twenty (16.5%) patients were found to harbor more than one FLT3 mutant allele, and the median was 2 mutant alleles (range, 2-5). Fragments of corresponding length were detected using cloning and sequence analysis in 4 of these patients. In 16 patients, the FLT3 mutant/wt ratio exceeded 2; thus the intensity of the wt allele was half or less of the intensity of the mutant allele (Figure 2D). This might indicate a loss of the wt allele in these patients.
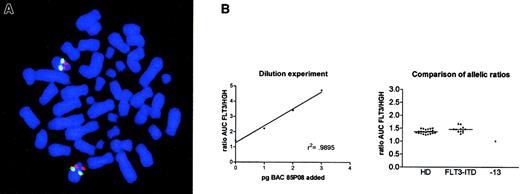
To analyze this phenomenon in more detail, we performed FISH. Metaphase preparations of 9 patients with FLT3-ITD showing a strong overrepresentation of the mutant allele (ie, mutant/wt ratio greater than 2) were screened for chromosomal loss at 13q12 using a BAC clone, which contained the FLT3 genomic region. None of these samples displayed loss of the FLT3 locus at 13q12 (Figure3A). To exclude a deletion of only theFLT3 gene, a probe corresponding to the cDNA sequence was used that yielded ambiguous results because of low signal intensity. To clarify this, we used a PCR-based assay comparing the relative genomic amplification of FLT3 and the human growth hormone gene,HGH, a single copy gene localized on chromosome 17. The results are summarized in Figure 3B. We did not find any difference in the FLT3/HGH allelic ratio between healthy persons (n = 22) and patients with a mutant/wt FLT3 allelic ratio greater than 2 (n = 12). In contrast, we were able to demonstrate such a difference in a patient with monosomy 13 proven by conventional and spectral karyotyping analysis. This might indicate that the observed loss of the wt-FLT3 allele was not due to allelic deletion but to the consequence of homologous recombination.
Analysis of allelic loss in FLT3-ITD–positive samples.
(A) FISH analysis of a patient with an allelic ratio of 3.65. Red signals denote the BAC 85P08 labeled with SpectrumRed, and the green signals indicate a commercial DNA FISH probe (LSI 13) containing the entire RB1 gene and regions telomeric to RB1 labeled with SpectrumGreen. Original magnification, × 125. (B) Results of the PCR assay to detect allelic loss at the FLT3 locus. The left graph denotes the results of the spiking experiment. A linear increase in the FLT3/HGH ratio was obtained after the addition of 1 to 3 pg BAC 85P08 DNA containing the FLT3 genomic locus,r2 = 0.9895. The right graph shows the comparison of the allelic ratios obtained in healthy donors (HD; n = 22), patients with FLT3 wt/mutant ratios greater than 2 (FLT3-ITD; n = 12), and an AML patient with monosomy 13 (−13).
Analysis of allelic loss in FLT3-ITD–positive samples.
(A) FISH analysis of a patient with an allelic ratio of 3.65. Red signals denote the BAC 85P08 labeled with SpectrumRed, and the green signals indicate a commercial DNA FISH probe (LSI 13) containing the entire RB1 gene and regions telomeric to RB1 labeled with SpectrumGreen. Original magnification, × 125. (B) Results of the PCR assay to detect allelic loss at the FLT3 locus. The left graph denotes the results of the spiking experiment. A linear increase in the FLT3/HGH ratio was obtained after the addition of 1 to 3 pg BAC 85P08 DNA containing the FLT3 genomic locus,r2 = 0.9895. The right graph shows the comparison of the allelic ratios obtained in healthy donors (HD; n = 22), patients with FLT3 wt/mutant ratios greater than 2 (FLT3-ITD; n = 12), and an AML patient with monosomy 13 (−13).
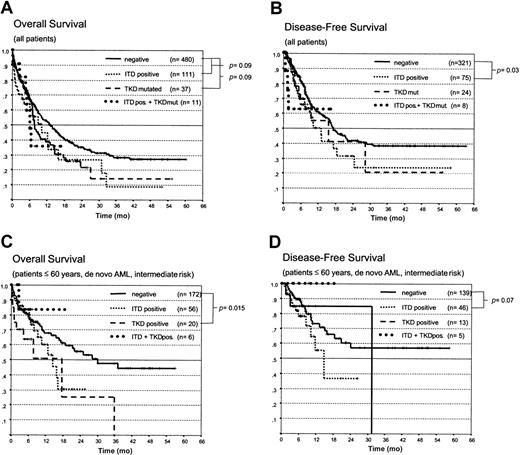
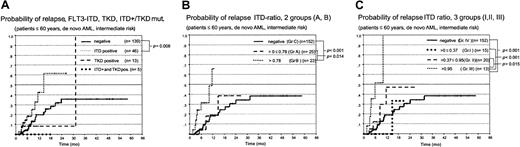
First, we correlated the presence of the different types of FLT3 mutations (ITD, TKD, or ITD+TKD) with the clinical outcome in 640 patients treated in the AML SHG96 trial between 1996 and April 2000. Patients with AML-M3 were analyzed separately because they had been treated according to a different protocol (APL 199326). Patients treated by SCT were censored at the time of transplantation to eliminate the potential bias induced by this intensified treatment. Detection of an FLT3 aberration had no influence on the remission rate (CR rate in patients with de novo AML younger than 60, intermediate risk cytogenetics, FLT3-ITD negative, 66.8% (n = 217); in patients with FLT3-ITDs, 71.2% (n = 66). As shown in Figure4A-D, the presence of an FLT3-ITD or a TKD mutation was associated with inferior OS and DFS for the entire group and for the group of patients younger than 60 with de novo AML/RAEB-t and intermediate-risk cytogenetics. However, these differences were statistically significant only for the DFS in patients with ITD mutations and the OS in young patients with TKD mutations. No significant difference was seen in patients with both types of mutations; however, this may be a result of the small sample size. The presence of an FLT3-ITD mutation was associated with a statistically increased risk for relapse (Figure 5A).
Kaplan-Meier analysis of the OS and DFS in patients with AML with different FLT3 mutations.
Comparison of (A) OS and (B) DFS in all patients with FLT3-ITD mutations, TKD mutations, both types of mutations (ITD+ and TKD mut.) and those with wt-FLT3. (C) OS and (D) DFS in patients younger than 60 with de novo AML and intermediate-risk cytogenetics.
Kaplan-Meier analysis of the OS and DFS in patients with AML with different FLT3 mutations.
Comparison of (A) OS and (B) DFS in all patients with FLT3-ITD mutations, TKD mutations, both types of mutations (ITD+ and TKD mut.) and those with wt-FLT3. (C) OS and (D) DFS in patients younger than 60 with de novo AML and intermediate-risk cytogenetics.
Probability of relapse in patients with FLT3 mutations and different mutant/wt allelic ratios.
Kaplan-Meier analysis of the probability of relapse in AML patients (60 and younger, de novo AML, and intermediate-risk cytogenetics). (A) ITD, TKD, and both mutations (ITD+ and TKD mut.). (B) Grouped according to a mutant/wt ratio below (group A) or above (group B) the median of 0.78 compared to FLT3-ITD–negative patients (group C). (C) Probability of relapse in patients with a mutant/wt ratio 0.37 or lower (group 1), 0.37 to 0.95 (group 2), and greater than 0.95 (group 3) compared to FLT3-ITD–negative patients (group 4).
Probability of relapse in patients with FLT3 mutations and different mutant/wt allelic ratios.
Kaplan-Meier analysis of the probability of relapse in AML patients (60 and younger, de novo AML, and intermediate-risk cytogenetics). (A) ITD, TKD, and both mutations (ITD+ and TKD mut.). (B) Grouped according to a mutant/wt ratio below (group A) or above (group B) the median of 0.78 compared to FLT3-ITD–negative patients (group C). (C) Probability of relapse in patients with a mutant/wt ratio 0.37 or lower (group 1), 0.37 to 0.95 (group 2), and greater than 0.95 (group 3) compared to FLT3-ITD–negative patients (group 4).
The patient with t(8;21) and FLT3-ITD is still in remission after 24 months. In addition, in the group of patients at poor risk, no difference was observed between FLT3-ITD– and TKD-positive and -negative patients in either OS or DFS. No significant difference was seen in the OS and DFS between patients with FLT3-ITD and those without this aberration in patients with t(15;17).
We next analyzed the impact of the FLT3 mutant/wt ratio. Patients were grouped according to a FLT3 ratio below or above the median of 0.78 (Figure 6A). As shown in Figure 6for the DFS in patients with AML who were younger than 60 years and had intermediate-risk cytogenetics, we observed highly significant shorter OS and DFS in patients with ratios above the median than in patients with ratios below this value and compared with FLT3-ITD patients (Figure 6B-C). No significant difference in OS and DFS was observed between the latter 2 groups. The probability of relapse was significantly increased in these patients (Figure 5B). Cox regression analysis showed a relative risk of relapse of 1.6 (95% confidence interval, 1.204-2.169; P = .001) in patients whose mutant/wt ratio was greater than 0.78 compared with patients without FLT3 aberrations. To further characterize these risk groups, we also looked for patients whose ratio was below the 25thpercentile (ratio ≤ 0.37; group 1) compared with patients whose ratio was between the 26th and 74thpercentile (ratio > 0.37 ≤ 0.95; group 2) and those above the 75th percentile (ratio > 0.95; group 3) and the FLT3-ITD patients (group 4) (Figure 6D). Here again, patients with the highest ratio (group 3), strongly arguing for allelic loss, had a highly significant shorter OS and DFS than the other 3 groups. Fifteen patients with FLT3 mutant/wt ratios greater than 2 had the worst clinical course; 14 of 15 died by 12 months. The surviving patient underwent allogeneic blood stem cell transplantation.
Kaplan-Meier analysis of the OS and DFS in patients with AML, younger than 60, and with intermediate-risk cytogenetics.
(A) Distribution of the mutant/wt allelic ratio and risk groups defined. (B) OS and (C) DFS for patients with a mutant/wt ratio below (group A) or above (group B) the median of 0.78 compared to FLT3-ITD–negative patients (group C). (D) DFS for patients with a mutant/wt ratio 0.37 or lower (group 1), 0.37 to 0.95 (group 2), and greater than 0.95 (group 3) compared with FLT3-ITD–negative patients (group 4).
Kaplan-Meier analysis of the OS and DFS in patients with AML, younger than 60, and with intermediate-risk cytogenetics.
(A) Distribution of the mutant/wt allelic ratio and risk groups defined. (B) OS and (C) DFS for patients with a mutant/wt ratio below (group A) or above (group B) the median of 0.78 compared to FLT3-ITD–negative patients (group C). (D) DFS for patients with a mutant/wt ratio 0.37 or lower (group 1), 0.37 to 0.95 (group 2), and greater than 0.95 (group 3) compared with FLT3-ITD–negative patients (group 4).
We also compared the clinical variables in the different groups. Patients with high mutant/wt ratios were found to have significantly higher WBC and BM blast counts than patients with low ratios, but these groups did not differ significantly with respect to age, FAB subtype, or karyotype characteristics.
Multivariate analysis was performed to investigate whether FLT3 aberrations represented an independent prognostic factor. We included several known risk factors in the model (age, cytogenetics, WBC, sAML) and FLT3 aberrations (ITD, TKD, and ITD+TKD mutations) and the FLT3-ITD ratio. As shown in Table 4, though FLT3 mutations were not associated with OS and DFS on multivariate analysis, a high mutant/wt ratio was found to represent an independent prognostic factor with odds ratios between 1.8 and 8.
Multivariate analysis of clinical and biologic variables
| . | All patients . | Patients 60 years or younger . | ||||||
|---|---|---|---|---|---|---|---|---|
| OS . | DFS . | OS . | DFS . | |||||
| P . | OR (95% CI) . | P . | OR (95% CI) . | P . | OR (95% CI) . | P . | OR (95% CI) . | |
| Age, y (≤ 60 vs > 60) | < .001 | 1.8 (1.4-2.3) | < .001 | 2.8 (1.9-4) | — | — | — | — |
| Karyotype (good vs intermediate and poor) | < .001 | .24 (.1-.5) | .004 | .26 (.1-.6) | .001 | .2 (.1-.5) | .02 | .2 (.04-.8) |
| WBC (≤ 20 × 109 WBC/L vs > 20 × 109WBC/L) | NS | — | NS | — | .05 | 1.4 (1-2) | NS | — |
| Status | ||||||||
| De novo | — | 1 | — | 1 | — | 1 | 1 | |
| AML from pre-existing MDS | .011 | 1.4 (1.1-1.9) | NS | — | .007 | 1.8 (1.2-2.9) | NS | — |
| tAML | .021 | 1.8 (1.1-2.9) | .005 | 3.1 (1.4-6.8) | .032 | 2 (1.1-3.9) | NS | — |
| FLT3 status | ||||||||
| wt | — | 1 | — | 1 | — | 1 | 1 | |
| ITD+ | NS | — | NS | — | NS | — | NS | — |
| TKD mut | NS | — | NS | — | NS | — | NS | — |
| ITD+/TKD mut | NS | — | NS | — | NS | — | NS | — |
| FLT3 mut/wt ratio 1 | ||||||||
| FLT3 ITD—vs mut/wt ratio greater than .784-150 | .002 | 1.8 (1.2-2.5) | < .001 | 3.2 (1.8-5.5) | NS | — | < .001 | 4.2 (2.1-8.3) |
| FLT3 mut/wt ratio 2 | ||||||||
| FLT3 ITD—vs mut/wt ratio greater than .954-150 | .001 | 2.3 (1.4-3.7) | < .001 | 8 (3.8-16.6) | NS | — | < .001 | 6.9 (3.3-14.8) |
| . | All patients . | Patients 60 years or younger . | ||||||
|---|---|---|---|---|---|---|---|---|
| OS . | DFS . | OS . | DFS . | |||||
| P . | OR (95% CI) . | P . | OR (95% CI) . | P . | OR (95% CI) . | P . | OR (95% CI) . | |
| Age, y (≤ 60 vs > 60) | < .001 | 1.8 (1.4-2.3) | < .001 | 2.8 (1.9-4) | — | — | — | — |
| Karyotype (good vs intermediate and poor) | < .001 | .24 (.1-.5) | .004 | .26 (.1-.6) | .001 | .2 (.1-.5) | .02 | .2 (.04-.8) |
| WBC (≤ 20 × 109 WBC/L vs > 20 × 109WBC/L) | NS | — | NS | — | .05 | 1.4 (1-2) | NS | — |
| Status | ||||||||
| De novo | — | 1 | — | 1 | — | 1 | 1 | |
| AML from pre-existing MDS | .011 | 1.4 (1.1-1.9) | NS | — | .007 | 1.8 (1.2-2.9) | NS | — |
| tAML | .021 | 1.8 (1.1-2.9) | .005 | 3.1 (1.4-6.8) | .032 | 2 (1.1-3.9) | NS | — |
| FLT3 status | ||||||||
| wt | — | 1 | — | 1 | — | 1 | 1 | |
| ITD+ | NS | — | NS | — | NS | — | NS | — |
| TKD mut | NS | — | NS | — | NS | — | NS | — |
| ITD+/TKD mut | NS | — | NS | — | NS | — | NS | — |
| FLT3 mut/wt ratio 1 | ||||||||
| FLT3 ITD—vs mut/wt ratio greater than .784-150 | .002 | 1.8 (1.2-2.5) | < .001 | 3.2 (1.8-5.5) | NS | — | < .001 | 4.2 (2.1-8.3) |
| FLT3 mut/wt ratio 2 | ||||||||
| FLT3 ITD—vs mut/wt ratio greater than .954-150 | .001 | 2.3 (1.4-3.7) | < .001 | 8 (3.8-16.6) | NS | — | < .001 | 6.9 (3.3-14.8) |
NS indicates not significant.
For these analyses separate models were used.
Discussion
We have analyzed the frequency of FLT3 gene mutations in 979 patients with AML. The overall incidence of FLT3 tandem mutations was 20.4% (200 of 979), which is consistent with numbers reported by other groups.12 In the same cohort of patients, point mutations of the activation loop of the second tyrosine kinase domain (AA 835/836) were identified in 7.7% (75 of 979). Taken together, constitutive activation of FLT3 was present in 26.4% (258 of 979) of these patients, which indicates that FLT3 is an important target of mutational activation in adult AML.
Comparison of FLT3 mutational data and the clinical and cytogenetic findings revealed several parallels between the 2 types of mutation. FLT3-ITD and TKD mutations were associated with similar clinical features—that is, both aberrations were associated with significantly higher WBC and higher numbers of bone marrow blasts (Table 1). As a novel finding, our data indicate that mutational FLT3 activation was significantly increased in patients with AML FAB M5, among whom approximately 40% carried a mutated FLT3 gene. FLT3 is persistently expressed during monocyte differentiation,38and the addition of FLT3-L is needed for optimal differentiation of monocytes from CD34+ cells.39 Thus, constitutive activation of FLT3 might be associated with monocytoid differentiation. In contrast, FAB subtypes M2 and M6 were significantly less frequently associated with FLT3 activation, which is also in line with the expression patterns of this protein during normal hematopoietic differentiation. With respect to the cytogenetic data, ITD and TKD mutations were approximately 3 times more prevalent in patients with normal karyotype than in patients with cytogenetic alterations. Most patients with cytogenetic aberrations showed a low number of FLT3-ITD mutations (Table 2). As an exception, a higher number of FLT3-ITDs and TKD mutations (30.2% and 19%, respectively) was observed in patients with t(15;17), a frequency similar to or even higher than that observed in the entire group. Two other groups reported incidences of 20.3%40 and 28.6%41FLT3-ITD–positive patients with t(15;17). Thus, there appears to be an increased incidence of FLT3-aberrations in patients with the PML-RAR α rearrangement. The rate of FLT3-ITDs in 10 patients with t(6;9), 9 of whom showed this mutation, was unexpectedly high. Given the low frequency of the t(6;9) of approximately 1%,42 the association appears highly significant and may give clues to the genetic background of the FLT3 mutations (see below).
Our results on the prognostic relevance are in line with other reports14-16 showing that the presence of an FLT3-ITD mutation is associated with worse clinical response. However, the prognostic impact of FLT3-ITDs in our study was less pronounced than in other reports and reached statistical significance only for DFS. Several factors might be responsible for this observation. The overall follow-up time in our study is only 12.2 months (range, 0-60.3 months). Because autologous or allogeneic SCT could influence the outcome, patients treated with transplantation were censored at the time of transplantation, which further shortened the follow-up time. The fact that 140 patients underwent transplantation might affect the analysis because these patients are not eligible for the long-term outcome.
Differences might also be attributed to the assessment of FLT3-ITDs. In the present study, we regarded any additional FLT3 signal as ITD+. However, using a quantitative assay based on Genescan analysis after PCR with fluorescently labeled primers, we found considerable heterogeneity of the mutant/wt ratio in 121 FLT3-ITD+ patients analyzed, with a median ratio of 0.78 (range, 0.03 to 32.56). When we looked for the prognostic relevance of this ratio, we saw that a high ratio of the ITD allele compared to the wt allele conferred a negative prognostic impact with respect to OS and DFS (Figure 6). However, because of our results, the poor outcome was not confined to patients with clear loss of the wt allele, which was already visible on standard agarose gels, but appeared to increase dramatically when the ratio of the mutant/wt FLT3 exceeded 1. These patients had a significantly increased rate of relapse; even more important, recurrent disease developed rapidly, with a median OS time of 8 months in patients otherwise considered at intermediate risk. Multivariate analysis showed that a high mutant/wt ratio was an independent prognostic factor for worse OS and DFS (Table 4). In contrast, as shown in Figure 6, patients with low allelic ratios did not have significantly different OS and DFS than patients without FLT3-ITD mutations. When other groups were chosen—patients with the lowest quartile (ratio 0.37 or lower), intermediate 50% (ratio 0.38-0.95), and highest quartile (ratio 0.95 or higher)—this discrimination was even more evident (Figure 6D). The results presented here are well in line with data published during the review of this manuscript. Whitman et al43 reported data indicating that FLT3 allelic loss occurs in approximately 10% of patients with FLT3-ITD mutations and that patients with allelic loss have a significantly inferior outcome. Thus, besides the information on the presence of a FLT3 aberration, quantitative determination of the ratio between mutant and wt FLT3 alleles might be of major prognostic importance.
In contrast to allelic loss, the presence of both types of mutations (ITD and TKD) in a patient was not associated with a different clinical course. However, the precise prognostic relevance of FLT3-TKD mutations remains open. Although FLT3-TKD mutations had a similar effect on WBC counts and BM blasts and tended to be associated with lower OS and DFS, this association was not significant in most patients. In addition to the reasons discussed regarding the follow-up, we did not analyze the transforming activity of the different TKD mutations, which may differ considerably, as has been shown for the fms tyrosine kinase.44 Clearly, this must be done to elucidate the clinical importance of this change.
Evidence for allelic loss at 13q12 with loss of the wt FLT3 allele (ie, mutant/wt ratio greater than 2) has recently also been reported by others.45 However, we did not find any evidence of genomic deletion in FISH assays using a 100-KBp BAC fragment corresponding to chromosomal region 13q12. To further clarify this issue, we performed differential PCR, which also failed to show significant differences in the allelic ratios between normal diploid samples and the patients.
There are several potential explanations for this observation. First, the deletion could have involved only parts of the FLT3gene, which stretches over a total of 35 kb. Alternatively the wtFLT3 gene could have been deleted by homologous recombination, a mechanism of loss of heterozygosity frequently observed in solid tumors.46 The substitution of the wt FLT3 allele by the mutant would explain why we did not observe a genomic deletion of this locus. The finding that FLT3 aberrations were abundant in patients with normal karyotype according to conventional cytogenetics and less frequent in patients with typical aberrations indeed points to a novel mechanism of leukemia development in this group of patients.
Kiyoi et al20 postulated that the palindromic DNA sequence between codons 593 and 602 of FLT3 favors the formation of secondary structures, thus leading to the tandem mutation. Sequence analysis in 101 mutated FLT3 sequences in our study indicates that the main cluster of the mutations lies between codons 591 and 601, which supports this hypothesis. Nevertheless we observed a substantial number of patients without involvement of this stretch or the insertion of nucleotides not matching FLT3 sequences. This points to a more complex background of the FLT3 aberrations. Furthermore, 20 (16.5%) of the patients with FLT3-ITDs contained several clonally unrelated mutations, which indicates that FLT3 aberrations are potentially not the causative event but the result of an inherent genetic instability of the leukemic cells due to an as yet undefined defect. In this context, the association of FLT3 mutations with 2 translocations, t(15;17)/PML-RARA and t(6;9)/DEK-CAN, observed in our study deserves attention. FLT3-ITDs were observed in 90% of patients with DEK-CAN fusion (Table2). The can gene on chromosome 9 codes for a part of the nucleoporin complex.47,48 In contrast, the role of the DEK protein is largely unknown. Meyn et al49 have shown that the carboxy-terminal 35 amino acids of DEK are able to revert the genetic instability of ataxia-telangiectasia-mutated (ATM) cells, indicating that DEK might function in controlling genetic stability. The DEK-CAN fusion protein lacks these terminal 35 amino acids. More recently, DEK has been shown to be important in the maintenance of chromatin structure and seems to be an essential part of the replication machinery.50 A significant increase of FLT3-ITD and TKD mutations was also observed in patients with t(15;17). The PML gene involved in t(15;17) is an interferon-inducible gene that encodes a RING-finger protein typically concentrated within nuclear structures called PML nuclear bodies or PML oncogenic domains.51 Overexpression of the PML protein induces growth arrest; however, the precise function of the protein is still not fully understood. Recently, the PML protein was found to interact with the Bloom syndrome gene, BLM, a RecQ DNA helicase whose absence induces genomic instability and high levels of sister-chromatid exchange, indicating that PML might also be involved in controlling genetic stability.52 Thus it is tempting to speculate that alterations of PML, DEK, or other proteins involved in chromosomal stability might be relevant in the development of FLT3-ITD.
In summary our data indicate that FLT3 mutations are a common alteration in adult AML. Patients with the FLT3-ITD mutation and a loss of the wt-FLT3 allele seem to have a worse prognosis. In this regard, we believe that the use of fluorophore-labeled primer molecules and denaturing gel electrophoresis offers several advantages compared with conventional gel analysis. This mode of detection enables the unambiguous separation of single or multiple mutant FLT3 fragments. In addition, it allows calculation of a ratio between the mutant and the wt FLT3 alleles that might be more accurate and sensitive in the detection of allelic loss and that might add prognostic information to the qualitative detection of FLT3 alterations. This may help to identify high-risk patients who should potentially be offered more intensive treatment options. Clearly, the prognostic value of this ratio has to be analyzed in future prospective trials.
We thank all participating centers of the SHG AML-96 study (“”). S. Soucek and S. Freund provided excellent help in statistical analyses. The full-length cDNA clone of FLT3 was a kind gift of Dr O. Rosnet. We thank M. Hartwig, U. Löwel, M. Neumann, P. Grassmel, and C. Grosse for their skillful technical assistance. We thank Prof Joachim Deeg and Dr Michael Maris (Fred Hutchinson Cancer Research Center, Seattle, WA) for critically reading the manuscript.
The following is a list of participating centers and physicians: D. Huhn, O. Knigge (Universitätsklinikum Charite, Berlin), R. Kolloch, U. Krümpelmann (Krankenanstalten Gilead, Bielefeld), K.-H. Pflüger, T. Wolff (Evang. Diakonissenanstalt Bremen), H.-H. Heidtmann (St Joseph-Hospital, Bremerhaven), F. Marquard (Allgemeines Krankenhaus, Celle), F. Fiedler, R. Herbst (Krankenhaus Küchwald, Chemnitz), M. Gramatzki, G. Helm (Universitätsklinikum, Erlangen), J.-G. Saal (Malteser Krankenhaus, Flensburg), H.-G. Höffkes, M. Arland (Städtisches Klinikum, Fulda), E. Faßhauer (St Elisabeth-Krankenhaus, Halle), R. Kuse (Allgemeines Krankenhaus St Georg, Hamburg), H. Schmidt, K. Buhrmann (Kreiskrankenhaus, Hameln), H. Dürk (St Marien-Hospital, Hamm), M. Burk (Klinikum Stadt, Hanau), A.-D. Ho, R. Weber-Nordt (Universitätsklinikum, Heidelberg), A. A. Fauser (Klinik f. Hämatologie/Onkologie und KMT, Idar-Oberstein), H. Link, F.-G. Hagmann (Westpfalzklinikum, Kaiserslautern), G. Köchling (Kreiskrankenhaus, Leer), K.-P. Schalk (St Vincent-Krankenhaus, Limburg/Lahn), S. Fetscher (Städtisches Krankenhaus Süd, Lübeck), T. Wagner (Universitätsklinikum, Lübeck), H. Bodenstein, J. Tischler (Klinikum Minden, Minden), H. Pohlmann, N. Brack (Städtisches Krankenhaus München-Harlaching, München), H. Wandt, K. Schäfer-Eckart, T. Denzel (Städtisches Klinikum, Nürnberg), B. Seeber (Klinikum Offenbach, Offenbach), F. Hirsch (Kreiskrankenhaus, Offenburg), T. Geer, H. Heißmeyer (Diakonie-Krankenhaus, Schwäbisch-Hall), J. Labenz (Ev. Jung-Stilling-Krankenhaus, Siegen), J. Kaesberger (Diakonissen-Krankenhaus, Stuttgart), W. E. Aulitzky, L. Leimer (Robert-Bosch-Krankenhaus, Stuttgart), M. R. Clemens, R. Mahlberg (Mutterhaus der Borromaerinnen, Trier), R. Schwerdtfeger (Deutsche Klinik für Diagnostik, Wiesbaden), R. Engberding, R. Winter (Stadtkrankenhaus, Wolfsburg), M. Sandmann (Klinikum St Antonius, Wuppertal), H. Rückle-Lanz, M. Wilhelm (Universitätsklinikum, Würzburg).
Supported in part by grants from the Deutsche Krebshilfe (70-2210-Eh5) (G.E.). Supported by the Kompetenznetzwerk Akute und Chronische Leukämien, sponsored by the BMBF.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Christian Thiede, Medizinische Klinik und Poliklinik I, Universitätsklinikum Carl Gustav Carus der Technischen Universität, Fetscherstrasse 74, 01307 Dresden, Germany; e-mail: thiede@mk1.med.tu-dresden.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal