Abstract
The receptors for human granuloctye-macrophage colony-stimulating factor (GM-CSF), interleukin-3 (IL-3), and IL-5 are composed of a ligand-specific α-chain (eg, α–GM-CSF receptor [α-GMR]) and a common β-subunit (β-GMR). Ligand binding is believed to induce assembly or conformational changes in preformed complexes containing more than one α- and β-subunit in the activated receptor complex. To analyze the function of a splice variant of β-GMR with a truncation in the intracellular domain (β-GMRIT), BaF-3 cells expressing human α-GMR plus β-GMR were transfected with β-GMRIT. In these cells, coexpression of β-GMRIT inhibits GM-CSF–mediated survival and proliferation in a GM-CSF concentration–dependent manner. To analyze the effect of cytoplasmic assembly of truncated and full-length intracellular β-GMR sequences, β-GMR and β-GMRIT were coexpressed with different chimeric α/β-GMR constructs. Whereas both β-GMR and β-GMRIT generate high-affinity GMR complexes in the presence of α/β-GMR, β-GMRITinhibits while β-GMR supports proliferation and cell survival mediated by α/β-GMR. Correspondingly, β-GMR, but not β-GMRIT, generates functional GMR complexes when coexpressed with a defective α/β-GMR construct. These data indicate that β-GMRIT can inhibit survival and mitogenic signaling of the wild-type GMR and demonstrate that recruitment of alternatively spliced receptor subunits may regulate the function of heteromeric cytokine receptors.
Introduction
The receptors for human granulocyte-macrophage colony-stimulating factor (GM-CSF) (GMRs), interleukin-3 (IL-3) (IL-3Rs), and IL-5 (IL-5Rs) are heteromeric receptors composed of a ligand-specific α-chain (α-GMR, IL-3Rα, IL-5Rα) and a common β-subunit (β-GMR or βc).1-4 Whereas the α-chains bind their respective ligands with low affinity, β-GMR does not bind any ligand by itself, but confers high-affinity binding in the presence of α-GMR, IL-3Rα, or IL-5Rα. Ligand binding by the α-chain is believed to induce or stabilize the assembly with β-GMR, leading to a ligand-activated complex of α- and β-receptor subunits.5-8 Preformed complexes of α-GMR and β-GMR have been reported in transfected cell lines and in primary myeloid cells even in the absence of GM-CSF.8 Amino acids involved in GM-CSF/α-GMR and GM-CSF/β-GMR interactions have been identified (reviewed in Bagley et al7), and the structure of the activation domain of β-GMR bound to an antagonist has recently been described.9 However, the stoichiometry of the GMR in the presence or absence of GM-CSF has not yet been defined. Data from mutagenesis, immunoprecipitation, cross-linking, and functional studies suggest at least a 2:2:2 configuration of α-GMR to β-GMR to GM-CSF in the activated GMR-complex.7,10-12 This configuration is believed to be stabilized by ligand-receptor interactions as well as by disulfide-linked receptor heterodimerization.11
The molecular mechanisms mediating intracellular signaling are not completely understood. However, β-GMR was found to be essential for survival and mitogenic signaling in all systems studied so far.13,14 The β-GMR constitutively associates with Janus kinase 2 (JAK2)15 and may link receptor activation to transphosphorylation and activation of JAK2 either by cytoplasmic assembly of receptor subunits or by conformational changes in preformed receptor complexes. In addition to its role in activation of JAK2,16 β-GMR is a substrate for tyrosine kinase activity and may be involved in the assembly of signaling complexes via phosphotyrosine/src homology 2 (SH2) or phosphotyrosine/phosphotyrosine binding (PTB)–domain interactions.17-20 Furthermore, Ser585 in β-GMR interacts with 14-3-3 proteins in a phosphoserine-dependent manner.21
At least 2 distinct cytoplasmic regions of β-GMR involved in different signaling cascades have been identified: (1) a membrane proximal domain (β-GMR 455-517), which is required for JAK2 activation and c-myc induction, and (2) a membrane distal region (β-GMR 626-763), which is involved in activation of the Ras-pathway.13 14 In addition, β-GMR–mediated activation and/or phosphorylation of the PI-3K/Akt-pathway, signal transducer and activator of transcription 1 (STAT1), STAT3, STAT5, Vav, Cbl/CrkL, SH2-containing tyrosine phosphatase 1 (SHP-1), and SHP-2, as well as SH2-containing inositol phosphatase (SHIP), have been described.
In 1998, Gale et al22 reported a splice variant of β-GMR lacking a 104-nucleotide exon in myeloid cell lines as well as in normal and primary leukemic cells (β-GMRIT). Loss of the exon leads to a frameshift in the sequence encoding the membrane-proximal cytoplasmic region. The β-GMRIT forms a truncated protein with the extracellular region, the transmembrane region, and the first 23 cytoplasmic amino acids (amino acids 451-473), including the box-1 motif required for interaction with JAK223 in common with β-GMR followed by 23 unique amino acids. When coexpressed with α-GMR, β-GMRITmediates high-affinity binding of GM-CSF and is capable of binding and activating JAK2 in transfected COS-7 cells. However, β-GMRIT does not confer a mitogenic or survival signal upon stimulation with GM-CSF when transfected into α-GMR–expressing cytotoxic T-lymphocyte line 2 (CTLL-2) cells.22
To analyze the function of β-GMRIT in the presence of wild-type α-GMR plus β-GMR, we transfected the murine IL-3–dependent pro-B cell line BaF-3 with expression plasmids encoding α-GMR, β-GMR, and β-GMRIT. Upon expression of functional human GMR, BaF-3 cells can be grown in either murine (m) IL-3 or human (hu) GM-CSF, thus allowing the analysis of cytokine-dependent mitogenic and survival signaling. In addition, we used different chimeric receptors composed of the extracellular and transmembrane regions of α-GMR and the cytoplasmic region of β-GMR5 24 to analyze the cytoplasmic assembly of full-length and truncated β-GMR sequences in transfected BaF-3 cells. We demonstrate that β-GMRIT acts as a dominant-negative variant of β-GMR for survival and mitogenic signaling. These data suggest the regulation of heteromeric cytokine receptor function by recruitment of alternatively spliced receptor subunits.
Materials and methods
Plasmids
Generation of transgenic BaF-3 cell lines
BaF-3 cells were grown in RPMI 1640 supplemented with 10% fetal calf serum (FCS) and 5% WEHI-conditioned medium (WEHI-CM) for the supply of murine IL-3.5,24 For transfection, 1.5 × 106 cells were electroporated at 350 V and 950 μF at 4°C in the presence of 10 μg transgenic plasmid. BaF-3/α-GMR plus β-GMR cells were grown in 20 ng/mL huGM-CSF in the presence of 500 μg/mL G418 and 1.4 μg/mL puromycin and were cotransfected with 20 μg pcDNA3-β-GMRIT and 1 μg pZeoSV to generate triple transfectants. After electroporation, cells were suspended in culture medium with WEHI-CM for 48 hours and 104 cells per well were then seeded in 96-well tissue-culture plates for selection. Parental BaF-3 cells were transfected with pcDNA3 encoding the chimeric α/β-GMR and selected with huGM-CSF in the absence of mIL-3 as described.5,24BaF3 cells transfected with pZeoSV plasmids either without transgene or encoding both β-GMR isoforms were selected in the presence of Zeocin (1 mg/mL) (Invitrogen) and WEHI-CM. Generation of BaF-3 α/β-GMR-ΔE cells has been described earlier.24
Conventional reverse-transcriptase–polymerase chain reaction
RNA was extracted from BaF-3 cells by means of Trizol reagent (Life Technologies, Rockville, MD). For reverse-transcriptase–polymerase chain reaction (RT-PCR), 3 μg RNA was reverse-transcribed in a 50-μL reaction mix containing 100 U Moloney murine leukemia virus RT, 1 μg oligo d(T) primer, 1 mM each deoxynucleoside 5′-triphosphate (dNTP), 10 mM dithiothreitol, and 40 U RNase inhibitor (RNase Block) (Stratagene, La Jolla, CA) in appropriate buffer conditions. We subjected 4 μL of the RT reaction to PCR with 35 cycles of denaturation (95°C, 1 minute), annealing (62°C, 1 minute), and extension (72°C, 2 minutes). The reaction mix contained 50 pmol both sense and antisense primer, 0.48 mM each dNTP, and 1.5 mM MgCl2 in a total volume of 20 μL. After 5 minutes of denaturation, 2.5 U Taq Polymerase (Perkin-Elmer, Weiterstadt, Germany) was added. To amplify both α-GMR and chimeric α/β-GMR sequences, a set of primers from the extracellular domain of α-GMR was chosen (sense, CGGATCTGCGAACAGTGG; antisense, GCGGACGTCTGCAGCTCTG), resulting in an amplicon of 822 base pairs (bp) (primer set A). We achieved messenger RNA (mRNA) detection of both β-GMR isoforms by amplification of a membrane-spanning sequence, resulting in an amplicon of 529 bp for β-GMR and 425 bp for β-GMRIT, respectively, as described earlier (sense, GCACCGGCT ACAACGGGACCT; antisense, CAGGTAGGGCCCATTGAAGTC) (primer set B).25 Intracellular β-GMR sequences resulting from α/β-GMR, β-GMR, or β-GMRIT were amplified with a third set of primers yielding a 386–bp fragment from α/β-GMR and β-GMR, and a 282–bp amplicon from β-GMRIT (sense, TACGGGTACAGGCTGCG; antisense, CAG GTAGGGCCCATTGAAGTC) (primer set C).
TaqMan RT-PCR
The differential quantitation of β-GMR and β-GMRIT was performed on an ABI Prism 7700 sequence detector (Applied Biosystems, Foster City, CA) with 40 cycles of a 2-step PCR (15 seconds of denaturation at 95°C and 60 seconds of annealing and extension at 63°C).25 The sequences for isoform-specific primers and for TaqMan probes and the validation of the differential quantitation have been described in detail.25 Standard curves were generated from 50 to 105 β-GMR and β-GMRIT plasmid molecules, respectively, with correlation coefficients of at least 0.98 for each analysis.
Surface expression of transfected GMR-constructs
Surface expression of α-GMR, α/β-GMR, or either β-GMR isoform were analyzed by fluorescence-activated cell sorting (FACS) analysis. For this analysis, 5 × 105 cells were incubated with a monoclonal antibody directed against the extracellular domain of α-GMR (GM-CSFRα S-20; dilution 1:100) (Santa Cruz Biotechnology, Santa Cruz, CA) or β-GMR (GM-CSFRβ S-16, dilution 1:10; Santa Cruz Biotechnology) for 60 minutes on ice followed by incubation with a fluorescein isothiocyanate (FITC)–labeled goat antimouse antibody (Dianova, Hamburg, Germany). Untransfected BaF-3 cells and transfected cells incubated with the secondary antibody served as controls.
Proliferation assay
GM-CSF–dependent proliferation was analyzed by means of the CyQuant Cell Proliferation Assay Kit (Molecular Probes, Eugene, OR) according to the manufacturer's instructions. Briefly, 1 × 104 to 1 × 105 cells/mL were grown in 96-well tissue-culture plates in a total volume of 200 μL with increasing amounts of huGM-CSF. After 48 to 72 hours, cells were harvested by centrifugation, lysed by freezing and subsequent incubation in lysis buffer, and stained with CyQuant GR5. The fluorescence was measured by means of an enzyme-linked immunosorbent assay reader at 540 nm. Untransfected BaF-3 cells were always included as controls. In parallel, cell numbers and viability were determined by trypan blue staining.
Apoptosis
For quantification of apoptosis, BaF-3 clones were grown at 1 × 105 cells/mL in the presence of either varying concentrations of huGM-CSF or 5% WEHI-CM. After 48 hours, cells were washed twice and stained with annexin V and propidium iodide (PI) with the use of the annexin V/FITC kit from Bender Med Systems (Vienna, Austria). Antihuman CD8–FITC (Becton-Dickinson, San Jose, CA) was used as control, and annexin V– and PI-positive cells were quantitated by FACS analysis.
Detection of β-GMRIT by immunoprecipitation and immunoblotting
We suspended 15 × 106 BaF-3/α/β-GMR and BaF-3/α/β-GMR plus β-GMRIT cells (Nos. 1 through 5) as well as BaF-3/α/β-GMR-ΔE and BaF-3/α/β-GMR-ΔE plus β-GMRIT (Nos. 1, 3, and 4) in lysis buffer as described.5 The lysates were incubated with 5 μL affinity-purified anti–β-GMRIT polyclonal antiserum (Gale et al22), kindly provided by Dr R. Gale, for 4 hours at 4°C. Immune complexes were precipitated with protein G–sepharose beads, washed 4 times in lysis buffer, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and transferred to Protran nitrocellulose membranes (Schleicher & Schuell, Dassell, Germany). The membranes were blocked with 5% dry milk plus 3% bovine serum albumin (BSA) in phosphate-buffered saline and incubated with the polyclonal anti–β-GMR antiserum N20 at a 1:200 dilution (Santa Cruz Biotechnology). The β-GMRIT was visualized by chemiluminescence by means of the Renaissance kit (NEN Life Science, Boston, MA).
Equilibrium binding of 125I–GM-CSF
BaF-3 clones grown in WEHI-CM were washed 3 times, and 1.5 × 106 to 5 × 106 cells were incubated for 90 minutes on ice with 125I–GM-CSF (NEN Life Science Products) in a total volume of 200 μL binding buffer (25 mM Hepes, 5% BSA, in RPMI 1640) in the presence or absence of 1 μM unlabeled GM-CSF. The GM-CSF molecular weight of 14 000 d was used to calculate molar concentrations with 10 ng/mL corresponding to about 0.7 nM. Specific 125I–GM-CSF binding was analyzed at concentrations between 0.01 nM and 5 nM. For analysis of GM-CSF binding at concentrations of 1 nM or higher, 125I–GM-CSF was diluted with unlabeled GM-CSF, resulting in decreased specific activity. After incubation, the cells were pelleted and the supernatant was saved for analysis. The pellet was resuspended in 500 μL cold binding buffer, layered over 700 μL FCS, and spun at 14 000g for 2 minutes. After aspirating the supernatant, radioactivity of both the cell pellet and the supernatant from the first incubation was measured in a Berthold MAG 315 γ-counter. The equilibrium dissociation constant (KD) and the density of binding sites were analyzed by means of Scatchard analysis, and the results were confirmed in a second analysis with the LIGAND program (Biosoft, Cambridge, United Kingdom).
Results
Coexpression of β-GMRIT with wild-type GMR inhibits GM-CSF–dependent survival and mitogenic signaling
To analyze the function of β-GMRIT in the context of wild-type GMR, we transfected BaF-3 cells expressing human α-GMR and β-GMR (BaF-3/α-GMR plus β-GMR) with the cDNA encoding β-GMRIT. After culture in mIL-3 and Zeocin and analysis of transgene expression by conventional and isoform-specific TaqMan RT-PCR,25 4 independent clones were selected for further analysis. As shown in Figure 1A and Table1, both β-GMRIT and β-GMR mRNA expression could be detected in all 4 triple-transfected BaF-3/α-GMR plus β-GMR plus β-GMRIT clones. The ratio of β-GMRIT ranged from 83.6% to 95.1% of total β-GMR mRNA. FACS analysis with a monoclonal antibody against an extracellular epitope identical for both β-GMR and β-GMRIT revealed a slightly increased staining after transfection of β-GMRIT(Figure 1B and data not shown). To determine the impact of β-GMRIT on GMR-mediated mitogenesis, GM-CSF–dependent proliferation of BaF-3/α-GMR plus β-GMR and triple-transfected BaF-3/α-GMR plus β-GMR plus β-GMRIT cells was analyzed in the absence of mIL-3. Coexpression of β-GMRITinhibited GMR-mediated proliferation at low concentrations in all 4 BaF-3/α-GMR plus β-GMR plus β-GMRIT clones with approximately 5-fold more GM-CSF needed for proliferation as compared with BaF-3/α-GMR plus β-GMR cells (Figure2). However, at 100 ng/mL huGM-CSF, all clones showed maximal proliferation, even with expression of β-GMRIT. In addition, when cultured for 3 days at low concentrations of huGM-CSF, triple-transfected cells showed diminished proliferation as compared with BaF-3/α-GMR plus β-GMR cells, whereas at concentrations of 20 ng/mL huGM-CSF no inhibition of cell proliferation was detectable (data not shown).
mRNA and protein expression of GMR-constructs in transfected BaF-3 cells.
The mRNA expression of GMR-constructs in BaF-3/α-GMR plus β-GMR cells after cotransfection of β-GMRIT. Primer set A was used for amplification of α-GMR by conventional RT-PCR as described in “Materials and methods.” The β-GMR and β-GMRITwere detected with primer set C, resulting in amplicons of 386 bp and 282 bp, respectively. The α plus β indicates BaF-3/α-GMR plus β-GMR cells, and Nos. 1 through 4 indicate independent BaF-3/α-GMR plus β-GMR plus β-GMRIT clones. (B) Surface expression of GMR-constructs in transfected BaF-3 cells. BaF-3/α-GMR plus β-GMR (panel Bi) and BaF-3/α-GMR plus β-GMR plus β-GMRIT cells (panel Bii) were stained with an anti–α-GMR (black) and an anti–β-GMR antibody recognizing both isoforms of β-GMR (gray) followed by FACS analysis. The interrupted line represents controls stained with the secondary antibody only.
mRNA and protein expression of GMR-constructs in transfected BaF-3 cells.
The mRNA expression of GMR-constructs in BaF-3/α-GMR plus β-GMR cells after cotransfection of β-GMRIT. Primer set A was used for amplification of α-GMR by conventional RT-PCR as described in “Materials and methods.” The β-GMR and β-GMRITwere detected with primer set C, resulting in amplicons of 386 bp and 282 bp, respectively. The α plus β indicates BaF-3/α-GMR plus β-GMR cells, and Nos. 1 through 4 indicate independent BaF-3/α-GMR plus β-GMR plus β-GMRIT clones. (B) Surface expression of GMR-constructs in transfected BaF-3 cells. BaF-3/α-GMR plus β-GMR (panel Bi) and BaF-3/α-GMR plus β-GMR plus β-GMRIT cells (panel Bii) were stained with an anti–α-GMR (black) and an anti–β-GMR antibody recognizing both isoforms of β-GMR (gray) followed by FACS analysis. The interrupted line represents controls stained with the secondary antibody only.
Ratio (%) of β-granulocyte-macrophage colony-stimulating factor receptor with a truncation in the intracellular domain (GMRIT) to β-GMR plus β-GMRIT in transfected BaF-3/α-GMR plus β-GMR plus β-GMRITcells
| Clone no. . | β-GMRIT/ (β-GMR + β-GMRIT), % . | SD, % . |
|---|---|---|
| 1 | 83.6 | 7.9 |
| 2 | 95.1 | |
| 3 | 85.8 | 6.9 |
| 4 | 92.3 | 4.1 |
| Clone no. . | β-GMRIT/ (β-GMR + β-GMRIT), % . | SD, % . |
|---|---|---|
| 1 | 83.6 | 7.9 |
| 2 | 95.1 | |
| 3 | 85.8 | 6.9 |
| 4 | 92.3 | 4.1 |
Data were determined by quantitative TaqMan reverse-transcriptase polymerase chain reaction. Numbers represent mean values and SDs in percentages. Since clone No. 2 was analyzed only twice, no SD was calculated for it.
Inhibition by β-GMRIT of proliferation mediated by α-GMR plus β-GMR.
BaF-3/α-GMR plus β-GMR (black square) and BaF-3/α-GMR plus β-GMR plus β-GMRIT cells (Nos. 1 through 4) were cultured with increasing concentrations of huGM-CSF, as indicated, or in the presence of mIL-3 (10 ng/mL). GM-CSF–mediated proliferation was normalized in comparison with proliferation induced by mIL-3 (100%). The background-corrected fluorescence intensity in response to mIL-3 was between 112 and 133, and background fluorescence without cytokine stimulation was lower than 5 relative fluorescence units. Untransfected cells did not exhibit any huGM-CSF–dependent proliferation.
Inhibition by β-GMRIT of proliferation mediated by α-GMR plus β-GMR.
BaF-3/α-GMR plus β-GMR (black square) and BaF-3/α-GMR plus β-GMR plus β-GMRIT cells (Nos. 1 through 4) were cultured with increasing concentrations of huGM-CSF, as indicated, or in the presence of mIL-3 (10 ng/mL). GM-CSF–mediated proliferation was normalized in comparison with proliferation induced by mIL-3 (100%). The background-corrected fluorescence intensity in response to mIL-3 was between 112 and 133, and background fluorescence without cytokine stimulation was lower than 5 relative fluorescence units. Untransfected cells did not exhibit any huGM-CSF–dependent proliferation.
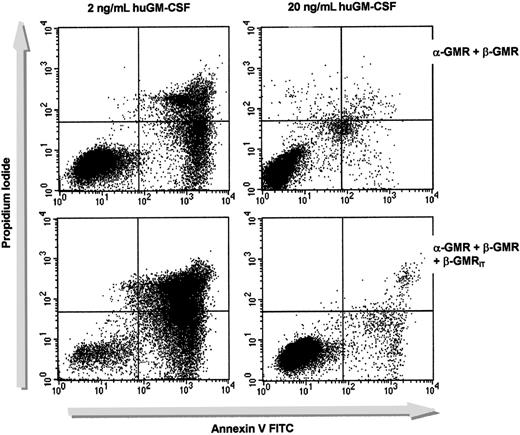
We next analyzed the effects of β-GMRIT expression on GM-CSF–dependent cell survival. BaF-3/α-GMR plus β-GMR and BaF-3/α-GMR plus β-GMR plus β-GMRIT (no. 1) were cultured in the presence of either 2 ng/mL or 20 ng/mL huGM-CSF without mIL-3 for 2 days, and the proportion of apoptotic cells was determined by annexin V staining (Figure 3). BaF-3/α-GMR plus β-GMR cells showed 3.4% annexin V–positive cells when grown in 20 ng/mL huGM-CSF. This rate increased to 33.6% when cells were incubated in 2 ng/mL huGM-CSF. While BaF-3/α-GMR plus β-GMR plus β-GMRIT exhibited a similar amount of annexin V–positive cells at 20 ng/mL (4.4%), a significantly higher rate of apoptosis (84.1% annexin V–positive cells) was detected in cells cultured in 2 ng/mL huGM-CSF. Comparable results were found with all other clones of triple-transfected BaF-3 cells (data not shown).
The β-GMRIT inhibits cell survival mediated by α-GMR plus β-GMR.
BaF-3/α-GMR plus β-GMR (upper panels) and BaF-3/α-GMR plus β-GMR plus β-GMRIT cells (no. 1, lower panels) were grown in either low (2 ng/mL, left) or high (20 ng/mL, right) concentrations of huGM-CSF. Cells were incubated with PI and annexin V after 48 hours and analyzed by FACS. Annexin V–positive cells were considered apoptotic. When the cells were cultured in WEHI-CM, the rate of apoptosis was equal in all clones tested.
The β-GMRIT inhibits cell survival mediated by α-GMR plus β-GMR.
BaF-3/α-GMR plus β-GMR (upper panels) and BaF-3/α-GMR plus β-GMR plus β-GMRIT cells (no. 1, lower panels) were grown in either low (2 ng/mL, left) or high (20 ng/mL, right) concentrations of huGM-CSF. Cells were incubated with PI and annexin V after 48 hours and analyzed by FACS. Annexin V–positive cells were considered apoptotic. When the cells were cultured in WEHI-CM, the rate of apoptosis was equal in all clones tested.
Coexpression of β-GMR or β-GMRIT with a functional chimeric α/β-GMR generates high-affinity GMR-complexes in BaF-3 cells
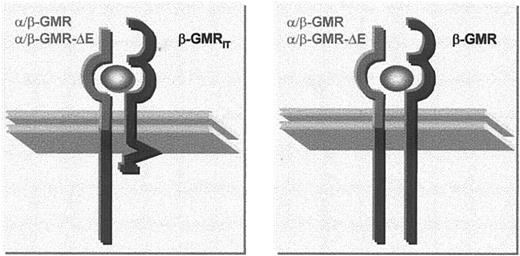
To focus on the effects of cytoplasmic assembly of full-length and truncated β-GMR sequences into GMR-complexes, β-GMRITwas coexpressed with a functional chimeric α/β-GMR that was created by fusing the extracellular and transmembrane regions of α-GMR to the cytoplasmic domain of β-GMR.5 The α/β-GMR binds GM-CSF with low affinity and is capable of mediating survival and mitogenic signaling even in the absence of β-GMR. However, coexpression of β-GMR with α/β-GMR in BaF-3 cells allows proliferation in GM-CSF concentrations about 100-fold lower than in cells lacking β-GMR.5 Upon GM-CSF binding, α/β-GMR is thought to interact with the extracellular domain of either β-GMR or β-GMRIT. This extracellular interaction results in cytoplasmic assembly of β-GMR sequences (from α/β-GMR and β-GMR) or to hetero-oligomerization of truncated chain (from β-GMRIT) with the wild-type (from α/β-GMR) sequences as schematically depicted in Figure 4.
Intracellular configuration of GMR complexes.
The images present configurations of cytoplasmic β-GMR sequences in cells expressing chimeric α/β-GMR–constructs and β-GMRIT or β-GMR.
Intracellular configuration of GMR complexes.
The images present configurations of cytoplasmic β-GMR sequences in cells expressing chimeric α/β-GMR–constructs and β-GMRIT or β-GMR.
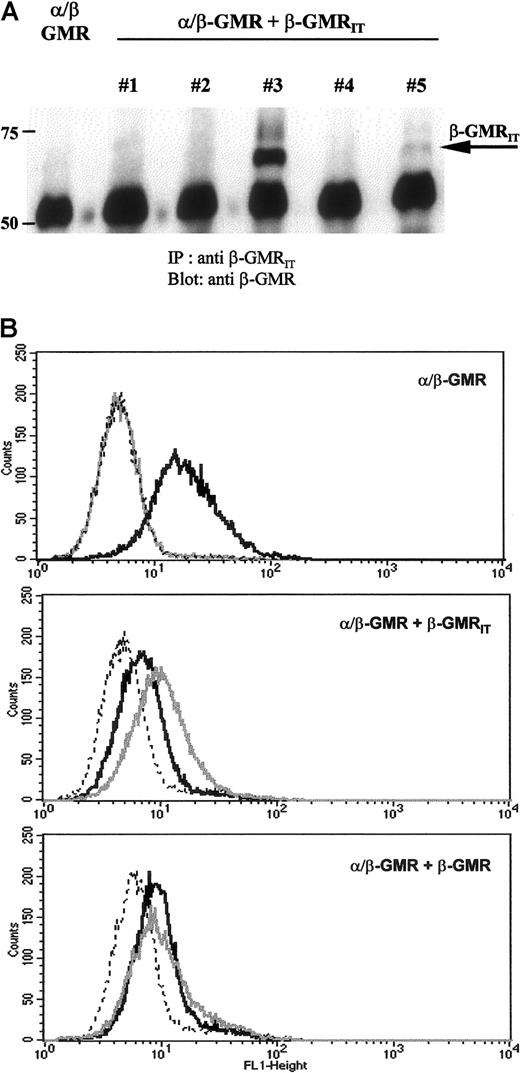
BaF-3/α/β-GMR cells were cotransfected with either β-GMR or β-GMRIT and cultured in medium supplemented with mIL-3 and Zeocin. Five independent BaF-3/α/β-GMR plus β-GMRIT clones and 2 independent BaF-3/α/β-GMR plus β-GMR clones were selected and analyzed by conventional RT-PCR. The β-GMRIT and the β-GMR mRNA expression were detectable in all BaF-3/α/β-GMR plus β-GMRIT and BaF-3/α/β-GMR plus β-GMR clones, respectively (data not shown). Protein expression of β-GMRIT in BaF-3/α/β-GMR plus β-GMRIT clones was analyzed by immunoprecipitation and Western blotting as well as by FACS analysis. As shown in Figure5A, β-GMRIT protein expression was heterogeneous among BaF-3/α/β-GMR plus β-GMRIT clones. High expression was observed in clone no. 3. Clone no. 5 harbored only moderate levels of the protein, and no detectable expression was detected in clones no. 1, 2, and 4. These data were confirmed by FACS analysis, which also demonstrated high expression of β-GMRIT in clone no. 3, moderate expression in clone no. 5, and no detectable surface expression in clones no. 1, 2, and 4 (Figure 5B and data not shown).
Protein expression of α/β-GMR, β-GMR, and β-GMRIT in transfected BaF-3 cells.
(A) Immunoprecipitates of β-GMRIT from BaF-3/α/β-GMR (left) and BaF-3/α/β-GMR plus β-GMRIT cells (clones no. 1 through 5) were immunoblotted with a monoclonal antibody against the extracellular domain of β-GMR and β-GMRIT. The molecular weight is given in kilodaltons on the left, and the migration of β-GMRIT is indicated by the arrow. (B) BaF-3/α/β-GMR (top), BaF-3/α/β-GMR plus β-GMRIT(no. 3, middle), and BaF-3/α/β-GMR plus β-GMR (bottom) cells were stained with an anti–α-GMR antibody recognizing α/β-GMR (black) and an anti–β-GMR antibody recognizing both β-GMR variants (gray) followed by FACS analysis. The interrupted line represents controls stained with the secondary antibody only.
Protein expression of α/β-GMR, β-GMR, and β-GMRIT in transfected BaF-3 cells.
(A) Immunoprecipitates of β-GMRIT from BaF-3/α/β-GMR (left) and BaF-3/α/β-GMR plus β-GMRIT cells (clones no. 1 through 5) were immunoblotted with a monoclonal antibody against the extracellular domain of β-GMR and β-GMRIT. The molecular weight is given in kilodaltons on the left, and the migration of β-GMRIT is indicated by the arrow. (B) BaF-3/α/β-GMR (top), BaF-3/α/β-GMR plus β-GMRIT(no. 3, middle), and BaF-3/α/β-GMR plus β-GMR (bottom) cells were stained with an anti–α-GMR antibody recognizing α/β-GMR (black) and an anti–β-GMR antibody recognizing both β-GMR variants (gray) followed by FACS analysis. The interrupted line represents controls stained with the secondary antibody only.
The GM-CSF–dependent interactions of α/β-GMR with β-GMR or β-GMRIT were studied by analyzing the equilibrium-binding characteristics of BaF-3/α/β-GMR, BaF-3/α/β-GMR plus β-GMR, and BaF-3/α/β-GMR plus β-GMRIT (no. 3) cells after incubation with 125I–GM-CSF. Scatchard plot analysis revealed only low-affinity binding sites for 125I–GM-CSF in BaF-3/α/β-GMR cells with KD between 387 pM and 3981 pM (mean, 1800 pM) and between 537 and 5300 receptors per cell (mean, 2611) (Table 2). In contrast, for both BaF-3/α/β-GMR plus β-GMR and BaF-3/α/β-GMR plus β-GMRIT cells, high-affinity receptors for GM-CSF were revealed with KD 200 pM (α/β-GMR plus β-GMR) and KD 218 pM (α/β-GMR plus β-GMRIT), respectively. In addition, BaF-3/α/β-GMR plus β-GMRIT cells also exhibited low-affinity binding of GM-CSF with an estimated KD of 2570 pM.
Equilibrium binding of 125I–GM-CSF to BaF-3 cells transfected with GMR constructs
| Clone . | KD, pM . | BMax, pM . | Receptor/cell . |
|---|---|---|---|
| α/β-GMR | 1800 | 43.1 | 2611 |
| α/β-GMR + β-GMR | 200 | 10.9 | 504 |
| α/β-GMR + β-GMRIT | 218 | 12.5 | 603 |
| 2570 | 59.6 | 2872 |
| Clone . | KD, pM . | BMax, pM . | Receptor/cell . |
|---|---|---|---|
| α/β-GMR | 1800 | 43.1 | 2611 |
| α/β-GMR + β-GMR | 200 | 10.9 | 504 |
| α/β-GMR + β-GMRIT | 218 | 12.5 | 603 |
| 2570 | 59.6 | 2872 |
Data represent mean values of 4 (α/β-GMR) and 2 (α/β-GMR + β-GMR) independent experiments; α/β-GMR + β-GMRIT cells (clone 3) were analyzed once.
KD indicates equilibrium dissociation constant; BMax, density of binding sites.
Coexpression of β-GMRIT with a functional chimeric α/β-GMR inhibits GM-CSF–dependent survival and mitogenic signaling
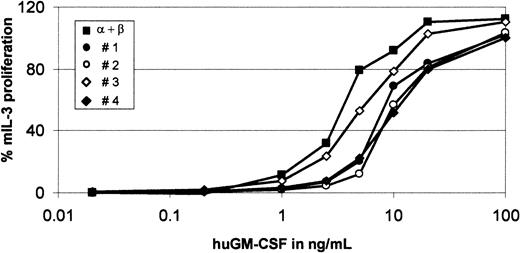
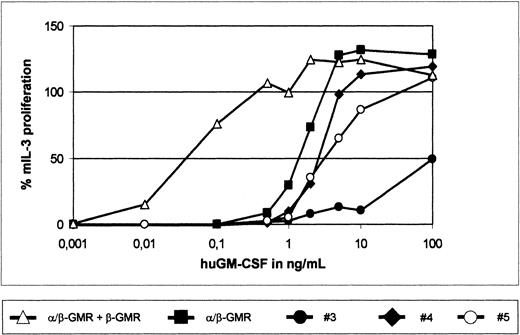
To study the impact of β-GMR and β-GMRIT on α/β-GMR signaling, the GM-CSF–dependent proliferation of BaF-3/α/β-GMR, BaF-3/α/β-GMR plus β-GMR, and BaF-3/α/β-GMR plus β-GMRIT cells (clones no. 3, 4, and 5) was analyzed. As demonstrated earlier,5BaF-3/α/β-GMR plus β-GMR cells required about 100-fold lower concentrations of huGM-CSF for proliferation as compared with BaF-3/α/β-GMR cells (Figure 6). In contrast, proliferation of BaF-3/α/β-GMR plus β-GMRITclones required higher concentrations of huGM-CSF as compared with the parental BaF-3/α/β-GMR cells, with an inverse correlation of β-GMRIT protein expression and GM-CSF responsiveness (Figures 5A, 6). In addition, when grown at different concentrations of huGM-CSF for 3 days, BaF-3/α/β-GMR plus β-GMR and BaF-3/α/β-GMR cells showed maximal expansion similar to cultures stimulated with mIL-3 at 1 ng/mL and 10 ng/mL of huGM-CSF, respectively. In contrast, BaF-3/α/β-GMR plus β-GMRITclone no. 5 required 100 ng/mL to reach cell expansion comparable to that induced by mIL-3. Interestingly, at the huGM-CSF concentrations tested, clone no. 3, which has the highest β-GMRITexpression level (Figure 5A), never approached more than 50% of that induced by mIL-3 (data not shown).
The β-GMR supports but the β-GMRITinhibits proliferation mediated by α/β-GMR.
BaF-3/α/β-GMR (black square), BaF-3/α/β-GMR plus β-GMR (triangle), and BaF-3/α/β-GMR plus β-GMRIT (clones no. 3 through 5) cells were seeded at increasing concentrations of huGM-CSF, as indicated, or in the presence of mIL-3 (10 ng/mL). GM-CSF–mediated proliferation was normalized in comparison with proliferation induced by mIL-3 (100%). The background-corrected fluorescence intensity in response to mIL-3 was between 106 and 133, and background fluorescence without cytokine stimulation was lower than 4 relative fluorescence units.
The β-GMR supports but the β-GMRITinhibits proliferation mediated by α/β-GMR.
BaF-3/α/β-GMR (black square), BaF-3/α/β-GMR plus β-GMR (triangle), and BaF-3/α/β-GMR plus β-GMRIT (clones no. 3 through 5) cells were seeded at increasing concentrations of huGM-CSF, as indicated, or in the presence of mIL-3 (10 ng/mL). GM-CSF–mediated proliferation was normalized in comparison with proliferation induced by mIL-3 (100%). The background-corrected fluorescence intensity in response to mIL-3 was between 106 and 133, and background fluorescence without cytokine stimulation was lower than 4 relative fluorescence units.
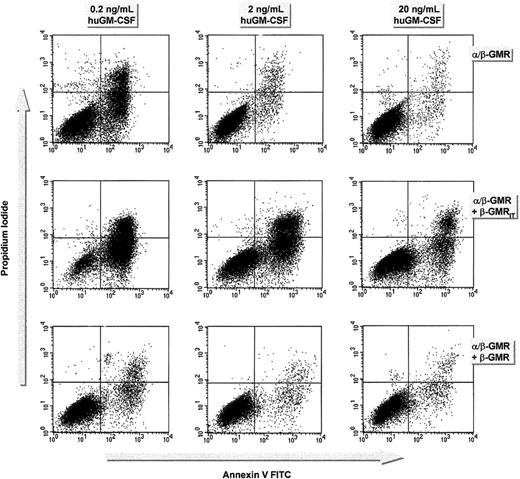
To analyze the effects of β-GMR or β-GMRIT expression on α/β-GMR–mediated cell survival, BaF-3/α/β-GMR, BaF-3/α/β-GMR plus β-GMR, and BaF-3/α/β-GMR plus β-GMRIT cells were cultured in the presence of different concentrations of huGM-CSF without mIL-3 for 2 days. The proportion of apoptotic cells was then determined by annexin V staining. At concentrations of 0.2 ng/mL, 2 ng/mL, and 20 ng/mL huGM-CSF, the proportion of annexin V–positive cells was 33.4%, 4.7%, and 3.4% for BaF-3/α/β-GMR; 88.5%, 54.3%, and 21% for BaF-3/α/β-GMR plus β-GMRIT cells (no. 3); and 9%, 4.6%, and 4.6% for BaF-3/α/β-GMR plus β-GMR, respectively (Figure7).
Effect of β-GMR and β-GMRIT on α/β-GMR–mediated cell survival.
The α/β-GMR–mediated cell survival is supported by β-GMR but inhibited by β-GMRIT. BaF-3/α/β-GMR (top), BaF-3/α/β-GMR plus β-GMRIT (clone no. 3, middle), and BaF-3/α/β-GMR plus β-GMR cells (bottom) were grown in 0.2 (left), 2 (middle), or 20 ng/mL huGM-CSF (right). Cells were incubated with PI and annexin V after 48 hours and analyzed by FACS. Annexin V–positive cells were considered apoptotic. When the cells were cultured in WEHI-CM, the rate of apoptosis was equally low (less than 6%) in all clones tested. Data represent 1 out of 3 experiments.
Effect of β-GMR and β-GMRIT on α/β-GMR–mediated cell survival.
The α/β-GMR–mediated cell survival is supported by β-GMR but inhibited by β-GMRIT. BaF-3/α/β-GMR (top), BaF-3/α/β-GMR plus β-GMRIT (clone no. 3, middle), and BaF-3/α/β-GMR plus β-GMR cells (bottom) were grown in 0.2 (left), 2 (middle), or 20 ng/mL huGM-CSF (right). Cells were incubated with PI and annexin V after 48 hours and analyzed by FACS. Annexin V–positive cells were considered apoptotic. When the cells were cultured in WEHI-CM, the rate of apoptosis was equally low (less than 6%) in all clones tested. Data represent 1 out of 3 experiments.
The β-GMR but not β-GMRIT can complement the function of the defective α/β-GMR-ΔE chimera
We next asked whether β-GMRIT, like β-GMR, was able to generate a functional GMR when coexpressed with the defective chimeric α/β-GMR-ΔE construct. In α/β-GMR-ΔE, deletion of a glutamic acid residue initially introduced into α/β-GMR results in loss of GM-CSF–dependent signaling capacity.24 However, α/β-GMR-ΔE can mediate proliferation and survival upon stimulation with huGM-CSF and β-GMR coexpression, or upon activation with a monoclonal anti–α-GMR antibody. Therefore, BaF-3/α/β-GMR-ΔE cells were cotransfected with β-GMR or β-GMRIT. Twelve independent BaF-3/α/β-GMR-ΔE plus β-GMR clones and 11 BaF-3/α/β-GMR-ΔE plus β-GMRITclones were isolated. Both β-GMRIT and β-GMR mRNA expression were detected in all BaF-3/α/β-GMR-ΔE plus β-GMRIT clones studied (No. = 11) and in all 3 BaF-3/α/β-GMR-ΔE plus β-GMR clones analyzed (data not shown). In addition, protein expression of β-GMRIT in BaF-3/α/β-GMR-ΔE and in BaF-3/α/β-GMR-ΔE plus β-GMRIT cells (nos. 1, 3, and 4) was detectable by immunoprecipitation and Western blotting in all BaF-3/α/β-GMR-ΔE plus β-GMRIT clones studied, but not in BaF-3/α/β-GMR-ΔE cells (data not shown).
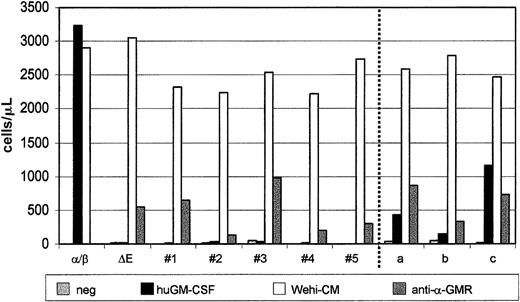
BaF-3/α/β-GMR, BaF-3/α/β-GMR-ΔE, BaF-3/α/β-GMR-ΔE plus β-GMR, and BaF-3/α/β-GMR-ΔE plus β-GMRIT cells were cultured for 3 days alone or in the presence of mIL-3, huGM-CSF (20 ng/mL), or the anti–α-GMR monoclonal antibody GM-CSFRα S-20. All clones grew similarly in the presence of mIL-3 (Figure8). However, only BaF-3/α/β-GMR cells showed similar expansion after stimulation with either mIL-3 or huGM-CSF. All BaF-3 clones expressing α/β-GMR-ΔE, alone or in combination with β-GMR or β-GMRIT, could be grown in the presence of the monoclonal anti–α-GMR antibody. However, whereas BaF-3/α/β-GMR-ΔE and BaF-3/α/β-GMR-ΔE plus β-GMRIT cells did not proliferate and survive in the presence of huGM-CSF, BaF-3/α/β-GMR-ΔE plus β-GMR clones showed huGM-CSF–dependent cell expansion similar to that induced by the anti–α-GMR antibody.
The β-GMR but not β-GMRIT confers GM-CSF–dependent proliferation when coexpressed with the defective α/β-GMR-ΔE chimera.
In this experiment, 1 × 105 BaF-3/α/β-GMR (α/β), BaF-3/α/β-GMR-ΔE (ΔE), BaF-3/α/β-GMR-ΔE plus β-GMRIT (clones no. 1 through 5), and BaF-3/α/β-GMR-ΔE plus β-GMR (clones a through c) cells were cultured in the presence of WEHI-CM as a source of mIL-3, huGM-CSF (20 ng/mL), or the monoclonal anti–α-GMR S-20 antibody as indicated. Viable cells were counted after trypan blue staining on day 3.
The β-GMR but not β-GMRIT confers GM-CSF–dependent proliferation when coexpressed with the defective α/β-GMR-ΔE chimera.
In this experiment, 1 × 105 BaF-3/α/β-GMR (α/β), BaF-3/α/β-GMR-ΔE (ΔE), BaF-3/α/β-GMR-ΔE plus β-GMRIT (clones no. 1 through 5), and BaF-3/α/β-GMR-ΔE plus β-GMR (clones a through c) cells were cultured in the presence of WEHI-CM as a source of mIL-3, huGM-CSF (20 ng/mL), or the monoclonal anti–α-GMR S-20 antibody as indicated. Viable cells were counted after trypan blue staining on day 3.
Discussion
We analyzed the expression and function of an alternatively spliced variant of the common β-chain of the human receptors for GM-CSF, IL-3, and IL-5 (β-GMRIT) in the mIL-3–dependent BaF-3 cell line. The β-GMRIT is translated into a molecule truncated C-terminal of the cytoplasmic box 1 motif required for interaction with JAK2.22,23 We used ectopic expression of different GMR constructs to study GMR-mediated mitogenic and survival signaling. It is important to note that this model cannot be used to analyze signaling involved in cellular differentiation.26,27 We found high relative β-GMRIT mRNA expression in all analyzed BaF-3 clones coexpressing α-GMR, β-GMR, and β-GMRIT. Among these clones, 4 with the highest mRNA levels were selected. A dominant negative effect of β-GMRIT on mitogenic and survival signaling mediated by the wild-type GMR (α-GMR plus β-GMR) has been observed in these cells (Figures 2, 3). However, the inhibition of receptor function by β-GMRIT depended on the GM-CSF concentration, suggesting that, even in the presence of high β-GMRIT mRNA levels, a sufficient number of functional GMR complexes could be activated at higher concentrations of the ligand in this system. It remains unclear to what extent these data are applicable to other cell types. It is conceivable that the effects of β-GMRIT are underestimated in the BaF-3 model since BaF-3 cells can proliferate upon expression of various GMR mutants that are defective to induce proliferation in other cell lines, such as WT19 or CTLL-2.28 29
If GM-CSF/GMR complexes display a 2:2:2 configuration of α-GMR to β-GMR to GM-CSF,7,10-12 and if α-GMR, β-GMR, and β-GMRIT are expressed in the same cell, 3 types of GMR complexes differing only in the β-GMR components may be formed: the functional complex with 2 β-GMR molecules, a nonfunctional complex with 2 β-GMRIT molecules, and a hybrid configuration of unknown function consisting of 1 molecule β-GMR and β-GMRIT each. In order to analyze the effects of cytoplasmic assembly of full length with truncated β-GMR sequences into GMR complexes thought to occur in the third GMR configuration, we expressed both functional and defective chimeric α/β-GMR constructs together with β-GMR or β-GMRIT in BaF-3 cells (Figure4). The functional chimeric α/β-GMR and the defective α/β-GMR-ΔE differ only by a single glutamic acid at the fusion of the α-GMR and β-GMR sequences.5,24 The α/β-GMR-ΔE, but not α/β-GMR, requires coexpression of β-GMR for GM-CSF–mediated signaling.24 BaF-3/α/β-GMR cells bind GM-CSF with low affinity,5 but coexpression of α/β-GMR with either β-GMR or β-GMRIT generates high-affinity GMR complexes (Table 2). In spite of almost identical high-affinity GM-CSF binding for double-transfected clones, BaF-3 cells coexpressing α/β-GMR plus β-GMR proliferated at approximately 100-fold lower concentrations of huGM-CSF, whereas BaF-3 cells coexpressing α/β-GMR plus β-GMRIT required higher concentrations of GM-CSF for proliferation as compared with parental BaF-3/α/β-GMR cells (Figure 6). Furthermore, the inhibition of α/β-GMR function correlates with β-GMRIT protein expression (Figures 5A, 6), and most likely depends on the ratio of receptor subunits available at the cell surface. In addition, GM-CSF–dependent survival mediated by α/β-GMR is enhanced by β-GMR and inhibited by β-GMRIT (Figure 7). Furthermore, only β-GMR can induce GM-CSF–dependent proliferation in transfected BaF-3 cells expressing defective α/β-GMR-ΔE (Figure 8). These data suggest that complexes of truncated and full-length β-GMR cytoplasmic sequences are unable to transduce mitogenic and survival signals in transfected BaF-3 cells.
Alternative splicing has been described for many cytokine receptors. This often leads to soluble receptor variants (eg, α-GMR, IL-4Rα, IL-5Rα, IL-6Rα, erythropoeitin receptor [EpoR], or G-CSFR30,31) or to membrane-bound variants with different cytoplasmic sequences (eg, α-GMR, IL-5Rα, c-Mpl, or EpoR32-35). Recently, alternatively spliced isoforms of α-GMR were shown to mediate specific differentation signals in a leukemic cell line.36 In addition, Nakamura et al35,37 identified an intracytoplasmic truncated splice variant of the EpoR (EpoR-T) that behaves as a dominant-negative isoform and inhibits the function of EpoR in transfected BaF-3 cells in a way comparable to what is described here for β-GMRIT. In vivo, EpoR-T was found to be preferentially expressed in immature erythroid progenitor cells. Interestingly, diminished EpoR-T expression has been described in polycythemia vera, but not in patients with essential thrombocythemia or chronic myeloid leukemia, suggesting a potential role of EpoR-T expression in the pathophysiology of polycythemia vera.38 It is of interest to analyze the role of alternatively spliced cytokine receptors or receptor subunits in diseases characterized by altered cytokine receptor function or potential hypersensitivity to specific cytokines. For β-GMR, this may include myeloproliferative disorders and juvenile myelomonocytic leukemia39 as well as hypereosinophilic syndromes and, potentially, diseases with selective accumulation of eosinophils with allergic inflammation as observed in allergic asthma.40 41
Recent data demonstrate that variations in pre-mRNA splicing are often found in acute myeloid leukemia (AML) cells. For β-GMRIT, Gale et al22 reported consistently higher mRNA levels in primary AML cells than in normal neutrophils, bone marrow mononuclear cells, and CD34+ cells as determined by RNase protection or semiquantitative RT-PCR with an end-labeled primer. In preliminary studies using isoform-specific TaqMan RT-PCR for quantitation of β-GMRIT mRNA levels, we found more heterogeneous expression of β-GMRIT mRNA in primary AML, with samples expressing amounts lower than, equal to, or higher than those found in normal mononuclear cells (data not shown). However, extensive studies to analyze β-GMRIT mRNA and protein expression as well as functional studies on mitogenic and survival signaling are required to characterize the function of β-GMRIT for GM-CSF/IL-3/IL-5 receptor signaling in primary AML cells.
Alternative splicing and/or expression of alternatively spliced isoforms in AML cells have been described for signaling molecules such as SHP-142 and SMAD5,43 tumor susceptibility genes (TSGs) such as TSG101,44 cell surface receptors such as CD44,45 and enzymes involved in nuclear metabolism such as deoxycytidine kinase.46 Furthermore, the direct inhibition of pre-mRNA splicing and alterations of splicing profiles in transfected K562 cells by the TLS-ERG fusion protein found in AML with t(16;21) has recently been demonstrated.47 These data suggest that changes in pre-mRNA splicing may characterize a molecular defect in AML affecting many gene products besides β-GMR as described by Gale et al.22
Finally, our data regarding dimerization of full-length and truncated cytoplasmic β-GMR sequences offer an opportunity to further characterize the molecular role of β-GMR for intracellular signal transduction. The study by Gale et al22 and our data concerning defective oligomers containing full-length and truncated β-GMR sequences suggest a role, in addition to JAK2 activation, of full-length β-GMR dimers in intracellular signaling. The biochemical mechanisms involved and the role of β-GMRIT for receptor functions different from mitogenic or survival signaling should be further analyzed to characterize the regulation of cytokine receptor function in normal and malignant cells.
We thank Dr Rosemary Gale for providing the anti–β-GMRIT antiserum, Dietmar Klose for excellent secretarial help, and Michael A. Morgan for critical reading of the manuscript.
Supported in part by a grant of the Deutsche Forschungsgemeinschaft and the H.W. & J. Hector Stiftung.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Matthias Eder, Medizinische Hochschule Hannover, Zentrum der Inneren Medizin, Abteilung Hämatologie und Onkologie, Carl-Neuberg Strasse 1, D-30623 Hannover, Germany; e-mail:eder.matthias@mh-hannover.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal