Sporadic mutations in the thrombomodulin (TM) gene occur in patients with both arterial and venous thrombosis, but the effects of these mutations on expression and function are largely unexplored. Full-length wild-type TM complementary DNA (cDNA) was incorporated into vector pcDNA6 for transfection into COS-7 cells for transient expression. Mutagenesis was performed to create 7 TM mutants with natural mutations either previously identified (Ala25Thr, Gly61Ala, Asp468Tyr, Pro477Ser, Pro483Leu) or reported here (an 11-base pair [bp] deletion, del791-801, leading to STOP306, and a missense mutation, Arg385Ser). Four mutations were found to detrimentally affect the level of expression of the TM protein. Of the missense mutations, 3 had reduced expression compared to wild-type TM (100%), Arg385Ser (50.2% ± 5%, P < .001), Pro477Ser (76.8% ± 1%, P < .001), Pro483Leu (82.1% ± 8%, P < .007). No TM protein expression could be detected on the cell surface for mutation del791-801. The cofactor activity of TM in protein C activation was also evaluated. The Michaelis constant (Km) for wild-type thrombin-TM complex was 634 ± 6 nmol/L. Two mutants, with Arg385Ser and Pro477Ser, had increased (P < .0001) Km, 2967 ± 283 nM, and 2342 ± 219 nM, respectively, demonstrating impaired function of the thrombin-TM complex. This work presents biochemical evidence that certain (but not all) natural mutations in the TM gene reduce expression and impair function of the protein on the cell surface, and helps clarify the suggested contribution that these mutations might make to the risk of thromboembolic disease.
Introduction
Regulation of blood coagulation by natural anticoagulant mechanisms is essential to maintain blood fluidity. Two anticoagulant mechanisms are known to be important clinically, those involving antithrombin and protein C.1-3 Deficiencies of the proteins involved in these mechanisms are associated with risk of venous thrombosis. Thrombomodulin (TM) is an endothelial cell surface glycoprotein that acts as a receptor for thrombin. The thrombin-TM complex activates protein C to activated protein C (APC).5In addition, procoagulant and mitogenic activities of thrombin are abrogated on complex formation and fibrinolysis is down-regulated.6-11 It might be anticipated that TM deficiency could be associated with risk of thromboembolic disease. It was, however, only in recent years that evidence has been accumulated to support such a suggestion. Experimental evidence has been obtained from studies of mice genetically modified to inactivate or alter the TM gene. TM chimerism resulted in spontaneous, intravascular fibrin deposition that was dependent on age and the magnitude of the TM deficiency.12 Mice homozygous for a missense mutation in the TM gene (Glu382Pro) that abolishes anticoagulant function were shown to present increased fibrin deposition in their lungs after hypoxic challenge.13
Some evidence from clinical studies supports a role for TM in thrombotic disease. In a carefully set prospective study, the Atherosclerosis Risk in Communities (ARIC) study, soluble TM showed a strong, graded, inverse association with and was a good predictor of incident coronary artery disease (CAD). The levels did not correlate with major systemic markers of inflammation, a condition known to increase soluble TM levels by cleavage of the protein from the endothelial surface as a result of the activity of proteolytic enzymes. The levels did, however, correlate in their extreme quintiles with a marker of coagulation activation, prothrombin F1 + 2. It was suggested that in healthy individuals plasma concentrations of TM might reflect endothelial expression and anticoagulant function.14
A role for genetic variation in disease has been suggested by findings that mutations in the TM gene occur in patients with thrombosis. Since the first report,15 a number of mutations have been identified in patients with venous thrombosis and in their families.16-19 Furthermore, in a screening investigation of patients with myocardial infarction (MI), 3 mutations of the 5′ untranslated region of the TM gene were identified.20 Each of these latter mutations is in proximity to regulatory elements of the promoter.21 Only one mutation (G-33A), independently identified in a cohort of patients with venous thrombophilia, was associated with decreased transcriptional activity in reporter gene assay22 (G.K., unpublished data, August 1999). Furthermore, it was demonstrated that this mutation is associated with CAD in Asian patients.23 A recurring missense mutation in the TM coding sequence was also identified in the above screening investigation. This was a mutation predicting Ala25Thr substitution and its significance has been investigated subsequently in a large case-control study, Study of Myocardial Infarction Leiden (SMILE). The results of this study suggested an association between the mutation and the risk for MI in men under 50 years of age.24 Recently, we reported a private point mutation found in the same patient cohort, which was predicted to encode an elongated TM protein. This mutant protein was shown to have reduced cell surface expression both in vitro and in vivo.25 The ARIC study has recently suggested that the TM polymorphism Ala455Val may be associated with cardiovascular risk, but if so this is confined to black American racial groups.26 This polymorphism was found not to be associated with venous thrombophilia,27but has been suggested to be associated with varicose veins through linkage with promoter mutations.28
Although much of the above data are suggestive of a causal effect of the TM mutation in thromboembolic disease, there is incomplete understanding of the function of most of the identified mutations. Clarification of the functional consequences is important because most of the mutations are rare events, or occur in small families, and the clinical effects of the mutations are difficult to ascertain in these settings. In what follows the functional characterization of 5 previously reported and 2 yet unpublished mutations in the TM coding region will be presented.
Materials and methods
Mutations investigated
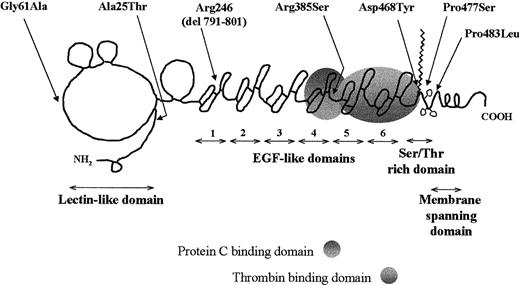
The location of the mutations investigated in this study relative to the domain structure of TM is depicted in Figure1. The substitution Ala25Thr (mutation G127A) was identified in a screening program of patients with MI.20,24 The same mutation has been identified in a patient with venous thrombophilia19 and in one patient with both venous and arterial thrombosis.29 Point mutations leading to substitutions Gly61Ala (G236C), Arg385Ser (G1209T), Asp468Tyr (G1456T), Pro477Ser (C1483T), Pro483Leu (C1502T) and to the premature STOP306 (del791-801) have been identified in screening programs involving patients with venous thrombophilia. Of these, Gly61Ala, Asp468Tyr, Pro477Ser, and Pro483Leu have been subject of prior reports of case histories.15,17,18 29 The case histories of the 2 individuals with the remaining mutations are presented below.
Schematic representation of TM mutations.
The location of the mutations within the domain structure of TM (the domain diagram of TM is based on one produced originally by Professor Bjorn Dahlback and is used here with permission).
Schematic representation of TM mutations.
The location of the mutations within the domain structure of TM (the domain diagram of TM is based on one produced originally by Professor Bjorn Dahlback and is used here with permission).
Case histories
Patient with TM del791-801 leading to STOP306.
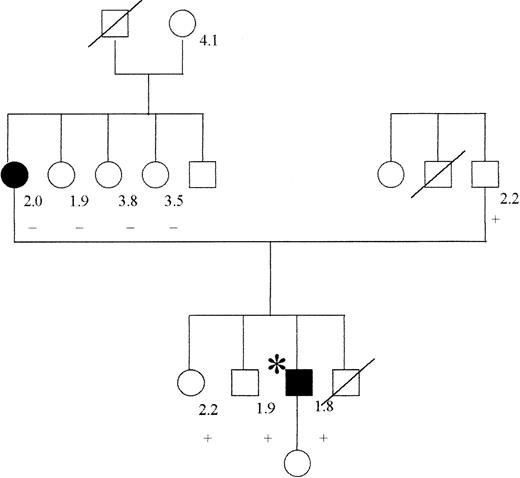
The male patient was born in 1968 and was first admitted to hospital, at the age of 22 years, presenting with cytomegalovirus hepatitis. After 2 weeks of immobilization, he developed pulmonary embolism, verified by lung perfusion-ventilation scintigraphy. Oral anticoagulant therapy was initiated, but stopped by the patient after 2 months. One and a half years later, he was readmitted and cerebral venous sinus thrombosis was diagnosed by angiography and magnetic resonance imaging (MRI). Four days later, in the course of the same admission, the patient experienced an episode of transient central facial palsy. MRI confirmed a minor, subcortical infarction. Extensive investigation of coagulation showed no abnormality other than APC resistance, with an APC ratio of 1.8 (normal values > 2.0), without the mutation G1691A in factor V. The mother of the propositus, who had suffered deep vein thrombosis (DVT) during pregnancy at the age of 26 years, also had APC resistance, but not the TM mutation. The family is 1 of the 50 original families described to have APC resistance.30 The father and 2 sisters, all asymptomatic, carried the TM mutation. One brother died in a car accident and could not be tested (see pedigree in Figure2).
Pedigree of patient with mutation TM del791-801.
The propositus is indicated by an asterisk; filled symbols indicate the presence of thrombosis, a crossed symbol indicates a deceased individual. The numbers at the right of the symbols represent the value of APC ratio, and the + or (minus) sign indicates the presence or absence of the TM mutation, respectively.
Pedigree of patient with mutation TM del791-801.
The propositus is indicated by an asterisk; filled symbols indicate the presence of thrombosis, a crossed symbol indicates a deceased individual. The numbers at the right of the symbols represent the value of APC ratio, and the + or (minus) sign indicates the presence or absence of the TM mutation, respectively.
Patient with Arg385Ser mutation.
The female patient was born in 1907 and investigated as part of a TM gene screening study of consecutive patients with venous thrombosis. She has suffered 3 episodes of DVT at the age of 76 and 85 years in the right calf and at the age of 87 years in the left calf, all documented by phlebography. At the age of 90 years, she suffered an MI, which led to cardiac insufficiency and death 2 months later. The patient had hypertension and non–insulin-dependent diabetes. Laboratory investigation of coagulation did not detect any other abnormality. Family investigation was not possible in this case.
Mutation detection
Blood collection, mutation detection, and identification methods have been described previously for most mutants.15,16,20Polymerase chain reaction (PCR) amplification of the TM gene fragments was also performed as previously described.15 16
Single-strand conformation polymorphism (SSCP) was performed with the GeneGel Clean 15/24 Kit and the GenePhor Electrophoresis Unit (Amersham Pharmacia Biotech, Uppsala, Sweden) essentially as described.15 Aberrant band patterns were found in fragments designated TM-E for the deletion mutant and TM-G for TM Arg385Ser. DNA samples from family members were then screened by PCR-SSCP using only the appropriate TM fragment.
DNA sequencing was performed from PCR products obtained using primers that amplified longer fragments of the TM gene than TM-E or TM-G using BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Perkin Elmer, Stockholm, Sweden). For the deletion mutation, PCR was performed with the primers tctcgatcacctacggcacc (sense) and tcgcacatgcacgagtagga (antisense) at 61°C annealing temperature. Other experimental conditions were as described.15 This generated a PCR product of 310 bp. DNA sequencing was then performed using primers TM6 + TM715 as sequencing primers. In the case of the Arg385Ser mutation, the PCR reaction and the DNA sequencing were performed on the same PCR product (358 bp) and primers acgtggatgactgcatactg (sense) and gtcgatgtccgtgcagatg (antisense), annealing temperature 57°C, with all other experimental conditions as described.15
Restriction analysis of PCR-amplified TM fragments was performed with appropriate enzymes. Restriction site analyses for the wild-type and the respective mutant TM genes revealed that restriction enzyme cleavage site(s) were predicted to change in both PCR fragments. In PCR fragment TM-E of the deletion mutant, one NspBII restriction site was removed. In TM-G of the Arg385Ser mutant, oneBanI restriction site was removed. Analysis with restriction enzymes was therefore used to demonstrate and confirm heterozygosity for the TM mutations in both patients and in the family members for the deletion mutant. The purified PCR product (8 μL) was mixed with 1 μL of the appropriate restriction enzyme and 1 μL digestion buffer according to the instructions from the manufacturer (NspBII; United States Biochemical, Cleveland, OH; BanI; New England Biolabs, Beverly, MA), followed by electrophoresis.
Plasmids for in vitro expression
Vector pRSVSVOTM31 was used as template. All mutations were created by PCR- based mutagenesis. In all cases the mismatch was incorporated in the appropriate oligonucleotide primer. Point mutations were introduced by the “megaprimer” method.32 To prepare the mutation with deletion of nucleotides 791-801, 2 primers flanking the site of the deletion were used in 2 separate PCR amplifications. The resulting products were ligated head-to-tail into the XmaI and DraIII sites of the vector. Other mutant vectors were prepared using standard approaches and details of primers can be obtained by application to the authors.
In the final stage of mutant complementary DNA (cDNA) preparation, the coding region of TM, either wild-type, or incorporating the mutations described, was amplified using primers which each added aPmeI restriction site to flank the coding sequence. The 1.8-kb PCR product was cloned into the PmeI sites of the vector pcDNA6V5/His (Invitrogen, de Schelp, The Netherlands), removing the V5 epitope and the His tag. The constructs were then used for expression of TM variants.
In the case of TM del791-801 it was necessary also to prepare vectors containing those sequences coding for the V5 epitope and His tag both in the mutant and wild-type vectors in-frame with the TM coding sequences. The mutant sequence, with the 11 nucleotides deleted between positions 791-801, was truncated adjacent to the new predicted STOP by removing 746 nucleotides of untranslated sequence. The resultant mutant cDNA and wild-type cDNA were cloned into vector pcDNA6BV5His in-frame with the V5 epitope and His tag.
All PCR reactions consisted of 200 ng of the appropriate template vector, 10 μL 10 times reaction buffer, 250 ng of each primer, 0.2 mM of each dNTP, 1 μL (5 U) Pfu polymerase. The cycling profile consisted of 30 cycles of 30 seconds denaturation at 94°C, 30 seconds annealing at 60°C to 65°C, and 3 minutes extension at 72°C. A final extension of 10 minutes followed the last cycle.
Cell culture, transfection, and selection of stable transfectants
COS-7 and CV-1 cells (CAMR, Whiltshire, United Kingdom) were propagated in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. Transfections were performed in 35-mm well plates, by the calcium phosphate method,33 as previously described.25 For transient transfection, COS-7 cells were cotransfected with 3 μg/well of circular expression plasmid and 2 μg/well vector pSEAP2Control (Clontech Laboratories, Palo Alto, CA). The latter vector encodes secreted alkaline phosphatase (SEAP), which was quantitated by use of chemiluminescent assay (Great Escape System, Clontech Laboratories) to assess transfection efficiency. Analysis of transient expression was performed 48 to 72 hours after transfection.
CV-1 cells were transfected by the same method with 5 μg/well of expression plasmid linearized with ScaI. Forty-eight hours after transfection, cells were reseeded in culture media supplemented with 10 μg/mL Blasticidin, the eukaryotic selection agent encoded by the plasmid pcDNA6. Stable transfectants were propagated as pools of colonies. Stably transfected cells were used only for the functional assay of TM (see below).
Western blotting
Forty-eight hours after transient transfection, COS-7 cell were washed twice with phosphate-buffered saline. Detergent lysates were prepared by addition of 150 μL lysis buffer: 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, with a cocktail of protease inhibitors (Boehringer-Mannheim, Lewes, United Kingdom). In the case of mutant TM del791-801, a whole cell lysate was also prepared by adding the Laemmli buffer (0.5M Tris-HCl, pH 6.8, 10% sodium dodecyl sulfate [SDS], 20% glycerol) directly to the cells.
The amount of TM variant expressed by the transiently transfected COS-7 cells was initially measured in the detergent lysates by enzyme-linked immunosorbent assay (ELISA) and the results were used to determine the amounts of antigen added to gels for Western blotting. Dilutions of mutants with low expression levels were also studied by Western analysis to ensure that these results had not arisen from altered epitopes recognized by the monoclonal antibodies used in the ELISA (mutants Arg385Ser, Pro477Ser, and Pro483Leu).
Samples were loaded on 12% NuPAGE Bis-Tris precast gels (Invitrogen). After electrophoresis and electroblotting, TM was immunodetected using sheep polyclonal antihuman TM antibody (American Diagnostica, Greenwich, CT) and horseradish peroxidase (HRP)–conjugated donkey antisheep antibody (Jackson Immuno Research Laboratories, West Grove, PA). For the deletional mutant and wild-type variant fused to V5/His, an HRP-conjugated monoclonal anti-V5 antibody (Invitrogen) was used. In all cases, the HRP was detected by enhanced chemiluminescence (ECL; Amersham, Little Chalfont, United Kingdom). Molecular weight markers (Amersham) were used to estimate the molecular weight of the proteins.
TM ELISA
Patient plasma prepared for assay as previously described25 and COS-7 cell lysates were prepared as detergent lysates. Samples were assayed for TM antigen using a commercially available ELISA (Immunbind TM, American Diagnostica) that uses 2 monoclonal antihuman TM antibodies. The capture antibody is directed against EGF1-EGF2 and the HRP-conjugated antibody is directed against EGF5-EGF6 (none of the missense mutations investigated is located in these regions). Duplicate transfections of COS-7 cells were performed on 3 separate occasions. For each transfection the amount of TM measured in the ELISA was normalized (divided) by the amount of SEAP secreted by the same cells, measured in the media.
Reverse transcription-PCR
Total RNA was extracted from COS-7 cells transiently transfected with wild-type or with TM del791-801 vectors using the RNeasy kit according to the manufacturer's instructions (Qiagen, Crawley, United Kingdom). The RNA was treated with DNAse (Stratagene, Amsterdam, The Netherlands) at a final concentration of 0.4 U/μL. cDNA was synthesized using random hexamers (Life Technologies, Glasgow, United Kingdom) and Sensiscript reverse transcriptase (Qiagen). Controls without reverse transcriptase were included for each sample to exclude DNA contamination. A 143-bp region of the TM gene (from 709 to 852) encompassing the deletion 791-801 was amplified using specific primers and the cDNA as template. The PCR was performed using the HotStart Taq polymerase Master Mix (Qiagen) according to the manufacturer's instructions. An initial denaturation of 15 minutes at 95°C was followed by 35 cycles of 30 seconds denaturation at 95°C, 15 seconds annealing at 65°C and 1 minute extension at 72°C, with a final extension at 72°C for 10 minutes. The products were electrophoresed on a 3% NuSieve GTG gel (FMC Bioproducts, Rockland, ME).
Immunocytochemistry
Indirect immunofluorescent staining and microscopical examination were performed as previously described.25
TM functional assay
A method described previously34 was modified to allow estimation of the Michaelis constant (Km) of protein C activation on the surface of transiently transfected COS-7 or on stably transfected CV-1 cells. Forty-eight to 56 hours before the assay, CV-1 cells were seeded in 96-well cell culture plates, 8 wells for each mutant. Alternatively, transiently transfected COS-7 cells were washed twice with ice-cold assay buffer (50 mM Tris-HCl, 2 mM CaCl2, 100 mM NaCl, 1% bovine serum albumin, pH 8.0) and scraped, and resuspended in 500 μL assay buffer, then divided in 50-μL aliquots. The cells (either growing in microwells or the 50-μL aliquots) were incubated at 37°C with fixed concentrations of thrombin (14.9 nmol/L) and increasing concentrations of protein C: 0, 100, 200, 300, 400, 800, 1000, 1200 nmol/L in a final volume of 100 μL. After the addition of 0.4 μM antithrombin and 13 IU/mL heparin, 50 μL of the reaction was assayed with 100 μL chromogenic substrate S2366 (200 mM final concentration). Absorption was determined at 405-nmol/L wavelength, on an iEMS type microtiter plate reader (Labsystems, Helsinki, Finland). The absorption values obtained in each well were converted to nanomoles APC generated per minute, using a reference curve constructed for each experiment. This was analyzed using EnzFitter for Windows version 2.0.6 (Biosoft) and Km was estimated for each TM variant. Note that for the mutants Pro477Ser and Arg385Ser, the derived Km values are approximate, due to the problem of saturating the thrombin-mutant TM complex with protein C. Purified proteins for these experiments were purchased from Enzyme Research Laboratories (Swansea, United Kingdom) and S2366 from Chromogenix (Molndal, Sweden).
Statistical methods
Means, variance, and SE were calculated for data sets and all results are expressed in the text as means ± SE. Significance was calculated using the t test for sample means with unequal variance. All operations were performed in Microsoft Excel (Microsoft, Redmond, WA).
Results
Mutation TM del791-801
The mutation del791-801 in the TM gene changes the reading frame from Arg246, is predicted to give rise to a premature STOP at codon 306, and encodes a variant, truncated protein (Figure3). The construct with the deleted 11 bp was transiently expressed in COS-7 cells. The efficiency of transfection was assessed by measuring the SEAP levels secreted in the media by the transfected cells.
The effect of mutation TM del791-801 on the TM protein sequence.
The normal sequence is at the top and the mutated sequence at the bottom of the rows. The unspaced sequence represents the nucleotides and the spaced one represents the amino acid translation. The deleted nucleotides are in bold italics and the new stop codon is underlined. The numbers at the top of each block indicate the amino acid position (numbering as in the mature protein).
The effect of mutation TM del791-801 on the TM protein sequence.
The normal sequence is at the top and the mutated sequence at the bottom of the rows. The unspaced sequence represents the nucleotides and the spaced one represents the amino acid translation. The deleted nucleotides are in bold italics and the new stop codon is underlined. The numbers at the top of each block indicate the amino acid position (numbering as in the mature protein).
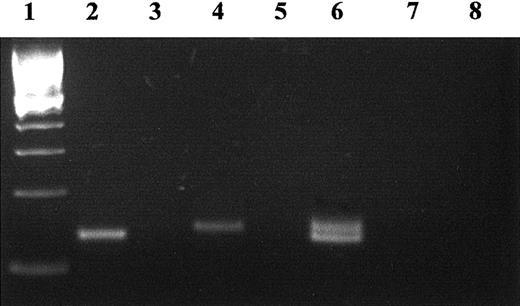
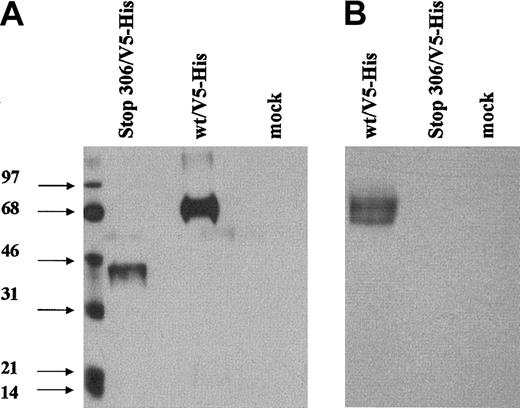
The reverse transcription (RT)–PCR assay of transfected cells demonstrated the presence of message in cells transfected with wild-type and mutant cell TMs (Figure 4), albeit with the expected size reduction of the PCR product of the mutant (from 143 to 132 bp). No protein C activation was detected on the surface of the cells transfected with the mutant TM vector, using the assay for functional thrombin-TM complex. Immunoblotting of detergent or whole cell lysates or of media using a polyclonal anti-TM antibody also failed to detect a variant form of the TM protein (not shown). The use of a polyclonal antibody should have enabled detection of any expressed truncated protein, because it would contain antibodies directed at several epitopes of the protein. However, to improve the prospect of detection of any mutant TM fragments, a readily detectable V5 tag epitope was attached to the truncated mutant TM protein and to the wild-type TM protein, respectively. Following transient expression in COS-7 cells and immunoblotting with anti-V5 antibody, a band of expected reduced size could be detected in whole cell lysates (Laemmli buffer) of cells transfected with the deletion mutant (Figure5). The same band could not, however, be detected in detergent lysates, which would not contain the cell organelles. Furthermore, the mutant could not be detected in the cell media. These results demonstrate cellular synthesis of the mutant truncated TM, but an inability to export the protein.
RT-PCR of the mRNA for mutant TM del791-801 and wild type from the transiently transfected COS-7 cells.
Total RNA was reverse transcribed with random primers and the cDNA used as a template for PCR amplification with 2 TM primers flanking the deleted region. The size of the normal product is 143 bp (lane 4). The deletion gives rise to a smaller, 132-bp product (lane 2). Both transcripts were detected in cells cotransfected with both TM variants (wild-type and mutant, lane 6). The 100-bp marker (Life Technologies) was loaded in lane 1. Negative controls without reverse-transcriptase (lanes 3, 5, 7) or without template (lane 8) were included.
RT-PCR of the mRNA for mutant TM del791-801 and wild type from the transiently transfected COS-7 cells.
Total RNA was reverse transcribed with random primers and the cDNA used as a template for PCR amplification with 2 TM primers flanking the deleted region. The size of the normal product is 143 bp (lane 4). The deletion gives rise to a smaller, 132-bp product (lane 2). Both transcripts were detected in cells cotransfected with both TM variants (wild-type and mutant, lane 6). The 100-bp marker (Life Technologies) was loaded in lane 1. Negative controls without reverse-transcriptase (lanes 3, 5, 7) or without template (lane 8) were included.
Western blot of the expressed del791-801 TM.
Transient transfections of COS-7 cells were performed using the wild-type or mutated TM expression vector with a V5 tag sequence attached adjacent to the STOP codons. Proteins in whole cell lysates prepared by addition of Laemmli buffer to cells (A) or in detergent extracts of the cells (B) were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) under nonreducing conditions. Immunodetection was performed using an anti-V5 tag antibody.
Western blot of the expressed del791-801 TM.
Transient transfections of COS-7 cells were performed using the wild-type or mutated TM expression vector with a V5 tag sequence attached adjacent to the STOP codons. Proteins in whole cell lysates prepared by addition of Laemmli buffer to cells (A) or in detergent extracts of the cells (B) were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) under nonreducing conditions. Immunodetection was performed using an anti-V5 tag antibody.
Levels of plasma TM determined by ELISA in the propositus and one sister who is a carrier of the mutation TM del791-801 were lower than in normal controls: 1.7 ng/mL and 1.6 ng/mL, respectively (normal range, 4.0-5.3 ng/mL for men and 2.7-4.8 ng/mL for women). These results are consistent with expression only from the normal TM allele in carriers of the deletion.
Missense mutations
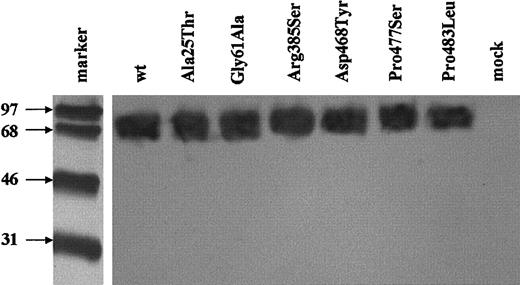
Western blotting of the detergent lysates confirmed that variant proteins with Ala25Thr, Gly61Ala, Arg385Ser, Asp468Tyr, Pro477Ser, and Pro483Leu of predicted sizes were expressed by the transiently transfected cells. No abnormality in the pattern of migration or the apparent molecular weight of the expressed mutated proteins could be detected when compared to the wild-type TM (Figure6).
Western blot of the expressed missense TM mutants.
SDS-PAGE under nonreducing conditions and immunoblotting of cell lysates expressing each of the mutants or the wild-type TM were carried out. Immunodetection was performed with sheep polyclonal anti-TM antibody followed by HRP-conjugated antigoat antibody and ECL detection. The molecular weight markers were reduced and following electroblotting were detected separately with streptavidin-HRP conjugate.
Western blot of the expressed missense TM mutants.
SDS-PAGE under nonreducing conditions and immunoblotting of cell lysates expressing each of the mutants or the wild-type TM were carried out. Immunodetection was performed with sheep polyclonal anti-TM antibody followed by HRP-conjugated antigoat antibody and ECL detection. The molecular weight markers were reduced and following electroblotting were detected separately with streptavidin-HRP conjugate.
The TM antigen levels after transfection were measured by ELISA. Transfection efficiency was assessed by measurement of secreted alkaline phosphatase (encoded by pSEAP Control vector, which was cotransfected into the cells) values in the conditioned media. The TM values obtained by ELISA measurement in each experiment were normalized for transfection efficiency and calculated as percent of the values obtained for the wild-type construct in the same experiment. Three missense mutations were found to reduce the expression of TM: Arg385Ser (50.2% ± 5%), Pro477Ser (76.8% ± 1%), and Pro483Leu (82.1% ± 8%; Table 1). No TM antigen was detectable in the media of transfected cells. These mutations that exhibited reduced levels are located outside the areas of the TM molecule against which the monoclonal antibodies of the ELISA were directed. However, to ensure that the reduction in the expression of TM measured by ELISA was not due to reduced affinity of the monoclonal antibodies used for their detection in the ELISA, dilutions of equal amounts of wild-type TM and of each of these TM variants, as measured by ELISA, were examined by immunoblotting with a polyclonal anti-TM antibody. The mutants could not be distinguished from the wild-type TM in terms of density of the TM bands. This argues against any monoclonal antibody–dependent assay artifact.
Cell surface expression and cofactor function of TM mutants
| TM mutant . | TM antigen* (% of wild-type) . | Significantly decreased (P) . | Km† . | Significantly increased (P) . |
|---|---|---|---|---|
| Wild-type | Reference | 634 ± 6 | ||
| Ala25Thr | 103 ± 9 | NS | 609 ± 20 | NS |
| Gly61Ala | 100 ± 8 | NS | 661 ± 119 | NS |
| Arg385Ser | 50.2 ± 5 | .001 | 2854 ± 249 | < .0001 |
| Asp468Tyr | 97.8 ± 9 | NS | 733 ± 202 | NS |
| Pro477Ser | 76.8 ± 1 | .001 | 2716 ± 565 | < .0001 |
| Pro483Leu | 82.1 ± 8 | .007 | 827 ± 178 | .03 |
| TM mutant . | TM antigen* (% of wild-type) . | Significantly decreased (P) . | Km† . | Significantly increased (P) . |
|---|---|---|---|---|
| Wild-type | Reference | 634 ± 6 | ||
| Ala25Thr | 103 ± 9 | NS | 609 ± 20 | NS |
| Gly61Ala | 100 ± 8 | NS | 661 ± 119 | NS |
| Arg385Ser | 50.2 ± 5 | .001 | 2854 ± 249 | < .0001 |
| Asp468Tyr | 97.8 ± 9 | NS | 733 ± 202 | NS |
| Pro477Ser | 76.8 ± 1 | .001 | 2716 ± 565 | < .0001 |
| Pro483Leu | 82.1 ± 8 | .007 | 827 ± 178 | .03 |
Results expressed as mean ± SE, n = 6.
Results expressed in nmol/L as mean ± SE, n = 6-10.
Microscopical examination of the cells expressing the missense mutations, following indirect immunofluorescent staining, failed to detect any impairment of intracellular protein trafficking or intracellular accumulation of TM (data not shown). Essentially all TM was distributed around the cell surface membrane following wild-type and mutant vector transfection.
The Km for activation of protein C by the thrombin-TM complex was determined for each TM variant. Increasing amounts of protein C were added to cells expressing TM preincubated with thrombin. Determinations were done initially on the surface of CV-1 cells, which were expressing the TM mutants in a stable fashion. Due to technical difficulties in maintaining the stable cell lines, additional determinations were repeated using COS-7 cells transiently expressing the TM wild-type and the mutants. The composite results for wild-type TM (634 ± 6 nmol/L) are compatible with previously reported results for the Km. The Km values for the mutants bearing the Ala25Thr, Gly61Ala, Asp468Tyr, and Pro483Leu substitutions were essentially identical to that of the wild-type TM. Two mutants, Arg385Ser (2854 ± 249 nmol/L) and Pro477Ser (2716 ± 565 nmol/L) were found to have considerably higher Km values than that of the wild-type TM (Table 1).
Discussion
Because TM is a membrane intercalated receptor, its deficiency is difficult to investigate with an approach based on phenotype. Application of direct gene screening to the TM gene in patients with both venous and arterial thrombosis has identified several sporadic mutations. These mutations are rare events and where families have been involved these have not been large. Consequently, the relationships between TM mutations and the thromboembolic event that led to their identification have been unclear. Although current experimental and clinical evidence supports a biologically plausible explanation for a role of these mutations in the development of thromboembolic disease, the effects of particular sequence changes on function have remained largely unexplored. In the present report we have investigated the expression and protein C activating function of the variant TM proteins encoded by 5 previously described and 2 novel naturally occurring mutations. We have demonstrated that 4 naturally occurring mutations of the TM gene may have a deleterious effect on the expression/function of TM.
The mutation del791-801 is an 11-bp deletion that is predicted to change the reading frame of the protein and cause premature translation termination after nucleotide 982. The predicted protein has an abnormal amino acid sequence from 246 (in the second EGF-like domain), resulting in a premature STOP at codon 306, which effectively deletes the last 4 EGF-like domains. An abnormal, truncated protein could be retained or degraded in the cell, or, because it lacks the transmembrane domain could be secreted. We have investigated these possibilities by performing transient transfection studies followed by RT-PCR and by immunoblotting. Detection of an appropriate-sized product by RT-PCR demonstrated that transcription of the mutated allele occurs. A mutant protein of reduced size was detected in whole cell lysates (prepared with the Laemmli buffer) but not in lysates lacking intracellular organelles (prepared with detergent buffer). It appears that the mutant protein may be present as an insoluble aggregate, or associated with the cytoskeleton. The position of the predicted premature STOP codon and the above in vitro findings suggest that expression and function of the TM on the endothelial cell surface of the patient will be 50%, and result solely from the normal allele. The suggestion is supported by the low levels of TM measured in the plasma from 2 carriers of mutation.
The TM Arg385Ser mutation results in an approximate 50% reduction in expression, coupled with an approximate 4-fold increase in Km. Although this mutant has not been studied before, substitution of Arg385 to Ala has been shown to reduce the cofactor activity of the mutant TM to about 40% of that of the wild type in an in vitro expression study.37 It was also shown that this reduction was not due to an alteration in the thrombin binding. Arg385 is the last amino acid in the fourth EGF-like domain of TM, a domain that has been shown to be necessary for efficient protein C activation.38,39 It is adjacent to the short loop connecting the EGF-like domains 4 and 5. This short interconnecting stretch is formed of 3 amino acids only, the most functionally important of these appears to be Met388. Both this residue and the short loop have been shown to critically affect TM cofactor activity.40,41 Recent crystallography studies predict that the loop is clamped to the TMEGF4 by hydrogen bonds and to TMEGF5 by major hydrophobic contacts. In addition to anchoring the 2 domains together, it directs TMEGF4 away from thrombin. Arg385 is one of the charged amino acids that comprises the TMEGF4 surface extending toward the active site groove of thrombin.42 The crystallography model also predicts direct contacts between the side chains of Arg385 and protein C residues that coordinate calcium binding, specifically Glu80.43 Such interactions have been implicated in the explanation for the unique calcium dependency of the thrombin-TM catalyzed activation of protein C.44 45 It is therefore not surprising to find that substitution of Arg385 by Ser in the present study also results in an increase in Km. This reduction in functional efficiency, coupled with its reduced level of secretion, suggests that the overall functional deficit associated with this mutation in vivo may approach that of del791-801.
The Pro477Ser mutation reduces TM expression about 23% and increases Km about 4-fold. Pro477 is conserved between mouse, bovine, and human TM, and most probably has an important role in shaping the secondary structure of the Ser/Thr rich domain. It is part of a proline triplet that forms a β-turn adjacent to the main chondroitin sulfate glycosaminoglycan (CS) attachment site (Ser 474).46 TM may exist as 2 glycoforms, one containing a CS attachment (+), and the other lacking this high molecular weight glycan (−). The CS+ form binds a second molecule of thrombin.47The CS− form has an increased (3.5-fold) Km in protein C activation.47 The core protein sequences directing O-linked glycosaminoglycan initiation by the enzyme xylosyltransferase are relatively poorly defined.49,50 A general feature is association of modified serine residues with adjacent glycine, as it is the case of the determined core sequence in TM, Ser472-Gly473-Ser474-Gly475, where Ser474 is the attachment site.46 Additional sequence or structural requirements (or both) must exist, because Ser/Gly dipeptides are numerous, but not all are substrates for the enzyme. Indeed it has been shown that the structure of the O-linked domain affects partitioning of TM in the 2 glycoforms. Substitution of the TM domain with that of decorin, mutation of the serines and threonines (except Ser474) to alanine, and deletion of the β-turn formed by Pro477-Pro478-Pro479, all decrease the ratio of TM with attached CS in COS-7 cells. This affected the cofactor activity of the TM in the protein C activation by thrombin.51 Our results are suggestive that mutation Pro477Ser has a similar influence. We were, however, unable to demonstrate a difference in the apparent molecular weight of this mutant by Western blotting. It has been shown that the efficiency of the CS attachment in COS-7 cells is low,51 and the sensitivity of the electrophoresis and Western blotting techniques may be too low to demonstrate small changes that nevertheless alter function. The change may be better reflected in the more sensitive cofactor assay. An alternative explanation may be that of a local conformational change caused by the mutation impeding thrombin binding. Again, the combination of decreased secretion of the mutant and its increased Km suggests an overall appreciable functional deficit in vivo.
Although TM mutant Pro483Leu exhibited a reduction in expression levels, about 18%, no functional alteration could be detected. As with the mutant TM Pro477, this residue is part of the Ser/Thr-rich domain. It lies only 6 amino acids downstream and it is likely that its substitution leads to a local conformational change, which slightly reduces the efficiency of intracellular trafficking to the membrane surface, the efficiency of the glycosylation, or the stability of the mutant protein.
No abnormality in cell surface expression or TM cofactor function could be found for TM mutants with the Ala25thr, Gly61Ala, and Asp468Tyr substitutions. Ala25Thr was associated with MI in the SMILE study.24 Failure to provide biochemical evidence to support a biologically plausible explanation for the statistical data are therefore intriguing. Either the association arose by linkage with another locus, or this was a chance finding, or the mutation mediates a currently unknown function. Ala25 and Gly61 are both in the lectinlike domain of TM, a domain of uncertain function. It has been suggested that the domain participates in the internalization of thrombin from the cell surface,52-54 but the quantitative importance of this function has been questioned.55 Unless another function of TM is mediated through the lectinlike domain, it is unlikely that these mutations would constitute an appreciable risk factor for thrombosis. There has been appreciable recent interest in the role of TM in mediating the activation by thrombin of thrombin-activatable fibrinolysis inhibitor (TAFI), a fibrinolytic inhibitor.9,10,56 However, it currently appears unlikely that mutations in the lectinlike domain can appreciably influence TAFI activation, as this appears to be determined predominantly by EGF-like domains 3-5 of TM.57 58
Mutation Asp486Tyr lies in the Ser/Thr-rich domain of TM. Although this domain has an important spacer function in positioning the thrombin biding site above the cell surface,31 the mutation does not appear to influence this role. Our findings are in agreement with a previous study that failed to show an effect of the mutation on function.50 In that report though, the Km for the wild type was divergent from the consensus value obtained in numerous reports. The lack of any identified functional defect with this mutant is compatible with inconsistent relationships between mutation, thromboembolic disease, and plasma levels of soluble TM.16-19
This work has demonstrated that certain mutations of the TM gene may have a detrimental effect on the expression and protein C activation cofactor function of the protein. These results suggest a plausible mechanism by which TM gene mutations might influence venous and arterial thrombotic disease. Decreased expression and function of TM will result in decreased effectiveness of the protein C anticoagulant pathway. This will be particularly marked in the case of TM del791-801, which results in no expression of intact TM from its allele. The other affected mutants (Arg385Ser, Pro477Ser, and Pro483Leu) have less severe reductions in expression/function that nevertheless could be important in pathophysiologic terms. It is currently unclear how critical the level of expression of TM is to regulation of coagulation in vivo. Although it is known that severe TM deficiency fails to protect against fibrin deposition in mice contained in normal and stressed environments, the clinical evidence for a role of these mutations in human thrombosis is still slender. Although the above findings of impaired expression/function, coupled with results from the experimental studies, provide biochemical support for reduced regulation of thrombin generation by the protein C anticoagulant pathway, the clinical consequences of heterozygous TM deficiency are uncertain. As with other genetic risk factors, the risk of thrombosis of a TM gene mutation in isolation may anyway be small and interaction with other genetic and acquired risk factors may be necessary to trigger a thrombotic event.
Supported by grants from the British Heart Foundation (PG/96065 and PG/98152 to D.A.L.), by grants from the Swedish Medical Research Council (grant K2000-71X-11194-06B), the Medical Faculty at the University of Lund (A.L.F. grant), the Swedish Heart-Lung Foundation and the research funds of the University Hospital in Lund (to A.K.Ö.), and Research funds from the University Hospital, Malmö, the Anna-Lisa and Sven-Eric Lundgrens Trust for Medical Research, the Crafoord Trust, the Foundation for the control of blood diseases, the Carin Trygger Trust (Swedish Medical Society), the Swedish Society for Medical Research and Carl-Bertil Laurells Nordic Fund (to B.Z.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Gabriella Kunz, Department of Haematology, Imperial College School of Medicine, Du Cane Rd, London, W12 0NN, United Kingdom; e-mail: g.kunz@ic.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal