To identify the regulatory elements controlling expression of the human CD4 (hCD4) gene in different cell types of the immune system, deletion and chimeric (human/murine) reporter genes were constructed and tested in transgenic (Tg) mice. Regulatory elements required for the proper hCD4 expression in the immature double-positive thymic T cells were identified in the enhancer and in the 3′ end of intron 1. Expression of hCD4 in macrophages is controlled by at least 2 sets of regulatory elements: one present in front of exon 1 and the second at the 5′ end of intron 1. The hCD4 elements required for expression on both myeloid and lymphoid CD8α+ dendritic cells (DCs) from lymph node and thymus were found to be different from those required for macrophage expression. The results indicate that expression of hCD4 in T cells, macrophages, and DCs is controlled by distinct regulatory elements.
Introduction
The CD4 cell surface receptor is expressed on many mouse or human cell populations, namely, on very early T-cell precursors (CD4LowCD44+CD25−), on immature double-positive (DP) CD4+CD8+ T cells, on mature single-positive (SP) CD4+CD8− T cells,1,2 and on a subpopulation of CD8α−dendritic cells (DCs).3,4 In humans, CD4 is also expressed in monocytes, macrophages, microglial cells, and some DCs.5,6 Several regulatory elements of the murine and human CD4 genes (mCD4, hCD4) have been identified and tested in transgenic (Tg) animal studies (reviewed by Ellmeier et al1 and Killeen and Littman2). We have exploited the structural homology, but the different patterns of expression, of the mCD4 and hCD4 genes and made chimeric mCD4/hCD4 transgenes, to map more precisely the regulatory regions of the hCD4 gene controlling its expression in macrophages and in DCs.
Study design
Tg mice
Constructs CD4A, CD4B, and CD4C were described previously.7 Other constructs were generated from the CD4C DNA by replacing the 2.6-kilobase pair (kbp) SacI fragment with the mouse promoter (CD4E), or the 9-kbpEcoRV-XbaI fragment with the murine silencer (0.5 kbp) (CD4F), or by deleting the 6-kbp EcoRV fragment (CD4H). Tg mice were generated as described elsewhere.7
Flow cytometry
Cell suspensions were prepared from lymphoid organs and stained with antibodies, as previously described.7 Peritoneal macrophages were plated overnight. Between 80% and 95% of them stained positive for Mac-1 (Figure 2B), Mac3, and F4/80 (data not shown). DCs were isolated from lymph node (LN)8 and thymus.9
Results and discussion
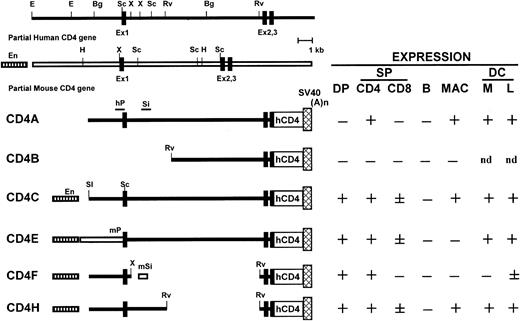
The Tg mice harboring 6 different DNAs were generated (Figure1). Northern blot analysis revealed that expression was highest in the thymus but was also detectable in the spleen and LNs, and was negative in all other organs tested (liver, heart, kidney, lung, intestine, muscle, brain; data not shown). This result is consistent with previous studies indicating that the human and mouse CD4 promoter/enhancer alone or in combination is sufficient to restrict expression to hematopoietic tissues.7 10-13 As expected, the level of expression varied among different founders carrying the same construct, an effect most likely related to the site of integration of the inoculated DNA.
Structure of the human/mouse CD4 transgenes and summary of expression data.
Symbols are as follows: human (black bar) and mouse (open bar) sequences; mouse enhancer (stippled bar), human and mouse promoters (hP, mP); and human and mouse silencer (Si, mSi). Restriction sites: E,EcoRI; Bg, BglII; H, HindIII; X,XbaI; RV, EcoRV; Sc, SacI; Sl,SalI. Expression in different subsets of cells is shown as + for expression, − for no expression, and ± for expression on a low number of cells. Double-positive (DP) CD4+CD8+ thymocytes; single-positive (SP) peripheral lymphocytes CD4+ and CD8+; B cells (B); macrophages (MAC); dendritic cells (DC); myeloid (M) and lymphoid (L) DCs; nd, not done.
Structure of the human/mouse CD4 transgenes and summary of expression data.
Symbols are as follows: human (black bar) and mouse (open bar) sequences; mouse enhancer (stippled bar), human and mouse promoters (hP, mP); and human and mouse silencer (Si, mSi). Restriction sites: E,EcoRI; Bg, BglII; H, HindIII; X,XbaI; RV, EcoRV; Sc, SacI; Sl,SalI. Expression in different subsets of cells is shown as + for expression, − for no expression, and ± for expression on a low number of cells. Double-positive (DP) CD4+CD8+ thymocytes; single-positive (SP) peripheral lymphocytes CD4+ and CD8+; B cells (B); macrophages (MAC); dendritic cells (DC); myeloid (M) and lymphoid (L) DCs; nd, not done.
Expression of hCD4 in DP CD4+CD8+T cells
Fluorescent-activated cell sorter (FACS) analysis showed that DP and SP CD4+ thymocytes of CD4E, CD4F, and CD4H Tg mice expressed the hCD4 reporter at high levels, similar to CD4C Tg mice7 (data not shown). Deletion of most of intron 1 (CD4F) or substitution of the human for the mouse promoter (CD4E) did not significantly change expression in the thymic T cells. Therefore, the CD4E and CD4F transgenes contain all the regulatory elements required for promoting CD4 expression in DP T cells. This result is in agreement with data from other groups who used similar human14,15 or mouse10 constructs. In contrast, other Tg mice harboring apparently similar mCD416,17 or hCD418,19 constructs did not express their reporter gene in the CD4+CD8+ DP T cells, while retaining expression in the SP CD4+ T cells. These discrepancies may reflect the presence or not of intronic element(s), just in front of exon 2, required for the expression in DP T cells and identified by Rushton and colleagues20 as the proximal promoter.
Silencing of the CD4 promoter in SP CD8+ T cells
In all Tg lines, including in most new lines (CD4E, F, H), expression of hCD4 in spleen and LNs was not detected on B cells and was restricted to T cells, almost exclusively on mature CD4+CD8− T cells (80%-90%, Table1, data not shown). The SP CD8+ T cells from CD4E and CD4H (about 10%-18%), but not from CD4F (<3%) Tg mice expressed hCD4 (Figure2A, Table 1, data not shown), indicating that switching the human with the mouse promoter (CD4E) or deleting a large portion of the intron 1 (CD4H, CD4F) had no effect on the expression on CD4+ T cells. These results also showed that the mouse silencer,10,11 in the context of the human promoter (CD4F), appears to be more effective in shutting off expression in CD8+ T cells than the human silencer14 (CD4H, CD4C). The human silencer may require additional collaborative sequences, not yet identified, to be fully operative.
hCD4 reporter gene expression on peritoneal macrophages and T cells from lymph nodes of hCD4 Tg mice
| Construct . | Founder . | hCD4 expression on specific populations (%) . | |||
|---|---|---|---|---|---|
| Peritoneal macrophages . | Lymph node . | ||||
| Mac1+ . | Class II+ . | CD4 SP . | CD8 SP . | ||
| CD4A/CD4 | 5 566 | 96.4 ± 0.8 | 97.1 ± 1.1 | 87.0 ± 3.3 | 3.4 ± 2.6 |
| 8 277 | 92.2 ± 3.5 | 92.3 ± 0.7 | 45.6 ± 3.8 | 4.3 ± 1.5 | |
| CD4C/CD4 | 8 951 | 92.8 ± 3.6 | 94.4 ± 1.8 | 81.0 ± 11.2 | 13.9 ± 2.0 |
| 8 956 | 92.5 ± 6.0 | 90.0 ± 3.6 | 94.2 ± 1.8 | 24.1 ± 11.2 | |
| CD4E/CD4 | 19 762 | 2.3 ± 1.8 | 2.3 ± 2.1 | ND | 11.2 ± 2.3 |
| 21 428 | 1.3 ± 1.4 | 1.9 ± 1.6 | 92.1 ± 6.7 | 18.0 ± 8.6 | |
| 21 429 | 2.8 ± 2.1 | 3.1 ± 2.8 | 39.7 ± 12.5 | 6.0 ± 2.1 | |
| CD4F/CD4 | 19 468 | 0.6 ± 0.6 | 0.5 ± 0.6 | ND | 4.0 ± 0.4 |
| 19 508 | 1.2 ± 1.6 | 3.2 ± 4.5 | 89.0 ± 9.6 | 3.34 ± 0.2 | |
| 19 523 | 0.8 ± 0.2 | 2.7 ± 1.4 | 50.9 ± 16.3 | 2.6 ± 1.0 | |
| CD4H/CD4 | 26 655 | 99.3 ± 1.2 | 97.9 ± 2.2 | 83.6 ± 2.7 | 5.4 ± 4.0 |
| 26 710 | 92.9 ± 5.1 | 89.8 ± 7.7 | 87.7 ± 1.6 | 14.8 ± 7.6 | |
| Construct . | Founder . | hCD4 expression on specific populations (%) . | |||
|---|---|---|---|---|---|
| Peritoneal macrophages . | Lymph node . | ||||
| Mac1+ . | Class II+ . | CD4 SP . | CD8 SP . | ||
| CD4A/CD4 | 5 566 | 96.4 ± 0.8 | 97.1 ± 1.1 | 87.0 ± 3.3 | 3.4 ± 2.6 |
| 8 277 | 92.2 ± 3.5 | 92.3 ± 0.7 | 45.6 ± 3.8 | 4.3 ± 1.5 | |
| CD4C/CD4 | 8 951 | 92.8 ± 3.6 | 94.4 ± 1.8 | 81.0 ± 11.2 | 13.9 ± 2.0 |
| 8 956 | 92.5 ± 6.0 | 90.0 ± 3.6 | 94.2 ± 1.8 | 24.1 ± 11.2 | |
| CD4E/CD4 | 19 762 | 2.3 ± 1.8 | 2.3 ± 2.1 | ND | 11.2 ± 2.3 |
| 21 428 | 1.3 ± 1.4 | 1.9 ± 1.6 | 92.1 ± 6.7 | 18.0 ± 8.6 | |
| 21 429 | 2.8 ± 2.1 | 3.1 ± 2.8 | 39.7 ± 12.5 | 6.0 ± 2.1 | |
| CD4F/CD4 | 19 468 | 0.6 ± 0.6 | 0.5 ± 0.6 | ND | 4.0 ± 0.4 |
| 19 508 | 1.2 ± 1.6 | 3.2 ± 4.5 | 89.0 ± 9.6 | 3.34 ± 0.2 | |
| 19 523 | 0.8 ± 0.2 | 2.7 ± 1.4 | 50.9 ± 16.3 | 2.6 ± 1.0 | |
| CD4H/CD4 | 26 655 | 99.3 ± 1.2 | 97.9 ± 2.2 | 83.6 ± 2.7 | 5.4 ± 4.0 |
| 26 710 | 92.9 ± 5.1 | 89.8 ± 7.7 | 87.7 ± 1.6 | 14.8 ± 7.6 | |
ND indicates not done.
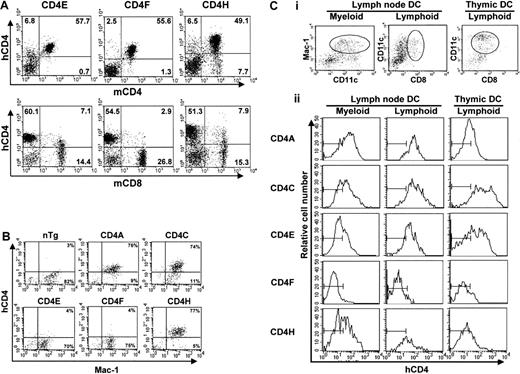
FACS analysis of hCD4 expression.
(A) LN cells. Double staining with antihuman CD4 (hCD4) and antimouse CD4 (mCD4) or mouse CD8 (mCD8) monoclonal antibodies was carried out. The percentage of T cells expressing hCD4 in a representative mouse for each line is shown. Note that the leakiness of the hCD4 expression on the mature CD8+ T cells was absent in the CD4F Tg line. (B) Peritoneal macrophages. Double staining of plated peritoneal macrophages was done with anti-hCD4 and anti–Mac-1 antibodies. The percentage of macrophages expressing Mac-1 and hCD4 is shown. The quadrant settings are based on unstained controls. (C) DCs. Triple staining of enriched LN DCs. Cells that were gated for CD11c+CD11b/Mac-1+ (myeloid) and CD11c+CD8α+ (lymphoid) expression (i) show expression of hCD4 (ii). The thymic DCs were all CD11c+CD8α+. The bars in each panel in ii show the staining of hCD4 on DCs from non-Tg control mice.
FACS analysis of hCD4 expression.
(A) LN cells. Double staining with antihuman CD4 (hCD4) and antimouse CD4 (mCD4) or mouse CD8 (mCD8) monoclonal antibodies was carried out. The percentage of T cells expressing hCD4 in a representative mouse for each line is shown. Note that the leakiness of the hCD4 expression on the mature CD8+ T cells was absent in the CD4F Tg line. (B) Peritoneal macrophages. Double staining of plated peritoneal macrophages was done with anti-hCD4 and anti–Mac-1 antibodies. The percentage of macrophages expressing Mac-1 and hCD4 is shown. The quadrant settings are based on unstained controls. (C) DCs. Triple staining of enriched LN DCs. Cells that were gated for CD11c+CD11b/Mac-1+ (myeloid) and CD11c+CD8α+ (lymphoid) expression (i) show expression of hCD4 (ii). The thymic DCs were all CD11c+CD8α+. The bars in each panel in ii show the staining of hCD4 on DCs from non-Tg control mice.
Transgene expression in peritoneal macrophages
In CD4A, CD4C, and CD4H Tg mice, hCD4 was expressed on over 85% of peritoneal macrophages (Mac-1+)7 (Figure2B, Table 1). However, macrophages from the CD4E and CD4F Tg mice showed very low or undetectable levels of expression. These results suggest that expression in macrophages requires the concomitant presence of 2 hCD4 sequences: one located in the 2.6-kbp promoter and a second (about 3.5 kbp) in the 5′ intron 1 region (Figure 1).
Transgene expression in DCs
Because macrophages and DCs can be derived from the same myeloid precursor and because the 2 major mouse subpopulations of DCs (“myeloid” CD11c+CD11b+CD8α−and “lymphoid” CD11c+CD11b−CD8α+) are thought to fulfill different functions in vivo,21 it was of interest to compare transgene expression in these DC subpopulations. This comparative analysis was carried out on purified DCs from peripheral LNs and from thymus. The data are shown for DCs expressing either a myeloid (CD11c+ CD11b+) or a lymphoid (CD11c+ CD8α+) phenotype.
In CD4A, CD4C, CD4E, and CD4H Tg mice, expression of hCD4 was detected in high proportion (80%) on both CD11b+ and CD8α+ LN DCs, as well as on thymic CD8α+ DC (Figure 2C). The CD4F Tg DC express the reporter gene at low or undetectable levels on few (1%-9%) myeloid DCs and on a significant proportion (11%-40%; depending on founder lines) of lymphoid DCs (Figure 2C). Thus, some DC regulatory elements appear to reside in the promoter, the larger remaining fragment in CD4F DNA. The mouse enhancer appears to be dispensable for expression in DC (CD4A), as in macrophages.7 The fact that the same hCD4 regulatory sequences allow expression in both myeloid and lymphoid murine DCs contrasts with the mCD4 expression that appears to be restricted to CD8α− subpopulations both in the spleen4and in LNs.3 Interestingly, the murine promoter, which was unable to direct expression in macrophages by itself, could nevertheless promote expression in DCs (CD4E). This result indicates a distinct requirement of these 2 populations for expression of thehCD4 gene and suggests that the myeloid-specific element is most likely present in the 3.5-kbp 5′ end intron 1 fragment remaining in CD4H.
This work confirms earlier studies in lymphoid T cells on the importance of some regulatory elements within the CD4 gene, the distal promoter,22-24 the proximal promoter,20 the proximal and distal enhancers,13,25,26 and the silencer,10,11 14and extends them significantly for macrophages and DCs.
We thank Ginette Massé, Nathalie Gauthier, Michel Ste-Marie, Michel Robillard, Stéphane Gagnon, and Karina Lamarre for their excellent technical assistance and Nathalie Tessier for her valuable help in the FACS analysis.
Supported by grants from the Medical Research Council to P.J. and Z.H. and from the National Cancer Institute to P.J.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Paul Jolicoeur, Laboratory of Molecular Biology, Clinical Research Institute of Montreal, 110 Pine Ave W, Montreal, QUE, H2W 1R7, Canada; e-mail: jolicop@ircm.qc.ca; or Zaher Hanna, Clinical Research Institute of Montreal, 110 Pine Ave W, Montreal, Quebec, H2W 1R7, Canada; e-mail: hannaz@ircm.qc.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal