Members of the Notch family encode transmembrane receptors that modulate differentiation, proliferation, and apoptotic programs of many precursor cells, including hematopoietic progenitors. Stimulation of Notch causes cleavage followed by translocation of the intracellular domain (NotchIC) to the nucleus, where it activates transcription of CBF1 responsive genes. The aim of this study was to elucidate the mechanisms leading to the overexpression of CD23, a striking feature of B-cell chronic lymphocytic leukemia (B-CLL) cells. By electrophoretic mobility shift assays, we identified a transcription factor complex (C1) that binds sequence specific to one known and 4 newly identified putative CBF1 recognition sites in the CD23a core promoter region. With the use of Epstein-Barr virus (EBV)–infected B cells as a model for CBF1 mediated CD23a expression, C1 was found to be EBV inducible. Supershift assays revealed that the nuclear form of Notch2 is a component of C1 in B-CLL cells, supporting a model in which NotchIC activates transcription by binding to CBF1 tethered to DNA. Transient transfection of REH pre–B cells with an activated form of Notch2 induced endogenous CD23a, confirming thatCD23a is a target gene of Notch2 signaling. Finally, reverse transcription-polymerase chain reaction and kinetic analysis demonstrated that the Notch2 oncogene is not only overexpressed in B-CLL cells but might also be related to the failure of apoptosis characteristic for this disease. In conclusion, these data suggest that deregulation of Notch2 signaling is involved in the aberrant expression of CD23 in B-CLL.
Introduction
B-cell chronic lymphocytic leukemia (B-CLL) is characterized by the relentless accumulation of monoclonal anergic B cells that coexpress CD5 and CD19 with faint to virtually undetectable amounts of surface immunoglobulins.1,2 The pathomechanisms underlying the disease primarily involve defects that prevent cell turnover because of apoptosis, rather than alterations in cell cycle regulation.1 2
One of the hallmarks of B-CLL cells is the overexpression of the transmembrane glycoprotein CD23,1-3 which undergoes spontaneous proteolysis, giving rise to soluble CD23 (sCD23) molecules.4 The serum concentration of sCD23 can be several hundred-fold higher than in healthy individuals and parallels the clinical stage of the disease.5 6
Two isoforms of CD23 exist, CD23a and CD23b, which are expressed from 2 different promoters.7 Expression of CD23a is restricted to B lymphocytes, whereas CD23b is found on a number of different hematopoietic cell types, predominantly after interleukin 4 (IL-4) treatment.8 In B-CLL cells, selective expression ofCD23a has been found to be concurrent to a state of cell survival, thereby providing a link between CD23a and the regulation of apoptosis.9
CD23 is also closely associated with Epstein-Barr virus (EBV)–mediated B-cell immortalization, because a naturally occurring EBV mutant (P3HR1), carrying a deletion of the EBNA2 gene, has lost its ability to induce CD23 expression and to transform primary B cells in vitro.10-12 EBNA2 activates the CD23a gene through a CBF1 repressor site located in the CD23a proximal promoter.13-15 Several lines of evidence strongly suggest that EBNA2 mimics Notch signaling by acting as a transcriptional activator after binding and masking the repression domain of CBF1.16-19
The Notch gene family encodes transmembrane receptors that modulate differentiation, proliferation, and apoptotic programs in response to extracellular ligands expressed on neighboring cells.20,21Ligand-mediated stimulation of Notch causes the proteolytic release of the intracellular domain (NotchIC), which then passes into the nucleus where it activates transcription of CBF1 responsive genes.22-25 Deregulation of this pathway by overexpression of a constitutively activated form of Notch not only diverts cell fate decisions but is also tumorigenic. For example, truncation ofNotch1 found in a subset of human T-cell leukemias leads to the expression of a ligand-independent oncogenic protein lacking the extracellular domain.26 The truncated Notch proteins consist primarily of the intracellular domain and localize predominantly in the nucleus. Enforced expression of Notch1IC in bone marrow stem cells causes T-cell leukemia in mice, indicating a causative role for Notch1 in T-cell oncogenesis.27
To elucidate the mechanisms leading to the up-regulation ofCD23a in B-CLL cells, we analyzed the CD23aproximal promoter for sequence-specific DNA-protein interactions. By electrophoretic mobility shift assays (EMSAs), we detected a transcription factor complex containing Notch2IC that binds to 5 different CBF1 responsive elements. Our data indicate thatNotch2 is overexpressed in B-CLL cells, suggesting that deregulation of Notch2 signaling is involved in the pathogenesis of this disease.
Materials and methods
Cell lines and culture
Peripheral blood mononuclear cells (PBMCs) from 6 patients with typical B-CLL (CD5+/CD19+/CD23++) and from one patient with atypical B-CLL with low CD23 expression (referred to as B-CLLCD23+/−) were isolated by using Ficoll-Hypaque (Pharmacia, Uppsala, Sweden) centrifugation. Tonsillar B cells were obtained after E-rosetting of T cells by using neuraminidase-treated sheep erythrocytes. The monocytes and macrophages were removed from lymphoid cells by adherence to plastic culture dishes. Resting peripheral blood B cells and Th cells from 3 healthy donors were obtained by immunoaffinity purification (CellPro, Bothell, WA). The purity of B-cell and T-cell preparations was higher than 95%, as determined by flow cytometry. Lymphoid cell lines were cultured in RPMI 1640 (BL41, BL41/B95-8; kindly provided by Dr G. Lenoir, Lyon, France) and Iscoves medium (REH). Culture media were supplemented with 10% heat-inactivated fetal calf serum, 2 mM glutamine, 100 U/mL penicillin, and 100 mg/mL streptomycin (all reagents were obtained from Gibco, Life Technologies, Paisley, United Kingdom). For the kinetic studies, B-CLL and peripheral blood B cells were cultured under serum-free conditions. Cells were collected after 1, 2, 3, and 5 days and processed for flow cytometry and reverse-transcription-polymerase chain reaction (RT-PCR) analysis.
Flow cytometry
Stainings were performed essentially as described by Willheim et al.28 Antibodies were purchased from Becton Dickinson (San Jose, CA), with the exception of anti-bcl-2 (Dako, Copenhagen, Denmark). Species- and isotype-matched irrelevant antibodies were used as controls. Flow cytometry was performed on a FACScan using CellQuest software (Becton Dickinson). Annexin V and propidium iodide staining was performed to estimate the percentages of cells undergoing apoptosis.
Sequence analysis of the CD23apromoter
Potential transcription factor binding sites in theCD23a proximal promoter were identified by using the matrix search program MatInspector V2.2 that is based on the transcription factor database TRANSFAC 4.0 (http://transfac.gbf.de).29
Preparation of nuclear extracts and EMSA
Nuclear extracts were prepared as described with minor modifications.30 Briefly, 15 × 106 cells were lysed in 1 mL hypotonic buffer (10 mM HEPES, pH 7.9; 1.5 mM MgCl2; 10 mM KCL) containing 0.15% NP-40 at 4°C for 10 minutes. The nuclear proteins were extracted from the nuclear fraction by resuspending the nuclei in 600 μL extraction buffer (300 mM KCl; 1.5 mM MgCl2; 20 mM HEPES, pH 7.9; 0.2 mM EDTA; 25% glycerin) at 4°C for 20 minutes with constant agitation. After removing the pellet by centrifugation, the supernatant was dialyzed against dialysis buffer (20 mM HEPES, pH 7.9; 100 mM KCl, 0.2 mM EDTA, 20% glycerol) for 45 minutes. EMSA probes (for sequence information see Figure 1) were Digoxigenin end-labeled (Roche, Mannheim, Germany). The binding reaction was performed by preincubating 3 μg nuclear proteins with 2.5 μg poly(dI-dC) in a buffer containing 10 mM HEPES, pH 7.9; 80 mM KCl, and 10% glycerol for 10 minutes at room temperature. All buffers were supplemented with 0.5 mM dithiothreitol, 0.2 mM phenylmethyl sulfonyl fluoride, and complete protease inhibitor cocktail (Roche, Mannheim, Germany). Competition experiments were performed by the addition of 2.5-, 5-, 25-, 125-fold excess of unlabeled probe EBV.Cp (5′-TGGTGTAAACACGCCGTGGGAAAAAATTTA-3′) and CBF1.2 (5′-CTCTAGTTCTCACCCAATTC-3′) to the binding reactions. All oligonucleotides were annealed with the respective reverse complementary strands. For immunodetection of DNA binding proteins, antisera were incubated with nuclear extracts 30 minutes at 4°C before addition of probe DNA. Anti-Pax5 (BSAP)-purified polyclonal immunoglobulin (Ig)G was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The Notch1IC (bTAN 20) and Notch2IC (C651.6DbHN) antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Science, Iowa City, IA. End-labeled probes (30 fmol) were added to the reaction mixture and incubated for an additional 15 minutes at room temperature.
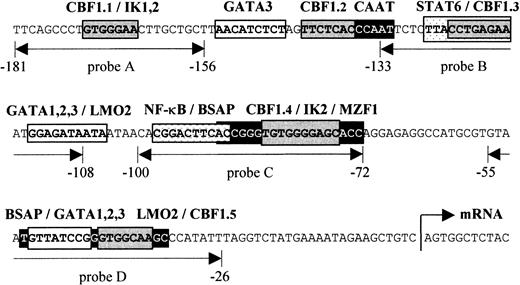
Schematic presentation of the human
CD23a proximal promoter. Sequence analysis of theCD23a core promoter region revealed a complex array of potential regulatory elements, including CBF1, STAT6, NF-κB, Pax5 (BSAP), Ikaros, GATA1-3, LMO2, and MZF1. EMSA probes are indicated as double-headed arrows below the sequence. The relative position of the transcriptional start site (arrow) is indicated according to Gene Bank (EMBL) accession no. X06049.
Schematic presentation of the human
CD23a proximal promoter. Sequence analysis of theCD23a core promoter region revealed a complex array of potential regulatory elements, including CBF1, STAT6, NF-κB, Pax5 (BSAP), Ikaros, GATA1-3, LMO2, and MZF1. EMSA probes are indicated as double-headed arrows below the sequence. The relative position of the transcriptional start site (arrow) is indicated according to Gene Bank (EMBL) accession no. X06049.
Transient transfection experiments
REH pre–B cells were transiently transfected by electroporation. Cells (1-2 × 106) were resuspended in 500 μL serum-free medium and were mixed with 10 μg plasmid DNA (pSG5-rN2IC and pCMV-green fluorescent protein [GFP], respectively) in 0.4-cm electroporation cuvettes. Electroporation was carried out after 5 minutes of incubation on ice on a BioRad gene pulser set at 950 μF and 280 V. Immediately after pulsing, cells were transferred to 4 mL culture medium and were incubated for 48 hours. Transfection efficiency was determined by quantification of GFP-positive cells by using flow cytometry analysis.
Determination of sCD23 concentration
Blood was obtained by venipuncture. After clotting, samples were centrifuged, and serum was stored at −20°C. The levels of sCD23 from sera and culture supernatants were measured by using the Cellfree CD23 enzyme-linked immunosorbent assay kit (Endogen, Woburn, MA).
RT-PCR analysis
Total RNA was extracted by using the RNAzol isolation system (CINNA/MRC) according to the manufacturer's instructions. One-step RT-PCR reactions (Titan; Roche, Mannheim, Germany) were performed on total RNA (50 ng per reaction) by using primer sequences as follows:Notch2, forward 5′-CTGGATGCAGGTGCAGATGCCAATGC-3′ and reverse 5′-GCAGAAGTCAACACGGTGCCTGGAGG-3′ (25 cycles; annealing temperature [AT], 65°C; product size, 540 base pair [bp]); CD23a, forward 5′-ATGGAGGAAGGTCAATATTCA-3′ and reverse 5′-GCATCATCACGCAGTCCTCGC-3′ (30 cycles; AT, 45°C; product size, 790 bp). Primer sequences and conditions for bcl-2 RT-PCR were performed according to Erlacher et al.31 The integrity of the isolated RNA and efficiency of complementary DNA synthesis was assessed by amplification of β-actin by using the following primers: forward 5′-GTGGGAATTCGTCAGAAGGACTCCTATGTG-3′ and reverse 5′-GAAGTCTAGAGCAACATAGCACAGCTTCTC-3′ (25 cycles; AT, 60°C; product size, 260 bp). Consequently, laser-scanning densitometry was performed, and the amount of RNA used for detection of the target genes was accordingly adjusted. PCR products were sequenced to verify specificity of amplified DNA.
Results
CD23a promoter sequence analysis
Figure 1 depicts the sequence upstream of the transcriptional initiation site of the CD23a gene that has been shown to be sufficient for an efficient expression of CD23a in B cells.7 This region corresponds also to a nuclease hypersensitive site found in B cells but not in T cells.32In most cases such hypersensitive sites reflect the binding of transcription factors to specific DNA sequences, resulting in nucleosome displacement. Instead of a canonical TATA box, 2 putative Pax5 (BSAP) binding sites between −29 bp and −90 bp could be predicted.33 The upper Pax5 site is situated close to a previously described NF-κB–like element (−89 to −98) that has been shown to cooperate with a STAT6 site (−117 to −126) in CD40/IL-4–mediated induction of the highly homologous murineCD23 promoter.34 In addition to one known CBF1 site (CBF1.1 in Figure 1),14,15 we were able to identify 4 putative CBF1 recognition sites matching the consensus (GTGG/AGAA) in at least 6 of 7 positions.35 Furthermore, by using the matrix search program MatInspector V2.2 (Research Group Bioinformatics, Braunschweig, Germany),29 several potential binding sites for Ikaros (IK1-2), GATA1-3, LMO2, and MZF1 could be predicted.
B-CLL–specific DNA-protein interactions in the CD23a proximal promoter
To identify transcription factors implicated in the up-regulation of the CD23a gene in B-CLL cells, EMSAs were performed with 4 oligonucleotide probes derived from the CD23a core promoter region (Figure 1). Nuclear extracts were prepared from freshly isolated B-CLL cells (n = 6, Table 1), from PBMCs (n = 3), and from tonsillar B cells (n = 1). The Burkitt lymphoma cell line BL41 infected with the EBV strain B95-8 (Table 1) served as a positive control for CBF1-mediated CD23aexpression.14 15 Nuclear proteins isolated from Th cells (n = 3) were included as a negative control for B-cell– and EBV-specific transcription factors.
Serum and supernatant levels and membrane expression of CD23
| . | sCD23 (U/mL) . | % CD19+/CD23+ . | % CD19+/CD23− . | % CD19−/CD23+ . |
|---|---|---|---|---|
| BL41 | 0* | 0 | 99.9 | 0 |
| BL41/B95-8 | 285* | 38.9 | 59.1 | 0 |
| PBMC HD (n = 3) | 112 ± 8 | 4.5 ± 0.6 | 1.8 ± 0.4 | 0.6 ± 0.2 |
| CLLCD23+/− | 120 | 31.8 | 57 | 0.9 |
| CLL 1 | 998 | 85.3 | 0.7 | 0.6 |
| CLL 2 | 1670 | 90.4 | 4 | 0.9 |
| CLL 3 | 2119 | 89.5 | 1.8 | 0.4 |
| CLL 4 | 2518 | 72.2 | 6 | 4.9 |
| CLL 5 | 3551 | 91.1 | 5.9 | 0.9 |
| CLL 6 | 4310 | 93.4 | 4.7 | 0.5 |
| . | sCD23 (U/mL) . | % CD19+/CD23+ . | % CD19+/CD23− . | % CD19−/CD23+ . |
|---|---|---|---|---|
| BL41 | 0* | 0 | 99.9 | 0 |
| BL41/B95-8 | 285* | 38.9 | 59.1 | 0 |
| PBMC HD (n = 3) | 112 ± 8 | 4.5 ± 0.6 | 1.8 ± 0.4 | 0.6 ± 0.2 |
| CLLCD23+/− | 120 | 31.8 | 57 | 0.9 |
| CLL 1 | 998 | 85.3 | 0.7 | 0.6 |
| CLL 2 | 1670 | 90.4 | 4 | 0.9 |
| CLL 3 | 2119 | 89.5 | 1.8 | 0.4 |
| CLL 4 | 2518 | 72.2 | 6 | 4.9 |
| CLL 5 | 3551 | 91.1 | 5.9 | 0.9 |
| CLL 6 | 4310 | 93.4 | 4.7 | 0.5 |
The soluble CD23 (sCD23) concentrations were determined by enzyme-linked immunosorbent assay in serum obtained from 3 representative healthy donors (HDs); data represent mean ± SEM. One patient with atypical B-cell chronic lymphocytic leukemia (B-CLL) (CLLCD23+/−) and 6 patients with typical B-CLL (CLL 1-6) enrolled in this study. The percentage of CD19+ and CD23+ cells from total cell populations were determined by using flow cytometry.
PBMC indicates peripheral blood mononuclear cell.
The sCD23 levels of BL41 and of BL41/B95-8 cells were measured in supernatants after a culture period of 48 hours.
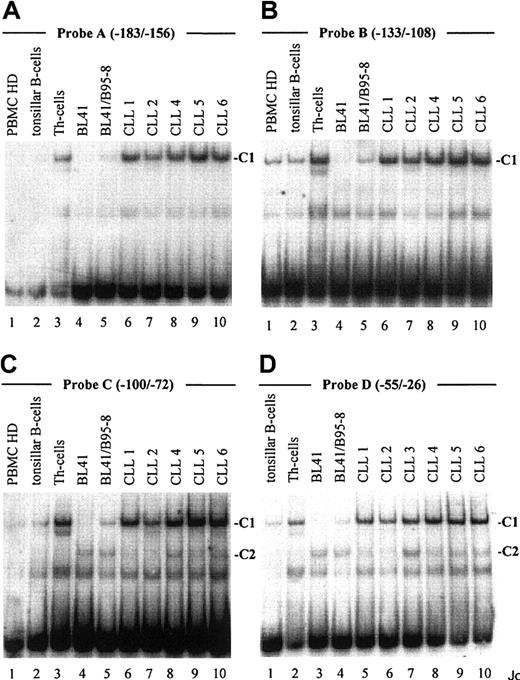
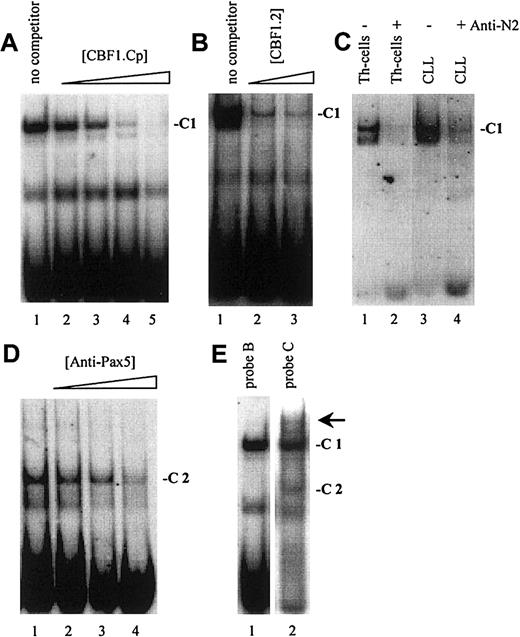
As demonstrated in Figure 2, EMSA with probe A to D, encompassing one known (Figure 1, probe A) and 3 newly identified putative CBF1 sites (Figure 1, probes B to D), visualized a prominent, slow-migrating DNA-protein complex designated as complex 1 (C1, Figure 2A-D). The significance of this complex was underlined by the fact that in all B-cell samples the intensity of C1 correlated with their respective levels of CD23a transcription. Most notable, C1 was found to be EBV inducible (Figure 2A-D, lanes 4 and 5), indicating that this complex is involved in CBF1-mediatedCD23a expression. To support this finding, EMSA was performed, including an unlabeled competitor oligonucleotide (CBF1.Cp) spanning a well-characterized CBF1 site derived from the EBV C promoter.15 C1 was completely abolished upon the addition of the competitor (Figure 3A), confirming that C1 binds sequence specific to the CBF1 recognition sites. An additional putative CBF1 site matching the consensus at 7 of 7 positions (CBF1 site 2; GTGAGAA, −137 to −143) was identified in inverted orientation closely juxtaposed to a putative CAAT-box (Figure1). A synthetic 20-bp unlabeled oligonucleotide comprising CBF1 site 2 (CBF1.2; −130 to −149) inhibited binding of C1 to probe A, indicating that C1 is also capable of interacting with CBF1 site 2 (Figure 3B).
An EBV-inducible transcription factor complex (C1) binds to 4 different oligonucleotide probes derived from the
CD23a proximal promoter. Labeled oligonucleotides spanning potential transcription factor binding sites (probe A-D, Figure 1) were analyzed in EMSA by using nuclear extracts from B-CLL cell samples (Table 1), PBMCs from healthy donors (representative for n = 3), tonsillar B cells, Th cells, BL41 cells, and BL41/B95-8 cells as indicated. Major observed complexes (C1 in A-D, C2 in C,D) are indicated. The sequences of the different oligonucleotides used as probes are given in Figure 1.
An EBV-inducible transcription factor complex (C1) binds to 4 different oligonucleotide probes derived from the
CD23a proximal promoter. Labeled oligonucleotides spanning potential transcription factor binding sites (probe A-D, Figure 1) were analyzed in EMSA by using nuclear extracts from B-CLL cell samples (Table 1), PBMCs from healthy donors (representative for n = 3), tonsillar B cells, Th cells, BL41 cells, and BL41/B95-8 cells as indicated. Major observed complexes (C1 in A-D, C2 in C,D) are indicated. The sequences of the different oligonucleotides used as probes are given in Figure 1.
Notch2 is a major part of C1 in B-CLL cells.
(A) A cold oligonucleotide (CBF1.Cp) spanning a well-characterized CBF1 site derived from the EBV C promoter competes with probe B (and also with probe A, C, D; data not shown) for binding to C1. (B) A 20-bp oligonucleotide (CBF1.2; −130 to −149) spanning CBF1 site 2 competes with probe A for binding to C1. (C) Supershift assay showing that Notch2IC is a component of C1 in B-CLL cells and in Th cells. (D) Including antibodies specific for Pax5 (BSAP) diminished the signal of C2. (E) A higher order complex (arrow) is visible with probe C (lane 2) but not with probe B (lane 1), indicating that C1 and C2 bind the shared DNA regions in a cooperative manner.
Notch2 is a major part of C1 in B-CLL cells.
(A) A cold oligonucleotide (CBF1.Cp) spanning a well-characterized CBF1 site derived from the EBV C promoter competes with probe B (and also with probe A, C, D; data not shown) for binding to C1. (B) A 20-bp oligonucleotide (CBF1.2; −130 to −149) spanning CBF1 site 2 competes with probe A for binding to C1. (C) Supershift assay showing that Notch2IC is a component of C1 in B-CLL cells and in Th cells. (D) Including antibodies specific for Pax5 (BSAP) diminished the signal of C2. (E) A higher order complex (arrow) is visible with probe C (lane 2) but not with probe B (lane 1), indicating that C1 and C2 bind the shared DNA regions in a cooperative manner.
Because B-CLL cells do not express detectable amounts of the EBV-encoded CBF1 activator EBNA2,36,37 we conducted supershift assays, including antibodies raised against the nuclear forms of Notch1 and Notch2,38 2 members of the Notch family known to be expressed in normal B cells.18,39 40Whereas antibodies specific for Notch1IC had no discernible effect on the formation or migration of C1 (data not shown), antibodies specific for Notch2IC led to a marked decrease in the intensity of C1 in Th cells and in B-CLL samples (Figure 3C). This result indicates that in B-CLL cells the nuclear form of Notch2 is a major part of the cellular activity that binds to the CBF1 recognition sites in theCD23a core promoter region.
EMSA with probe C and D (Figure 2C,D), spanning 2 predicted Pax5 sites, revealed the appearance of an additional complex, designated C2. This complex appeared in all B-cell samples (Figure 2C, lanes 4-10, and Figure 2D, lanes 3-10) but was absent in the Th-cell control (Figure2C, lane 3, and Figure 2D, lane 2). Incubation of nuclear extracts with Pax5 antibodies before the addition of probe C diminished the signal of C2 (Figure 3D), implying that this complex contains Pax5. A higher order complex was observed in all samples positive for both, C1 and C2 (Figure 3E), suggesting that these factors bind the shared DNA regions in a cooperative manner. The presence of 2 Pax5 binding sites in the −30 promoter region of the CD23a gene instead of a canonical TATA box argues for Pax5 (BSAP) as regulator of B-cell–specific CD23a expression as discussed for the CD19gene.41
Notch2 is overexpressed in B-CLL cells
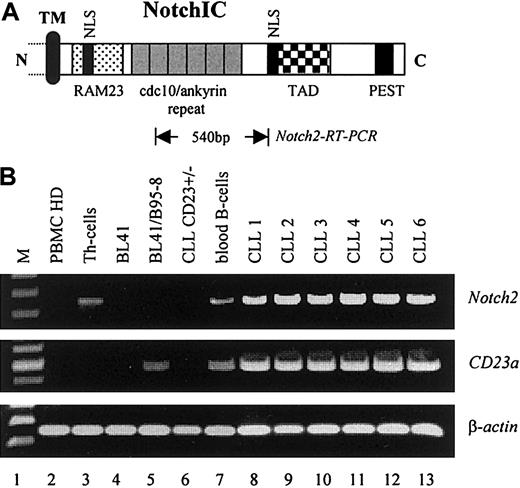
To examine whether elevated levels of Notch2IC in nuclear extracts from B-CLL cells were caused by increased transcription of the corresponding Notch2 gene, RT-PCR analysis was performed. As demonstrated in Figure 4,Notch2 was found to be consistently overexpressed in B-CLL cells (6 of 6) as compared with resting peripheral blood B cells from healthy donors (Figure 4B, lane 7, representative for n = 3) and to other B–non-Hodgkin lymphoma cell lines (Figure 4B, lanes 4 and 5). In total PBMCs from healthy donors, relative high levels ofNotch2 transcription were observed in purified Th cells (Figure 4B, lane 3) and in peripheral blood B cells (Figure 4B, lane 7) that are in accordance with published data.40 No signals for Notch1 were detected under these conditions (data not shown). Remarkably, neither Notch1 (data not shown) norNotch2 messages (Figure 4B, lane 6) were found in a rare case of B-CLL (B-CLLCD23+/−) in which the leukemic cells expressed low amounts of surface CD23 (Table 1). Because no detectable amounts of CD23a messenger RNA (mRNA) were found in this sample (Figure 4B, lane 6), it is likely that the surface expression of CD23 on these cells resulted from CD23b that is differently regulated.7 8 Figure 4B shows also the induction of theCD23a gene through EBV (lanes 4 and 5).
Notch2 is overexpressed in B-CLL cells.
(A) Relative position of the Notch2 sequence (Notch2-RT-PCR) used for mRNA detection. The intracellular domain of Notch is characterized by the RAM23 domain and 6 copies of a cdc10/ankyrin motif, known to bind CBF1 (TM, transmembrane domain; NLS, nuclear localization signals; TAD, transactivation domain; PEST, protein degradation signal). (B) RT-PCR analysis indicating that Notch2 is overexpressed in B-CLL samples where it correlates with the expression of CD23a(PBMCs, Th cells, and peripheral blood B-cells from healthy donors are representative for n = 3). β-Actin was used as an internal control.
Notch2 is overexpressed in B-CLL cells.
(A) Relative position of the Notch2 sequence (Notch2-RT-PCR) used for mRNA detection. The intracellular domain of Notch is characterized by the RAM23 domain and 6 copies of a cdc10/ankyrin motif, known to bind CBF1 (TM, transmembrane domain; NLS, nuclear localization signals; TAD, transactivation domain; PEST, protein degradation signal). (B) RT-PCR analysis indicating that Notch2 is overexpressed in B-CLL samples where it correlates with the expression of CD23a(PBMCs, Th cells, and peripheral blood B-cells from healthy donors are representative for n = 3). β-Actin was used as an internal control.
Notch2IC induces endogenousCD23a in B cells
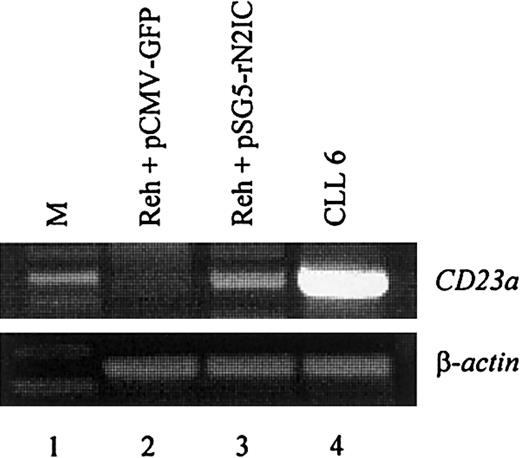
To directly demonstrate that CD23a is a target gene of Notch2 signaling in B lymphocytes, the CD23− pre–B-cell line REH42 was transiently transfected with a mammalian expression vector containing the complementary DNA coding for rat Notch2IC (kindly provided by Dr Diane Hayward).18 This vector leads to the expression of a ligand-independent constitutive-active Notch2IC protein lacking the extracellular domain.18 The transfection efficiency in the living cell population was 28% as determined by flow cytometry (not shown). After 2 days of incubation, RT-PCR analysis demonstrated that ectopic expression of Notch2IC induces CD23a transcription (Figure5, lane 3), whereas a GFP-encoding control plasmid had no effect (Figure 5, lane 2).
Notch2IC induces endogenous
CD23a in B cells. The CD23−pre–B-cell line REH was transiently transfected with pSG5-rN2IC, coding for the intracellular domain of rat Notch2. After 48 hours of incubation, total RNA was subjected to RT-PCR analysis that demonstrated that rNotch2IC induces CD23a transcription (lane 3). Lane 2 shows a control transfection experiment with pCMV-GFP. B-CLL cells (lane 4) served as positive control for CD23aexpression.
Notch2IC induces endogenous
CD23a in B cells. The CD23−pre–B-cell line REH was transiently transfected with pSG5-rN2IC, coding for the intracellular domain of rat Notch2. After 48 hours of incubation, total RNA was subjected to RT-PCR analysis that demonstrated that rNotch2IC induces CD23a transcription (lane 3). Lane 2 shows a control transfection experiment with pCMV-GFP. B-CLL cells (lane 4) served as positive control for CD23aexpression.
Expression of CD23a correlates with B-CLL cell survival
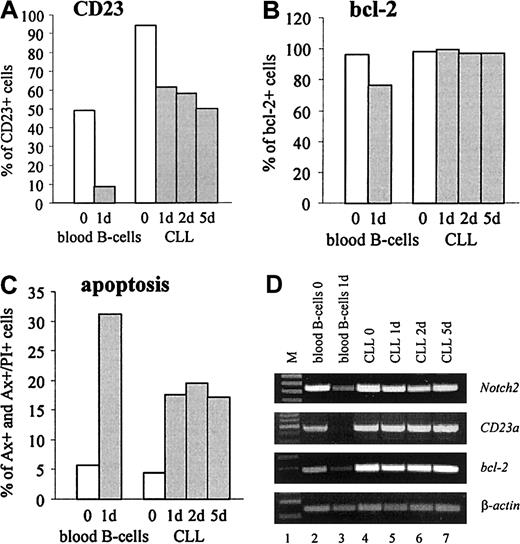
CD23 is detectable on virtually all resting B cells that coexpress surface IgM and IgD.4 In vitro, freshly isolated normal B cells rapidly lose both, CD23 protein and mRNA, suggesting that the expression of CD23 on resting B cells is not constitutive.4 To compare the expression ofCD23a in normal B cells with B-CLL cells, gene expression kinetic studies were performed. The results confirmed that after 24 hours of incubation purified normal B cells completely loseCD23a transcription (Figure6D, lane 3). Down-regulation ofCD23a was accompanied by a down-modulation ofNotch2, the antiapoptotic gene bcl-2, and by a rapid increase in cell death by apoptosis as determined by FACS analysis (Figure 6B,C) and by RT-PCR (Figure 6D, lane 3). In contrast, CD23a, bcl-2, and Notch2 remained high in B-CLL cells, and this finding was associated with high cell viability.
CD23a is differently regulated in B-CLL cells compared with normal B lymphocytes.
In vitro expression kinetic studies demonstrating that in B-CLL cells CD23a levels remain high during 5 days of incubation under serum-free conditions, whereas in normal B-cellsCD23a is down-regulated within 24 hours. (A-C) FACS analysis showing the percentage of CD23, bcl-2, and early and late apoptotic cells (Annexin V+ and Annexin V+/propidium iodide+, respectively). (D) RT-PCR analysis correlating the expression of Notch2 with CD23a andbcl-2.
CD23a is differently regulated in B-CLL cells compared with normal B lymphocytes.
In vitro expression kinetic studies demonstrating that in B-CLL cells CD23a levels remain high during 5 days of incubation under serum-free conditions, whereas in normal B-cellsCD23a is down-regulated within 24 hours. (A-C) FACS analysis showing the percentage of CD23, bcl-2, and early and late apoptotic cells (Annexin V+ and Annexin V+/propidium iodide+, respectively). (D) RT-PCR analysis correlating the expression of Notch2 with CD23a andbcl-2.
Discussion
The overexpression of the transmembrane glycoprotein CD23 is one of the major characteristics of B-CLL cells. Besides the prognostic potential of its soluble cleavage product, sCD23, which reflects tumor load and progression of disease,5,6 selective expression of the CD23a isoform is concurrent to a state of B-CLL cell survival,9 thereby providing a link betweenCD23a and the malfunction of apoptosis characteristic for this neoplastic B-cell type. The molecular basis underlying the up-regulation of CD23a on the gene level, however, has remained elusive.
In this report, we provide evidence that the Notch2 proto-oncogene plays a critical role in this process. Bandshift assays visualized a prominent transcription factor complex (C1) that binds sequence specific to one known and 4 newly identified putative CBF1 sites in theCD23a core promoter region.7,14,15 The significance of this complex was underlined because in all B-cell samples the intensity of C1 correlated with their respective levels ofCD23a transcription. Moreover, by using EBV-infected BL41 cells as a positive control for CBF1-mediated CD23aexpression,14 15 C1 was found to be EBV inducible. Supershift assays with antibodies raised against the nuclear forms of Notch1 and Notch2 pointed to the fact that Notch2IC is a component of C1 in B-CLL cells. Because transfection of REH pre–B cells with Notch2IC confirmed that CD23a is a target gene of Notch2 signaling, it is reasonable to conclude that Notch2 is involved in the overexpression of CD23a in B-CLL cells.
Considering that induction of CD23 through EBNA2 is an initiating event in EBV-driven B-cell immortalization,10-13 it is tempting to speculate that theNotch2 proto-oncogene plays an equally important role in the transformation process of B-CLL cells. In this context, Notch2 might not only account for B-CLL–specific CD23a expression but also for other phenotypic changes characteristic for this B-cell malignancy. In T cells, for instance, ectopic expression of Notch1IC results in the up-regulation of bcl-2 and renders thymomas resistant to glucocorticoid-induced apoptosis.43 Given these observations, the oncogenic nature of Notch receptors may reflect an inhibition of programmed cell death that would be consistent with the phenotype of B-CLL cells.1,2,20 21
Another characteristic feature of B-CLL lymphocytes that might be explained by Notch2 signaling is the low expression of surface immunoglobulins. Recently, it has been demonstrated that activated Notch1, which shares many functions with Notch2,18 acts as transcriptional suppressor of the Igμ gene, indicating that IgM expression is negatively controlled by the Notch signal transduction pathway.44 45
So far, however, the role of Notch2 signaling in B-cell differentiation and tumorigenesis is still not clear. In human fetal B cells, Notch2 expression is restricted to the late pre–B (CD19+Igμlow) compartment, suggesting that Notch2 confers an antiapoptotic signal when these cells undergo selection through the pre–B-cell receptor.39 In the current study, we show thatNotch2 as well as CD23a are also expressed in normal peripheral blood B cells that served as control because they were clustered next to B-CLL cells by genomic scale gene expression profiling.37 Recirculating blood B lymphocytes comprise naive and memory B cells and are characterized by their low proliferation rate and by their relative long life span. As compared with these cells, Notch2 was found to be consistently overexpressed in B-CLL cells, thereby explaining the up-regulation ofCD23a. Another clear difference between normal B cells and B-CLL lymphocytes was observed by in vitro kinetic studies showing that in normal B-cells CD23a is down-regulated within 24 hours, whereas in B-CLL cells CD23a levels remain high during 5 days of incubation. Interestingly, the CD23a levels correlate not only with Notch2 expression but also with the levels of the antiapoptotic gene bcl-2 together with enhanced cell viability pointing to a link between the Notch2/CD23a axis and the failure of apoptosis in B-CLL cells.
Several lines of evidence suggest that deregulation of Notch signaling locks cells into a less differentiated, proliferative state and inhibits apoptosis in certain leukemic cell lines.20 21Therefore, a structure-function analysis together with approaches to determine Notch2 function in vivo should provide further insights into the molecular mechanisms underlying the regulation of Notch2and its role in the pathophysiology of B-CLL.
We thank Drs M. Busslinger and T. Decker for critically reviewing the manuscript and Dr Diane Hayward for providing the pSG5-rN2IC plasmid.
Supported by grant 12210 from the Austrian Science Foundation, by the “Josefine Hirtl and Maria Buss Stiftung,” and by the Commission Oncology, Medical Faculty, University of Vienna.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Josef D. Schwarzmeier, University of Vienna, Dept of Internal Medicine I, Div of Hematology, Waehringer Guertel 18-20, A-1090 Vienna, Austria; e-mail:josef.schwarzmeier@akh-wien.ac.at.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal