The chemokine receptor, CCR5, is used as a human immunodeficiency virus coreceptor in combination with CD4 during transmission and early infection. CCR5 has been shown to be palmitoylated and targeted to cholesterol- and sphingolipid-rich membrane microdomains termed “lipid rafts.” However, the role of cholesterol and lipid rafts on chemokine binding and signaling through CCR5 remains unknown. We found that cholesterol extraction by hydroxypropyl-β-cyclodextrin (BCD) significantly reduced the binding and signaling of macrophage inflammatory protein 1β (MIP-1β) using CCR5-expressing CEM-NKR T cells. Reloading treated cells with cholesterol but not 4-cholesten-3-one, an oxidized form of cholesterol, restored MIP-1β binding to BCD-treated cells. Antibodies specific for distinct CCR5 epitopes lost their ability to bind to the cell surface after cholesterol extraction to varying degrees. Moreover, cells stained with fluorescently labeled MIP-1β extensively colocalized with the GM1 lipid raft marker while using anti-CCR5 antibodies; most of CCR5 on these cells only partially colocalized with GM1, suggesting that active ligand binding facilitates receptor association with lipid rafts or that raft association promotes a higher affinity conformation of CCR5. Together, these data demonstrate that cholesterol and lipid rafts are important for the maintenance of the CCR5 conformation and are necessary for both the binding and function of this chemokine receptor.
Introduction
Chemokine receptors have drawn much attention since their description as human immunodeficiency virus (HIV) coreceptors by several groups in 1996.1-4 Prior to that time, HIV tropism was defined as either macrophage or T-cell tropic, which corresponded to nonsyncytia- or syncytia-inducing viruses, respectively. Today, the classification of HIV tropism is defined by chemokine receptor usage of either CCR5, CXCR4, or both receptors, although usage of other chemokine receptors has been reported.2,5 Chemokine receptors are a family of 7 transmembrane-spanning G protein–coupled receptors that are differentially expressed by a number of immune and nonimmune cell populations. These receptors are quite specific and mediate immune cell responses to a family of soluble chemoattractant molecules, termed chemokines.6 CCR5 is the receptor for several CC chemokines, including RANTES, macrophage inflammatory protein 1α (MIP-1α), MIP-1β, and MCP-3. All of these ligands are capable of inhibiting HIV infection by CCR5.7 Although monocyte/macrophages and dendritic cells express high levels of CCR5 on their cell surface, this receptor can also be expressed in T cells, natural killer (NK) cells, and other cells. Targeting chemokine receptors to block HIV infection is beginning to show promise as an effective means to control viremia in infected individuals.8-11
“Lipid rafts” is a broad term for the collection of membrane microdomains enriched in cholesterol, sphingolipids, glycosylphosphatidylinositol-anchored proteins, and acylated signaling molecules.12 Lipid rafts are believed to be important signaling platforms enriched in many signaling proteins, including but not limited to src kinases, Gα subunit, H-Ras, linker for activation of T cells (LAT), and nitric oxide synthase (NOS). Signal transduction through the T- and B-cell receptors as well as the immunoglobulin E (IgE) receptor involves the recruitment of signaling assemblies to lipid rafts.13-15 CCR5 has been shown to be present in lipid rafts, colocalizing at the leading edge of migrating cells.16 This receptor has also recently been shown to be palmitoylated, which is one of the important modifications in lipid raft targeting of proteins.17-19
One important component in maintaining the higher lipid order of rafts is cholesterol. Circular multimeric sugar molecules, known as cyclodextrins, have been used to extract cholesterol from membranes, thereby disrupting lipid rafts and increasing overall membrane fluidity.20,21 The loss of cholesterol has been shown to have profound effects on the ability of several G protein–coupled receptors to bind their ligand. The oxytocin, cholecystokinin, galanin, and γ-aminobutyric acid receptors have all been found to require cholesterol for their function.21-23 However, a role for cholesterol in chemokine receptor function has not yet been reported. To address the question, we have examined the effects of cholesterol depletion by hydroxypropyl-β-cyclodextrin (BCD) and cholesterol restoration on CCR5 expression, ligand binding, and CCR5 signaling. Our data suggest that changes in CCR5 conformation are responsible for the inhibitory effects observed after BCD treatment of human T cells.
Materials and methods
Cell lines and reagents
CEM-NKR-CCR5 (referred to hereon as CEM-R5) cells were obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, originated by Dr Alexandra Trkola). These cells were grown in RPMI-1640 (Mediatech; Cellgro, Herndon, VA) supplemented with 10% heat inactivated fetal bovine serum (FBS) (Biosource, Rockville, MD), 10 mM HEPES, and antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin). Monoclonal antibodies (mAbs) to various epitopes on CCR5, namely clones CTC5, 45502.111, 45523.111, 45531.111, and 45549.111, were purchased from R & D Systems (Minneapolis, MN). Mouse IgG2a control, anti-CD4 (clone RPA-T4), and anti-CD45 (clone HI30) were purchased from Becton Dickinson Biosciences (San Diego, CA). Goat antimouse IgG (H + L) labeled with Alexa Fluor 488 (GAM-AF488) and cholera toxin B (CT-B) subunit labeled with Alexa Fluor 594 were purchased from Molecular Probes (Eugene, OR). Alexa Fluor 488 has a laser absorption and emission spectrum profile similar to fluorescein, whereas Alexa Fluor 594 has a profile similar to that of Texas Red.
BCD treatment
BCD, Trappsol (Cyclodextrin Technologies Development, Gainesville, FL), was dissolved in phophate-buffered saline (PBS) to the desired concentrations. To prepare cholesterol-loaded BCD (chol-BCD), BCD was dissolved in PBS to make 240 mM. Cholesterol (5-cholesten-3β-ol; 3β-hydroxy-5-cholestene) or 4-cholesten-3-one (Sigma, St Louis, MO) powder was added to the BCD solution at 1.16 mg/mL. The mixture was vortexed for 6 hours to completely dissolve the cholesterol and was subsequently sterilized with syringe filter by using a 0.22-μm filter unit. In cholesterol extraction studies, 8 × 106 suspension cells were washed with PBS and resuspended in 1 or 2 mL of 20 mM BCD/PBS or PBS alone. The cells were then incubated with BCD for 1 hour at 37°C before being washed with either PBS or RPMI. In cholesterol reloading studies, cells were incubated with chol-BCD in PBS at a concentration of 300 μM cholesterol for 30 minutes. To remove chol-BCD, cells were washed with at least 10 volumes of PBS and resuspended in PBS for further analysis.
Fluorokine ligand staining
Biotinylated MIP-1β (Fluorokine, R & D Systems) staining was performed according to R & D Systems protocols with slight modifications. Briefly, control or treated cells were resuspended in PBS at 4 × 106/mL. Cells (2 × 105) were mixed with 20 μL of 3.0 μg/mL biotinylated MIP-1β or 5.0 μg/mL biotinylated-soybean trypsin inhibitor (negative control) and then incubated at 4°C for 1 hour. Fluorescein-conjugated avidin (10 μg/mL) was added (10-20 μL) to the cells and incubated for an additional 30 minutes. at 4°C. After incubation, cells were washed with 1 × RDF-1 buffer (R & D Systems) and then fixed with 2% paraformaldehyde-PBS before being analyzed on a FACScan (Becton Dickinson).
Intracellular calcium mobilization
Measurement of calcium mobilization by chemokine stimulation was performed as previously described.24 Briefly, untreated or BCD-treated CEM-R5 cells (8 × 106/mL) were incubated in PBS with Ca++ and Mg2+ containing 5 μM Fura-2am for 30 minutes at room temperature. The cells were subsequently washed and then resuspended at 1 × 106/mL in PBS. A total of 2 mL of the cell suspension was placed in a continuously stirred cuvette at room temperature in an LS50B spectrophotometer (Perkin-Elmer, Wellesley, MA). Fluorescence was monitored at λex1 = 340 nm, λex2 = 380 nm, and λem = 510 nm. The data are presented as the relative ratio of fluorescence excited at 340 and 380 nm. Data were collected every 0.96 seconds. RANTES and MIP-1β (Pepro Tech, Rocky Hill, NJ) were tested at a final concentration of 1 μg/mL. Values were graphed as 30-second interval averages minus the background change in fluorescence ratio seen in untreated cells. The first 30-second interval of each set was set at 0. Error bars represent the SD within each 30-second interval.
Flow cytometry
CEM-R5 cells (1 × 106) in PBS containing 2% heat-inactivated FBS were added to 1 to 2 μg mAbs and incubated for 30 minutes on ice. Cells were washed with PBS, resuspended in 100 μL of 20 μg/mL GAM-AF488, and incubated on ice for 30 minutes. Cells were then washed with PBS and fixed with 2% paraformaldehyde in PBS, followed by analysis on a FACScan. For the prefixing experiments, cells were washed with PBS after BCD treatment and then fixed with 2% paraformaldehyde in PBS for 30 minutes on ice. After incubation, the cells were then washed with PBS, resuspended in PBS containing 2% FBS, and then incubated an additional 30 minutes on ice before staining with mAbs. Permeabilization experiments were performed by using Caltag Laboratories Fix and Perm Cell Permeabilization Kit (Burlingame, CA).
Immunomicroscopy
CEM-R5 cells (1 × 106) were washed in cold PBS, resuspended in 100 μL PBS containing 2% FBS and 20 μg/mL CT-B Alexa Fluor 596, and then incubated on ice for 30 minutes. Cells were then washed with PBS and subsequently stained for CCR5 by using 2 μg CCR5 mAb, CTC5, followed by GAM-AF488. The cells were washed with PBS and then fixed with 1% paraformaldehyde in PBS. After staining, the cells were placed into cytospin funnels and spun onto glass slides with the use of a cytospin centrifuge (Shandon, Pittsburgh, PA) at 1000 rpm for 2 minutes. Bound cells were layered with 30 μL 50% glycerol in PBS and covered with a glass coverslip. Images were acquired by Spot Advanced software on a Zeiss Axiovert S100 microscope under × 100 objective (Carl Zeiss, Thornwood, NY).
Confocal microscopy
Untreated or 10 mM BCD-treated CEM-R5 cells (1 × 106) were fixed in 1% paraformaldehyde in PBS for 30 minutes at 4°C, washed with PBS, and then resuspended in 100 μL PBS with or without 0.1% Triton X-100. Cells were then washed with PBS and subsequently stained for CCR5 by using 1 μg CCR5 mAb, 45531, for 30 minutes at 4°C. Cells were washed and resuspended in 100 μL GAM-AF488 plus 0.5 μg (10 μL) propidium iodide (PI) staining solution (BD Pharmingen, San Diego, CA) for 30 minutes at 4°C. The cells were washed with PBS and then fixed with 1% paraformaldehyde in PBS. The cells were cytospun onto slides as described above and then examined by a Zeiss LSM410 confocal microscope under × 63 objective under oil immersion. Images were processed by using Adobe Photoshop software for presentation.
Results
Cholesterol extraction inhibits MIP-1β binding and intracellular calcium mobilization
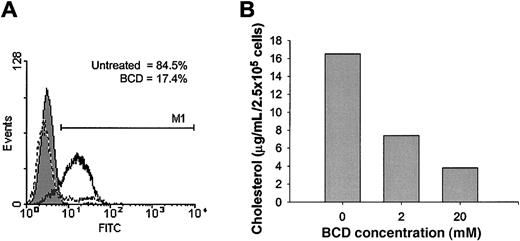
Recent studies have shown that the palmitoylation of CCR5 is essential for stable expression at the cell surface, most likely through localization with cholesterol-rich membrane lipid rafts.17,18 Membrane cholesterol, an important structural component of lipid rafts, has been shown to be important to the function of several G protein–coupled receptors. We wanted to determine if the expression and function of ligand binding to human CCR5 was similarly dependent on cholesterol. To examine this question, we treated the T-cell line, CEM-NKR, which was stably transfected with human CCR5 (CEM-R5)25 with BCD to extract cholesterol from the cell membrane. To measure ligand binding, we used a whole cell assay with a biotinylated form of MIP-1β, followed by binding of an avidin–fluorescein isothiocyanate (FITC) conjugate, and subsequent examination by using flow cytometric analysis. Cholesterol extraction from the cell membrane significantly impaired MIP-1β binding to cell surface CCR5 (Figure 1A). The percentage of positive cells for MIP-1β binding decreased from 84.5% for untreated cells to 17.4% for BCD-treated cells. Positive cells are defined by the M1 region with FITC fluorescence more than 98% of the mouse IgG2a control. Treatment of these cells with 20 mM BCD for 1 hour removed approximately 75% of total cellular cholesterol as measured by the Amplex Red Cholesterol Assay Kit (Figure 1B). Cells treated with BCD failed to demonstrate any levels of toxicity, as assessed by the release of cytoplasmic lactate dehydrogenase into the supernatant or by Trypan Blue exclusion (data not shown). Similar experiments were attempted with biotinylated RANTES; however, this ligand exhibited very low levels of binding. It seems quite possible that the biotinylation of RANTES may have altered the secondary structure of this ligand or masked some binding sites that normally interact with CCR5. These results clearly demonstrate a requirement for cholesterol in the cell membrane for MIP-1β binding to CCR5.
MIP-1β binding is inhibited by BCD treatment.
CEM-R5 cells were treated with BCD (A) and stained with biotinylated MIP-1β and detected with avidin-fluorescein conjugate as described in “Materials and methods.” Relative fluorescein (FITC) fluorescence is expressed on the x-axis. The results for biotinylated-negative control are graphed as a filled peak, MIP-1β binding to untreated cells is graphed as a solid line, and MIP-1β binding to BCD-treated cells are graphed as a dashed line. M1 indicates the range of cells with FITC fluorescence more than 98% of the negative control. One representative experiment of 4 is presented. (B) CEM-R5 cells were treated with the indicated concentrations of BCD for 1 hour. Cholesterol quantitation of cells was performed exactly according to Molecular Probes' Amplex Red Cholesterol Assay Kit. Results are expressed as cholesterol concentration per input of cells.
MIP-1β binding is inhibited by BCD treatment.
CEM-R5 cells were treated with BCD (A) and stained with biotinylated MIP-1β and detected with avidin-fluorescein conjugate as described in “Materials and methods.” Relative fluorescein (FITC) fluorescence is expressed on the x-axis. The results for biotinylated-negative control are graphed as a filled peak, MIP-1β binding to untreated cells is graphed as a solid line, and MIP-1β binding to BCD-treated cells are graphed as a dashed line. M1 indicates the range of cells with FITC fluorescence more than 98% of the negative control. One representative experiment of 4 is presented. (B) CEM-R5 cells were treated with the indicated concentrations of BCD for 1 hour. Cholesterol quantitation of cells was performed exactly according to Molecular Probes' Amplex Red Cholesterol Assay Kit. Results are expressed as cholesterol concentration per input of cells.
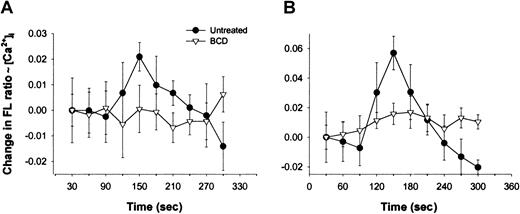
We next examined alterations in the function of CCR5 by measuring changes in intracellular calcium in response to ligand treatment of BCD-treated CEM-R5 cells. Recombinant human MIP-1β induced a small but significant rise in intracellular calcium in control CEM-R5 cells, as measured by the change in fluorescence of Fura-2 am. However, this MIP-1β response was inhibited in CEM-R5 cells treated with BCD (Figure 2A). Similarly, stimulation of control CEM-R5 cells with RANTES induced a more pronounced rise in intracellular calcium that was also inhibited by pretreatment with BCD (Figure 2B). Together, these results provide functional confirmation for the loss in surface binding of the CCR5 ligands, MIP-1β and RANTES, suggesting a loss in cell surface binding and/or signaling in BCD-treated cells.
Chemokine-induced intracellular calcium mobilization is inhibited by BCD treatment.
BCD-treated CEM-R5 cells, loaded with Fura-2, were stimulated with MIP-1β (A) or RANTES (B) at 60 seconds as described in “Materials and methods.” Results are graphed as the average change in fluorescence ratio (F340/F380) of each 30-second interval with error bars representing the SD.
Chemokine-induced intracellular calcium mobilization is inhibited by BCD treatment.
BCD-treated CEM-R5 cells, loaded with Fura-2, were stimulated with MIP-1β (A) or RANTES (B) at 60 seconds as described in “Materials and methods.” Results are graphed as the average change in fluorescence ratio (F340/F380) of each 30-second interval with error bars representing the SD.
Reloading T cells with cholesterol restores MIP-1β binding
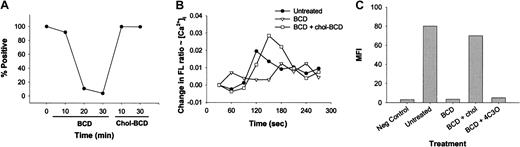
To confirm that the effects observed with BCD treatment were specifically associated with the removal of cholesterol, an analysis of MIP-1β binding to BCD-treated T cells for various times was performed, followed by reloading of fresh cholesterol into the cell membranes using chol-BCD complexes. We verified that the total cellular cholesterol levels were restored to normal (or above normal) levels through an assessment of total cholesterol by using the Amplex Red cholesterol assay (data not shown). The results in Figure3 demonstrate that MIP-1β binding is rapidly lost with BCD treatment, suggesting that MIP-1β binding is sensitive to cholesterol extraction. Reloading of cholesterol onto cell membranes restored MIP-1β binding to these treated cells (Figure 3A). Overall, these results confirm that reloading T cells with cholesterol restores MIP-1β binding activity and that cell death does not account for the initial loss in ligand binding. Moreover, BCD itself does not appear to exert any inhibitory activity as repletion of cholesterol into cell membranes also involves the use of BCD as a vehicle for cholesterol. Consequently, intracellular calcium mobilization by MIP-1β was also restored to normal when BCD-treated cells were reloaded with cholesterol (Figure 3B). To further verify the specificity of cholesterol reloading, we also reloaded BCD-treated cells with 4-cholesten-3-one, an oxidized form of cholesterol. These cells did not regain the ability to bind MIP-1β, unlike cholesterol-reloaded cells (Figure 3C).
Reloading of cholesterol restores MIP-1β binding and calcium mobilization to BCD-treated cells.
(A) CEM-R5 cells were treated for the indicated times with BCD before washing with PBS. Treated cells were then reloaded with chol-BCD for 10 or 30 minutes, and the level of MIP-1β binding was determined as described in “Materials and methods.” Results are expressed as percentage of positive. The data are representative of 2 independent experiments. (B) BCD-treated CEM-R5 cells were reloaded with chol-BCD and analyzed for intracellular calcium mobilization as described in “Materials and methods.” Results are graphed as the average change in fluorescence ratio (F340/F380) of each 30-second interval. (C) Binding of MIP-1β was analyzed after BCD treatment followed by reloading with cholesterol (chol-BCD) or 4-cholesten-3-one (4C3O). MFI of each population is expressed on the graph.
Reloading of cholesterol restores MIP-1β binding and calcium mobilization to BCD-treated cells.
(A) CEM-R5 cells were treated for the indicated times with BCD before washing with PBS. Treated cells were then reloaded with chol-BCD for 10 or 30 minutes, and the level of MIP-1β binding was determined as described in “Materials and methods.” Results are expressed as percentage of positive. The data are representative of 2 independent experiments. (B) BCD-treated CEM-R5 cells were reloaded with chol-BCD and analyzed for intracellular calcium mobilization as described in “Materials and methods.” Results are graphed as the average change in fluorescence ratio (F340/F380) of each 30-second interval. (C) Binding of MIP-1β was analyzed after BCD treatment followed by reloading with cholesterol (chol-BCD) or 4-cholesten-3-one (4C3O). MFI of each population is expressed on the graph.
BCD treatment of T cells decreases mAb binding to CCR5
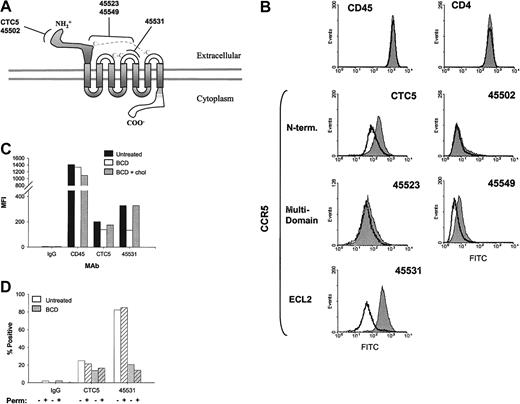
On the basis of the above results, we speculated that the loss in MIP-1β binding and subsequent signaling in BCD-treated T cells may be due to changes in CCR5 conformation. In an effort to analyze conformational changes in CCR5, we used a panel of anti-CCR5 antibodies with well-characterized binding epitopes. These antibodies were previously distinguished by their binding or lack of binding to CCR5/CCR2 chimeras with interchanged N-termini or extracellular loops (ECLs).26 More specifically, we analyzed the binding of 2 N-terminus–specific mAbs, CTC5 and 45502, an ECL2-specific mAb, 45531, and 2 multidomain-specific mAbs, 45523 and 45549 (Figure4A). In the current analysis, we attempted to determine if antibodies recognizing different regions of the CCR5 molecule would exhibit distinct alterations in binding after cholesterol extraction.
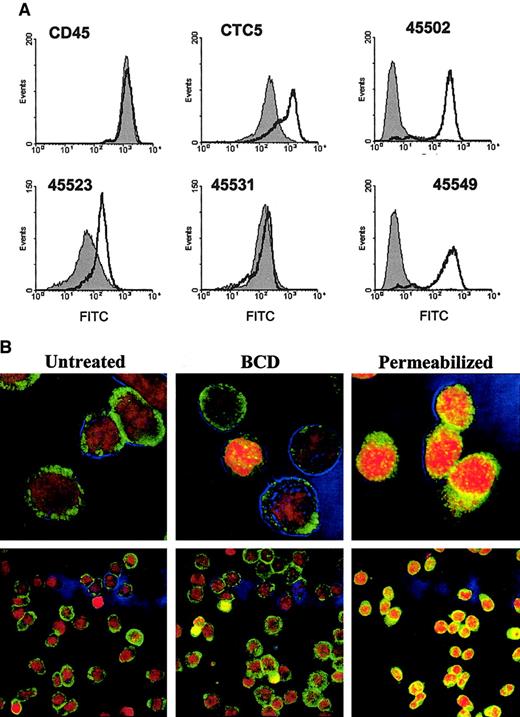
Anti-CCR5 mAb binding is decreased with BCD-treated cells.
(A) CCR5 epitopes recognized by mAbs, clones CTC5, 45502, 45523, 45531, and 45549, are diagrammed. (B) CEM-R5 cells were treated with BCD and subsequently analyzed by flow cytometric analysis as described in “Materials and methods.” The results for untreated cells are graphed as a filled peak, and BCD-treated cells are graphed as a solid line. (C) CEM-R5 cells treated with BCD were then reloaded with cholesterol and subsequently analyzed for binding of control IgG2a, anti-CD45, and anti-CCR5 mAbs, CTC5 and 45531. Results are expressed as MFI from one representative experiment of 3. (D) BCD-treated CEM-R5 cells were incubated with either CTC5 or 45531. Cells were subsequently washed, fixed, and permeabilized or simply treated with PBS, containing 2% FBS, and then stained with GAM-AF488. The percentage of positive is expressed on the graph. Negative control mouse IgG2a binding to nonpermeabilized untreated cells was gated as 2% positive. Crosshatched bars represent permeabilized cells and noncrosshatched bars represent nonpermeabilized cells.
Anti-CCR5 mAb binding is decreased with BCD-treated cells.
(A) CCR5 epitopes recognized by mAbs, clones CTC5, 45502, 45523, 45531, and 45549, are diagrammed. (B) CEM-R5 cells were treated with BCD and subsequently analyzed by flow cytometric analysis as described in “Materials and methods.” The results for untreated cells are graphed as a filled peak, and BCD-treated cells are graphed as a solid line. (C) CEM-R5 cells treated with BCD were then reloaded with cholesterol and subsequently analyzed for binding of control IgG2a, anti-CD45, and anti-CCR5 mAbs, CTC5 and 45531. Results are expressed as MFI from one representative experiment of 3. (D) BCD-treated CEM-R5 cells were incubated with either CTC5 or 45531. Cells were subsequently washed, fixed, and permeabilized or simply treated with PBS, containing 2% FBS, and then stained with GAM-AF488. The percentage of positive is expressed on the graph. Negative control mouse IgG2a binding to nonpermeabilized untreated cells was gated as 2% positive. Crosshatched bars represent permeabilized cells and noncrosshatched bars represent nonpermeabilized cells.
The results shown in Figure 4B demonstrate that all of our anti-CCR5 antibodies exhibited diminished binding to varying degrees on BCD-treated CEM-R5 cells. Shifts in the anti-CCR5+population curves can clearly be observed in Figure 4B. Table1 expresses the mean fluorescence intensity and percentage of positive gated T cells in this experiment. The control mAbs, anti-CD4 and anti-CD45, did not show any change in mean fluorescence intensity (MFI) and percentage positive. In terms of percentage reduction in MFI, all our anti-CCR5 mAbs exhibited a decrease in CCR5 binding, CTC5 (50.5), 45502 (24.7), 45523 (23.2), 45549 (40.0), and 45531 (85.7). As expected, BCD treatment failed to decrease the overall binding of HIV-1BaL gp120 to these cells, suggesting that CD4 remains intact and at functional levels on the cell surface (data not shown). Similarly, another study by Mañes et al27demonstrated that HIV-1IIIB recombinant gp120 binding to CD4+ MT2 cells was unaffected by BCD treatment. The differences in epitope-specific losses in anti-CCR5 binding suggest that the overall receptor conformation has been altered by the loss of cholesterol, whereas CD4 and CD45 epitopes remain unchanged.
Mean fluorescence intensity values and percentage positive for mAb staining on BCD-treated cells
| mAb specificity . | mAb clone . | MFI (% positive) . | |
|---|---|---|---|
| Control . | BCD . | ||
| Nonspecific | mIgG2b | 2.92 (2.10) | 3.18 (4.87) |
| CD4 | RPA-T4 | 403.9 (100) | 410.9 (99.9) |
| CD45 | HI30 | 1425 (100) | 1563 (100) |
| CCR5, N-terminal | CTC5 | 214.8 (100) | 106.3 (99.1) |
| CCR5, N-terminal | 45502 | 10.03 (69.0) | 7.55 (58.7) |
| CCR5, MD | 45523 | 56.8 (97.8) | 43.6 (97.8) |
| CCR5, MD | 45549 | 8.54 (77.2) | 5.16 (32.8) |
| CCR5, ECL2 | 45531 | 393.6 (99.5) | 56.1 (98.3) |
| mAb specificity . | mAb clone . | MFI (% positive) . | |
|---|---|---|---|
| Control . | BCD . | ||
| Nonspecific | mIgG2b | 2.92 (2.10) | 3.18 (4.87) |
| CD4 | RPA-T4 | 403.9 (100) | 410.9 (99.9) |
| CD45 | HI30 | 1425 (100) | 1563 (100) |
| CCR5, N-terminal | CTC5 | 214.8 (100) | 106.3 (99.1) |
| CCR5, N-terminal | 45502 | 10.03 (69.0) | 7.55 (58.7) |
| CCR5, MD | 45523 | 56.8 (97.8) | 43.6 (97.8) |
| CCR5, MD | 45549 | 8.54 (77.2) | 5.16 (32.8) |
| CCR5, ECL2 | 45531 | 393.6 (99.5) | 56.1 (98.3) |
MFI, mean fluorescence intensity; mAb, monoclonal antibody; BCD, hydroxypropyl-β-cyclodextrin; MD, multidomain; ECL, extracellular loop.
Cells were treated with 20 mM BCD or untreated in phosphate-buffered saline for 1 hour at 37°C. The cells were then washed and stained for flow cytometry as described in “Materials and methods.” The data presented here are from histograms in Figure 4.
To examine the specificity of cholesterol in these results, we once again reloaded BCD-treated T cells with chol-BCD and then examined anti-CCR5 mAb binding. Reloading T cells with cholesterol restored binding of the ECL2-specific mAb, 45531, to 100% of control, but the amino terminal–specific mAb, CTC5, binding was only restored to 87% (Figure 4C). Interestingly, anti-CD45 binding was not increased by reloading of cholesterol, suggesting that the changes in anti-CD45 binding seen with BCD treatment may be noncholesterol specific or may be affected in a way that cannot be restored by exogenous cholesterol.
We next examined if BCD treatment of T cells rendered them more susceptible to anti-CCR5 mAb-induced receptor internalization. As the staining of cells occurred at 4°C in our assays, we would expect very little internalization within our treatment groups. However, to directly examine this question, we stained BCD-treated cells with mAbs, fixed, permeabilized, and then added GAM-AF488. BCD treatment demonstrated no significant difference in mAb detection between permeabilized and nonpermeabilized cells, indicating that very little internalization had occurred over the treatment periods (Figure 4D).
Paraformaldehyde fixation prior to mAb staining enhances binding of some anti-CCR5 mAbs on BCD-treated cells
To confirm that CCR5 conformation was being altered and not internalized during BCD treatments, we mildly fixed the CEM-R5 cells with paraformaldehyde immediately after the BCD treatments. The results shown in Figure 5 and Table2 demonstrate an increase in the overall binding of the anti-CCR5 mAbs after BCD treatment of cells. The MFI of CTC5 on BCD-fixed cells increased 400% from the control-fixed population. More surprisingly, 45502 and 45549 MFI increased from 5.71 to 251 and from 5.8 to 241, respectively, resulting in a more than 40-fold increase in MFI after treatment/fixation. Interestingly, these 2 mAbs, 45502 and 45549, of all the CCR5 mAbs, have the least binding to control-fixed or unfixed CEM-R5 cells. In addition, there was also a 3-fold increase in the MFI of multidomain-specific mAb, 45523. These results were quite reproducible and specific, as there was no difference in alterations in the binding of the ECL2-specific mAb, 45531, between control-fixed and BCD-fixed cells. To visually confirm that the BCD-fixation treatments did not detect internalized receptors, we performed confocal microscopy on treated cells. We used the binding of a nonmembrane permeant DNA binding dye, PI, to evaluate membrane permeabilization on untreated, 10 mM BCD-treated, and 0.1% Triton X-100 treated CEM-R5 cells. We found that BCD treatment did not significantly increase the number of cells stained positive for PI, nor did it increase mAb 45531 staining of cytoplasmic receptors (Figure 5B). However, our positive control for permeabilization showed 100% cells nuclear stained with PI and cytoplasmic staining for 45531 mAb (Figure 5B).
Paraformaldehyde fixation prior to mAb staining enhances binding of certain epitope-specific mAbs to CCR5 on BCD-treated cells.
(A) BCD-treated CEM-R5 cells were fixed with paraformaldehyde immediately after treatment and then examined with the indicated antibodies using flow cytometric analysis. The results for untreated cells are expressed in a filled area peak and the BCD-treated cells are graphed using a solid line. (B) Untreated, 10 mM BCD-treated, or 0.1% Triton X-100 permeabilized cells were stained with anti-CCR5 (45531) and PI as described in “Materials and methods.” CCR5 staining is shown in green, nuclear PI staining is shown in red, and the blue outlines are phase-contrast overlays. Bright yellows or bright reds indicate permeabilized cells. The bottom picture shows a wider view of the cell population for each treatment. Magnification, × 630.
Paraformaldehyde fixation prior to mAb staining enhances binding of certain epitope-specific mAbs to CCR5 on BCD-treated cells.
(A) BCD-treated CEM-R5 cells were fixed with paraformaldehyde immediately after treatment and then examined with the indicated antibodies using flow cytometric analysis. The results for untreated cells are expressed in a filled area peak and the BCD-treated cells are graphed using a solid line. (B) Untreated, 10 mM BCD-treated, or 0.1% Triton X-100 permeabilized cells were stained with anti-CCR5 (45531) and PI as described in “Materials and methods.” CCR5 staining is shown in green, nuclear PI staining is shown in red, and the blue outlines are phase-contrast overlays. Bright yellows or bright reds indicate permeabilized cells. The bottom picture shows a wider view of the cell population for each treatment. Magnification, × 630.
Mean fluorescence intensity values and percentage positive for mAb staining on BCD-treated cells fixed with paraformaldehyde
| mAb specificity . | . | MFI (% positive) . | |
|---|---|---|---|
| mAb clone . | Control . | BCD . | |
| Nonspecific | mIgG2b | 3.54 (2.0) | 27.4 (30.9) |
| CD45 | HI30 | 1116 (100) | 1249 (100) |
| CCR5, N-terminal | CTC5 | 192 (98.7) | 766.9 (99.9) |
| CCR5, N-terminal | 45502 | 5.71 (15.7) | 250.6 (96.0) |
| CCR5, MD | 45523 | 57.69 (94.8) | 166 (99.7) |
| CCR5, MD | 45549 | 5.84 (22.1) | 241 (96.1) |
| CCR5, ECL2 | 45531 | 123.6 (97.9) | 132.8 (99.6) |
| mAb specificity . | . | MFI (% positive) . | |
|---|---|---|---|
| mAb clone . | Control . | BCD . | |
| Nonspecific | mIgG2b | 3.54 (2.0) | 27.4 (30.9) |
| CD45 | HI30 | 1116 (100) | 1249 (100) |
| CCR5, N-terminal | CTC5 | 192 (98.7) | 766.9 (99.9) |
| CCR5, N-terminal | 45502 | 5.71 (15.7) | 250.6 (96.0) |
| CCR5, MD | 45523 | 57.69 (94.8) | 166 (99.7) |
| CCR5, MD | 45549 | 5.84 (22.1) | 241 (96.1) |
| CCR5, ECL2 | 45531 | 123.6 (97.9) | 132.8 (99.6) |
MFI, mean fluorescence intensity; mAb, monoclonal antibodies; BCD, beta-cyclodextrin; MD, ; ECL, extracellular loop.
Cells were treated with BCD as described in “Materials and methods” and then fixed with 2% paraformaldehyde prior to staining with mAbs. The data presented here are from histograms in Figure 5.
MIP-1β binding colocalizes with the lipid raft marker, GM1
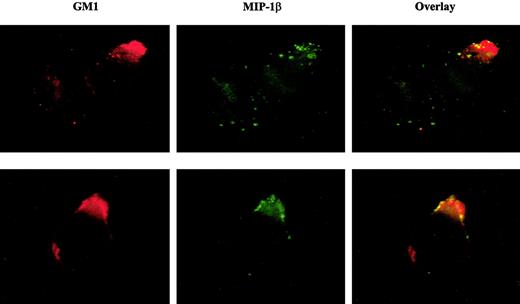
We next examined if MIP-1β binding to T cells corresponded to CCR5 localization within lipid rafts rather than nonraft CCR5. If cholesterol is essential to ligand binding, we suspected that lipid rafts might be the preferred site for MIP-1β binding, presuming that CCR5 outside of rafts may have less interaction with cholesterol. The results in Figure 6 show immunofluorescence microscopy images of CEM-R5 cells treated with biotinylated MIP-1β followed by avidin-FITC, and then overlaid with Alexa Fluor 596 CT-B conjugate. The ganglioside, GM1, has been extensively used as a specific marker for lipid rafts and is recognized by pentameric CT-B. A significant overlap in staining between MIP-1β and GM1 was observed on CEM-R5 cells, suggesting that functional CCR5 or higher affinity CCR5 preferentially localized to lipid rafts on the T-cell surface. Dense patches of MIP-1β bound to the cell surface would yield a brighter signal in raft areas in which CCR5 may be more dense. However, we want to point out that diffuse binding of MIP-1β in nonraft areas may not be as evident, given the diminished signal of such staining in nonraft less dense areas of the cell.
Fluorescently labeled MIP-1β colocalizes with cell membrane GM1.
CEM-R5 cells were stained simultaneously with CT-B–AF594 and biotinylated–MIP-1β followed by avidin-fluorescein (FITC). The MIP-1β binding is shown in green and the CT-B–AF594 staining (equivalent to Texas Red) is shown in red. Colocalization is indicated by a yellow and/or orange color in the overlay panel. Representative examples of 2 images are presented. Magnification, × 500.
Fluorescently labeled MIP-1β colocalizes with cell membrane GM1.
CEM-R5 cells were stained simultaneously with CT-B–AF594 and biotinylated–MIP-1β followed by avidin-fluorescein (FITC). The MIP-1β binding is shown in green and the CT-B–AF594 staining (equivalent to Texas Red) is shown in red. Colocalization is indicated by a yellow and/or orange color in the overlay panel. Representative examples of 2 images are presented. Magnification, × 500.
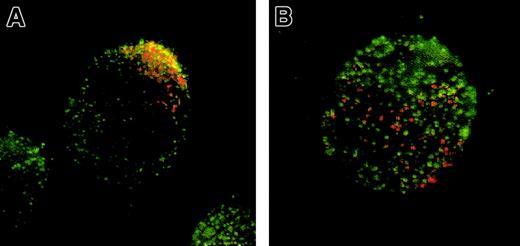
Anti-CCR5 staining partially colocalizes with CT-B labeled GM1
Previous raft isolation studies have shown only a partial partitioning of CCR5 to lipid raft fractions.16 We wanted to confirm and extend these results by using microscopy with our CEM-R5 cells. CT-B, as described earlier, recognizes ganglioside GM1 and partitions to lipid rafts. In our culture of CEM-R5 cells, about 25% of cells display a capped appearance for lipid rafts, visualized either as a crescent staining or a distinct uropod projection. In Figure7, we present images of capped cells compared with noncapped cells. Panels A and B were stained for GM1 with CT-B and CCR5 with mAb, CTC5. In capped cells, CCR5 appears to colocalize with GM1 (Figure 7A), but in noncapped cells GM1 remains in distinct microdomains that do not colocalize with CCR5 (Figure 7B). Our results demonstrate that normal CCR5 localization in noncapped cells appears to be in non-GM1 areas, presumably outside of rafts. However, binding of MIP-1β seems to be specific to capped cells in domains that are enriched in GM1. Together, these results support a role for cholesterol in the function and conformation of CCR5 in MIP-1β binding and signaling in T cells.
Anti-CCR5 staining partially colocalizes with GM1.
CEM-R5 cells were stained with CT-B–AF594 followed by staining with the anti-CCR5 mAb, CTC5, in combination with GAM-AF488 (A,B). AF488 fluorescence is shown in green and CT-B–AF596 fluorescence is shown in red. Panel A represents a subpopulation of capped cells observed within the stained cells, whereas panel B represents a subpopulation of noncapped cells. As in Figure 6, colocalization is indicated by yellow and/or orange color. Magnification, × 500.
Anti-CCR5 staining partially colocalizes with GM1.
CEM-R5 cells were stained with CT-B–AF594 followed by staining with the anti-CCR5 mAb, CTC5, in combination with GAM-AF488 (A,B). AF488 fluorescence is shown in green and CT-B–AF596 fluorescence is shown in red. Panel A represents a subpopulation of capped cells observed within the stained cells, whereas panel B represents a subpopulation of noncapped cells. As in Figure 6, colocalization is indicated by yellow and/or orange color. Magnification, × 500.
Discussion
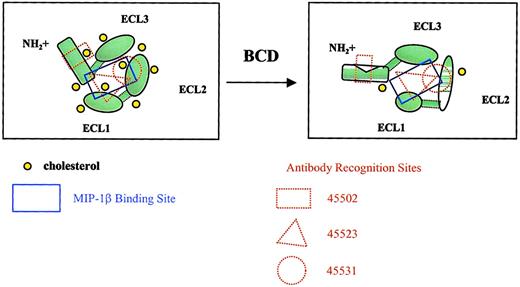
In this current report, we have provided evidence for an essential role for cholesterol in structural conformation and ligand binding activity of CCR5. MIP-1β binding was dramatically reduced by BCD treatment of cells; however, this binding was just as rapidly restored by reloading the treated cells with cholesterol. These functional alterations were directly attributable to membrane cholesterol and could not be simply explained by cell death or internalization. The rapid loss in MIP-1β binding within 10 to 20 minutes of BCD treatment strongly supports the sensitivity of CCR5 conformation rather than increased receptor internalization in response to cholesterol extraction. Several anti-CCR5 antibodies were capable of detecting CCR5 epitopes on the surface of BCD-fixed cells after 1 hour of treatment. This loss of CCR5 ligand binding after BCD treatment was also supported by a loss of intracellular calcium mobilization in response to MIP-1β and RANTES. A model for the loss of MIP-1β binding and receptor conformational change because of BCD treatment is depicted in Figure8.
Model for BCD effects on CCR5 conformation.
The diagrams show CCR5 from a top view, as if looking down onto a cell membrane. Cholesterol normally interacts with transmembrane domains of CCR5, providing a relatively rigid structure capable of binding MIP-1β. Cholesterol extraction by BCD alters the overall conformation of the receptor, changing the MIP-1β binding site and epitopes required for mAb recognition. The increase in membrane fluidity is also likely to increase protein fluidity, resulting in poor mAb binding. Individual loops may also have altered conformations as a result of cholesterol extraction. With fixation, protein rigidity is restored, which in turn restores mAb binding.
Model for BCD effects on CCR5 conformation.
The diagrams show CCR5 from a top view, as if looking down onto a cell membrane. Cholesterol normally interacts with transmembrane domains of CCR5, providing a relatively rigid structure capable of binding MIP-1β. Cholesterol extraction by BCD alters the overall conformation of the receptor, changing the MIP-1β binding site and epitopes required for mAb recognition. The increase in membrane fluidity is also likely to increase protein fluidity, resulting in poor mAb binding. Individual loops may also have altered conformations as a result of cholesterol extraction. With fixation, protein rigidity is restored, which in turn restores mAb binding.
Alterations in antibody binding to cell surface CCR5 after treatment with BCD provide interesting insights into the role of cholesterol in CCR5 conformational states. Although each of the antibodies used may be well characterized, very few antibodies to chemokine receptors recognize linear epitopes, borne out by the fact that most antibodies do not recognize denatured CCR5 in Western blot analysis.26 Lee et al26 found that even CTC5, which recognizes denatured CCR5 in Western blots, may not recognize a linear epitope because a 9–amino acid HA tag added to the N-terminus of CCR5 eliminates binding. The binding of 2 mAbs, 45502 and 45549, was dramatically increased (> 40-fold in MFI) when BCD-treated cells were fixed with paraformaldehyde. We suspect that these 2 mAbs recognize a form of CCR5 that has low levels of interaction with cholesterol. We believe that on any given cell membrane, CCR5 may exhibit different levels of cholesterol interaction. These antibodies may not only be recognizing different regions of the CCR5 molecule but also different conformation states based on their interactions directly with cholesterol or the interactions of CCR5 with cholesterol. For example, under normal conditions, there appears to be sufficient levels of cholesterol to maintain a high-cholesterol binding version of CCR5 (hi-chol-R5) in which the N-terminal–specific mAb, 45502, and the multidomain-specific mAb, 45549, would exhibit low levels of receptor binding and the ECL2-specific mAb, 44531, would exhibit a high level of binding. After BCD treatment, membrane fluidity is dramatically increased, thereby increasing CCR5 “floppiness” and decreasing binding of most of the antibodies. This finding would be supported by our data, demonstrating a significant loss in 45531 mAb binding that prefers hi-chol-R5. However, on paraformaldehyde fixation, the CCR5 molecule is no longer fluid and the low-chol-R5 version becomes preserved. This finding is supported by our results in Figure 5 and Table 2 that demonstrate a dramatic increase in binding of the low-chol-R5–recognizing mAbs, 45502 and 45549, even above that of the hi-chol-R5 mAb, 45531, under normal conditions. Overall, the conformational changes in CCR5 with BCD treatment are really too complex to be studied by simply examining changes in cell surface binding profiles by using several receptor-specific antibodies. However, we can conclude that the variations in epitope-specific antibody and ligand binding support the increased susceptibility to conformational changes of certain epitopes of CCR5 after cholesterol extraction.
Our immunofluorescence images provide 2 important findings. First, CCR5 and GM1 appear to colocalize in capped cells but are in distinct regions in noncapped cells. Cells exhibiting a “capped” phenotype demonstrated colocalization of CCR5 and GM1 similar to previous results using HELA and human breast carcinoma cells.17 27 These results suggest that cellular polarization or capping of suspension cells is required for CCR5 colocalization with GM1. Second, the majority of bound MIP-1β appeared to be colocalized to lipid rafts stained with CT-B, suggesting the presence of high-affinity CCR5 within the lipid rafts. Thus, it would appear that GM1-enriched lipid rafts are the primary sites for MIP-1β binding on human T cells. Moreover, receptor engagement with ligand may recruit CCR5 into GM1-enriched lipid rafts, resulting in colocalization and perhaps more efficient binding and signaling.
Multiple palmitoylation of cysteine residues is believed to be a mechanism to target transmembrane proteins to lipid rafts, likely because of the highly ordered lipid-packing properties of cholesterol and palmitoyl chains.12 CCR5 has been found to be palmitoylated at cysteine residues 321, 323, and 324 within the cytoplasmic tail, resulting in the formation of a fourth intracellular loop.17-19 Palmitoylation-deficient mutants of CCR5 that retain a similar binding capacity to MIP-1β compared with wild-type controls exhibited a diminished association with lipid rafts on transfected HELA cells compared with controls by confocal microscopic examination.17 Nevertheless, the presence of cholesterol alone in the membrane may be sufficient to provide proper conformation of palmitoylation-deficient CCR5 allowing MIP-1β binding. Interestingly, the T-cell tropic HIV coreceptor, CXCR4, possesses no palmitoylation sites, yet has been found to be colocalized with lipid rafts enriched in GM3 but not GM1.28 We have found that BCD treatment has similar effects on SDF-1α binding to CXCR4 that parallel our current findings with MIP-1β.42Although the colocalization of other chemokine receptors within lipid rafts has yet to be established, certain receptors, including CCR2b, CCR4, CCR6, CCR7, CCR8, CCR9, CCR10, CX3CR1, CXCR3, and CXCR5, possess potential palmitoylation sites in their carboxy-termini, suggesting possible raft association.
Transmission and early stages of HIV infection are predominated by CCR5-using viruses.29 Our results here have direct implications for the modulation of cholesterol in preventing HIV transmission. Recent studies have suggested that lipid rafts serve as the site for HIV fusion and infection based on interactions of gp120 with lipid raft components, such as glycolipids and CD4.30The inhibitory effects of cholesterol depletion on HIV infectivity are believed to be due to the disruption of lipid rafts, thereby preventing recruitment of chemokine receptors into receptor assemblies with CD4.27,31 We believe that lipid rafts and cholesterol may be playing a more significant role in HIV infection than serving as sites for protein recruitment. On the basis of the data presented here, cholesterol is not only necessary for lipid raft integrity but also appears to be important for CCR5 conformational stability. Recent studies have also implicated cholesterol-dense lipid rafts as the sites for concentration of SNAREs (solubleN-ethylmaleimide-sensitive factor attachment protein receptors), which are responsible for initiating fusion of exocytic vesicles with the cell membrane.32 Given these receptor-raft associations, it is not surprising that the HIV fusion process would require cholesterol-enriched patches dense with chemokine receptors. Similarly, fusion of the human T-cell leukemia virus type 1 (HTLV-1) may also be mediated through lipid rafts, as 4 mAbs that recognize lipid raft proteins were also found to block HTLV-1 syncytium formation.33 Other studies have demonstrated that lipid rafts are the areas of the cell membrane where newly formed virions of HIV assemble and bud.34 Several other viruses, including influenza, polio, measles, and Sendai virus, may similarly bud from lipid rafts.35-38 In fact, recent studies suggest that the ability to create “pseudotyped” virus particles that contain envelope molecules and structural proteins from different viruses is a consequence of the preferential budding of many virus types from lipid rafts.39 Localized targeting of raft function by cholesterol extraction may prove to be effective in preventing HIV transmission.
Overall, our results have numerous implications for chemokine receptor function outside of the HIV arena. Several inflammatory disease states have been associated with alterations in chemokine production and responses, including cardiovascular disease, asthma, multiple sclerosis, cancer, and arthritis.40 Modulation of the responses to chemokines could benefit individuals with many of these diseases. In addition, alterations in membrane cholesterol may hinder one's ability to mount an optimal immune response to a pathogenic or antigen challenge. Similarly, modifications of cholesterol as seen with cholesterol oxidation on immune cell populations may prevent optimal cellular recruitment into inflammatory sites or sites of vaccine administration. Our results with reloading 4-cholesten-3-one suggest that oxidized cholesterol may hinder the function of chemokine receptor by altering receptor conformation and inhibiting chemokine binding, similar to that seen with the galanin receptor.23 This finding may also apply to aging immune cells, as increases in lipid oxidation have been associated with process of aging.41Restoration of immune cells with normal nonoxidized cholesterol may potentially improve dampened chemokine and antigen responses in aging cells that may possess higher levels of oxidized cholesterol than cells from their younger counterparts. Understanding the membrane mechanisms and requirements involved in chemokine receptor function may lead to a number of possible therapeutic interventions to modulate immune responses and HIV pathogenesis.
We thank Christa Morris, Dr Robert Wersto, and Francis J. Chrest of the NIA Flow Cytometry Laboratory, and Magdalena Juhazsova of the NIA Confocal Imaging Facility for their assistance. We also thank Dr Eric Schaffer, Dr Valeria Coelho, and Deborah Nguyen for helpful discussions.
Prepublished online as Blood First Edition Paper, April 17, 2002; DOI 10.1182/blood-2001-11-0087.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Dennis Taub, National Institute on Aging, 5600 Nathan Shock Dr, Baltimore, MD 21224; e-mail: taubd@grc.nia.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal