The inhibitor-of-apoptosis protein survivin is expressed in most cancers and leukemias and during fetal development, but not in most normal adult tissues. Survivin expression was analyzed in umbilical cord blood (UCB) and adult bone marrow CD34+cells and in the factor-dependent MO7e cell line; also investigated was whether survivin expression was regulated by hematopoietic growth factors. Survivin messsenger RNA (mRNA) and protein were expressed in fresh UCB and marrow CD34+ cells. The combination of thrombopoietin, Flt3 ligand, and stem cell factor upregulated survivin expression in CD34+ cells within 24 hours; survivin expression was cell-cycle related and highest during G2/M, whereas growth-factor withdrawal resulted in decreased survivin expression. Cell-cycle fractionation of UCB CD34+ with Hoechst-33342/pyronin-Y demonstrated that survivin message was undetectable in freshly isolated G0 cells, but present in G1 cells. After cytokine stimulation, survivin mRNA and protein expression were observed in both G0 and G1 CD34+ cells as well as in cells that had progressed to S and G2/M phase, indicating that survivin expression is regulated in all phases of the cell cycle. This contrasts with the expression of survivin predominantly during G2/M in cancer cells. In CD34+ cells and MO7e cells, growth factor–mediated upregulation of survivin was associated with inhibition of apoptosis, and downregulation of survivin was coincident with increased apoptosis. Furthermore, an inverse correlation between survivin and active caspase-3 was observed in CD34+ cells. These findings demonstrate that survivin is not a cancer-specific antiapoptotic protein and plays a regulatory role in normal adult hematopoiesis.
Introduction
Apoptosis is an essential process for cell and tissue homeostasis, and apoptotic effector molecules and disordered apoptosis are involved in various diseases and neoplasias. Survivin is a newly described member of the inhibitor-of-apoptosis (IAP) family characterized by one or more highly conserved baculovirus IAP repeat (BIR) domains.1-4 In contrast to other members of the IAP family, which are widely expressed in human tissues,5,6survivin is expressed primarily in fetal but not adult tissues, and its expression is aberrantly enhanced in most cancers, including carcinomas of the lung, colon, pancreas, prostate, breast, and stomach, and in most hematopoietic malignancies.2 Furthermore, survivin is the only IAP whose expression is cell-cycle dependent.7Survivin suppresses apoptosis triggered by chemotherapeutic agents, tumor necrosis factor–α/Fas ligand–induced stimuli, or caspases 3, 7, and 9,8,9 and elevated expression correlates with poor prognosis in patients with solid tumors, acute leukemia, and lymphoma.3,4,10-16 Overexpression of survivin confers partial factor independence in interleukin-3 (IL-3)–dependent Baf/3 cells2 and promotes cell-cycle progression in hepatocarcinoma cells.17 Targeting of survivin by antisense or dominant-negative strategies in several transformed cell models results in induction of apoptosis.18-22 The enhancement of cell survival by survivin results from its binding to the mitotic spindle during G2/M in transformed cells,23 probably in a complex with Cdk4 and p21,24 which blocks caspase-3 activation.23,25,26 These findings have led to the belief that survivin overexpression may provide a survival or proliferation advantage for cancer cells and that, because of survivin's limited expression, disrupting survivin interactions may represent a novel cancer therapy.27
The pattern of survivin expression during development1suggests that it may be a general regulator of mitosis, preserving cell fidelity during cell division. However, little is known about regulation of survivin expression in normal adult cells, particularly by growth factors. Survivin is not expressed in most adult human tissues,2,18 but expression of the mouse survivin homolog, TIAP, is related to proliferation in thymus, testis, crypt cells, and activated T cells.26 Although survivin is not expressed in resting endothelial cells, survivin expression is induced in G2/M phase during angiogenesis stimulated by angiopoietin-1, basic fibroblast growth factor, and vascular endothelial growth factor.9,28 29 Since normal adult hematopoiesis is precisely regulated by growth factors that control cell survival and proliferation and since cytokine withdrawal initiates apoptosis, we investigated survivin expression and growth-factor regulation in normal hematopoietic cells and leukemic cells. The results reported here demonstrate that survivin is expressed in cord blood and normal adult bone marrow CD34+ cells. Furthermore, survivin messenger RNA (mRNA) and protein expression in CD34+ cells and granulocyte-macrophage colony-stimulating factor (GM-CSF)–dependent MO7e cells is regulated by hematopoietic cytokines. Upregulation of survivin message and protein is highest during G2/M of the cell cycle, but in contrast to transformed cells, its expression was found throughout the cell cycle. These findings demonstrate that survivin expression is not cancer-cell specific and may play an important role in normal hematopoietic cell survival and proliferation.
Materials and methods
Antibodies, cytokines, and reagents
Anti–human survivin monoclonal antibody (clone 91618.11), affinity-purified anti–human survivin polyclonal antibody (AF886), control mouse immunoglobulin (Ig)–G1, and recombinant human histidine-tagged survivin (survivin/HIS) were purchased from R&D Systems (Minneapolis, MN). Fluorescein isothiocyanate (FITC)–conjugated anti–mouse immunoglobulin and phycoerythrin (PE)–conjugated anti–human active caspase-3 polyclonal antibodies were from BD Pharmingen (San Diego, CA). Normal rabbit IgG was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). FITC–conjugated goat anti–rabbit IgG was from Caltag Laboratories (Burlingame, CA). FITC- and cy-chrome–conjugated anti-CD34 antibodies (BIRMA-K3) were from Dako (Carpinteria, CA). Anti–human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) monoclonal antibody (clone 6C5) was purchased from Biodesign International (Kennebunk, ME). Recombinant human GM-CSF, Flt3 ligand (FL), and thrombopoietin (Tpo) were provided by Immunex (Seattle, WA). Recombinant human stem cell factor (SCF) was a gift from Dr Karl Nocka, UCB Research (Cambridge, MA). The caspase inhibitor ZVAD-fmk was purchased from Biomol (Plymouth Meeting, PA).
Isolation of umbilical cord blood, bone marrow, and peripheral blood CD34+ cells
Normal umbilical cord blood (UCB) scheduled for discard after delivery was obtained with institutional review board approval and maternal consent. Low-density mononuclear cells were isolated on Ficoll-Paque (1.077 g/mL) (Amersham Pharmacia Biotech, Piscataway, NJ), washed with Iscoves modified Dulbecco medium (IMDM) (BioWhittaker, Walkersville, MD), and treated with erythrocyte lysis buffer (Puregene, Gentra Systems, Minneapolis, MN). Adherent cells were depleted on plastic, and CD34+ cells were isolated with a monoclonal anti–human CD34 antibody (Qbend/10) and 2 sequential positive selections with immunomagnetic beads (Miltenyi Biotech, Auburn, CA) according to the manufacturer's recommendations. The purity of CD34+ cells routinely exceeded 95% as determined by fluorescence-activated cell sorting (FACS) analysis (Becton Dickinson, San Jose, CA). Mononuclear cells unreactive with immunomagnetic beads were designated CD34−(lin+) cells and consisted primarily of lineage-positive cells.
Bone marrow was harvested from the iliac crest of healthy adult donors with a heparinized syringe, and peripheral blood from the same donor was collected in EDTA after informed consent. Mononuclear cells, CD34+ cells, and CD34− cells were isolated as described for UCB. In some experiments, peripheral blood mononuclear cell and neutrophil populations were isolated by means of PMN-One Step (Accurate Chemicals, Westbury, NY) according to the manufacturer's instructions.
Peripheral blood T-lymphocyte isolation and activation
Adult peripheral blood T lymphocytes were isolated from donors with Lympho-Kwik lymphocyte isolation reagents (One Lamda, Canoga Park, CA) according to the manufacturer's instructions. Isolated T cells were seeded at 1.0 × 106/mL in 12-well dishes precoated with 10 μg/mL anti–human CD3 antibody (BD Pharmingen) and incubated with 1 μg/mL anti–human CD28 antibody (BD Pharmingen) and 100 U/mL recombinant human (rh) IL-2 (Chiron, Emeryville, CA) for up to 72 hours.
Cell culture
CD34+ or CD34− cells were seeded at 1 to 2 × 105/mL in IMDM with 10% heat-inactivated fetal bovine serum (HI-FBS) (Hyclone Laboratories, Logan, UT) in the presence or absence of SCF (100 ng/mL), FL (100 ng/mL), and Tpo (100 ng/mL) in 96-well plates at 37°C, 5% CO2 in air. The GM-CSF–dependent human megakaryocytic leukemia cell line MO7e, which has been described elsewhere,30 31 was maintained in RPMI medium containing 20 ng/mL human GM-CSF, 20% HI-FBS, and penicillin-streptomycin. Exponentially growing cells were washed 3 times with RPMI and were factor-starved for 24 hours in RPMI with 1% bovine serum albumin (Sigma, St Louis, MO). After 24 hours, cells were harvested, washed, and suspended in RPMI plus 10% HI-FBS with or without 10 ng/mL GM-CSF plus 50 ng/mL SCF and cultured for an additional 24 hours. After incubation, cells were harvested and analyzed for survivin protein by intracellular flow cytometry and Western blots; survivin mRNA was analyzed by reverse-transcriptase polymerase chain reaction (RT-PCR); and cell cycle status and apoptosis were determined by flow cytometry by means of propidium iodide (PI) and FITC–annexin-V (BD Pharmingen), respectively. Human promyelocytic leukemia HL-60 cells were maintained in RPMI with 10% HI-FBS and used directly for intracellular flow cytometry and Western blot analysis.
Reverse-transcriptase polymerase chain reaction
Total RNA was harvested by means of the RNeasy kit (Qiagen, Valencia, CA) and incubated with 1 U DNase for 30 minutes at 37°C to eliminate genomic DNA. We reverse-transcribed 100 ng total RNA with the use of murine Moloney leukemia virus reverse transcriptase (Promega, Madison, WI). We used 10% of the complementary DNA (cDNA) for PCR. Since human survivin mRNA shows extensive homology with the complementary sequence of human EPR-1 mRNA,2 we designed primers to a sequence unique to survivin. The survivin primers, 5′-GAG CTG CAG GTT CCT TAT C-3′ and 5′-ACA GCA TCG AGC CAA GTC AT-3′, amplify a 431–base pair product encompassing nucleotides 917 through 1348. The generated survivin PCR product was verified by sequencing. The primers 5′-GAA GGT GAG GTC GGA GTC-3′ and 5′-GAA GAT GGT GAT GGG ATT TC-3′ were used to amplify GAPDH. PCR was performed with Pwo-polymerase (Roche Molecular Biochemicals, Indianapolis, IN). Reactions were carried out for 25, 30, 35, and 40 cycles consisting of 1 minute denaturing at 95°C, 1 minute annealing at 55°C, and 1 minute elongation at 72°C. The products were separated by electrophoresis in 2% agarose and visualized with ethidium bromide.
Western blot
Exponentially growing HL-60 cells; CD34+ cells cultured with Tpo, FL, and SCF; or MO7e cells cultured with or without GM-CSF and SCF or GM-CSF alone were washed and lysed in phosphate-buffered saline (PBS) containing 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 10 μg/mL phenylmethyl sulfonyl fluoride, and 1 mM sodium orthovanadate. Aliquots were kept on ice for 30 minutes and centrifuged at 10 000g for 20 minutes. Supernatant from each sample was denatured in 2 × SDS buffer at 95°C for 5 minutes, separated on 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gels, and transferred onto nitrocellulose membranes. The filters were incubated with anti–human survivin followed by horseradish peroxidase–conjugated anti–mouse or anti–rabbit immunoglobulin and developed by means of electrogenerated chemiluminescence (Amersham).
Intracellular staining and flow cytometry
For intracellular staining for survivin protein, cells were fixed with Cytofix/Cytoperm Solution (BD Pharmingen) and treated according to the manufacturer's instructions. Briefly, cells were fixed for 30 minutes on ice, washed with Perm/Wash buffer (BD Pharmingen), and suspended in 50 μL Perm/Wash buffer containing 5 μg/mL either isotype control, affinity-purified polyclonal anti–human survivin, or monoclonal anti–human survivin. After 30 minutes' incubation, unbound antibody was removed by washing in Perm/Wash buffer, and FITC-conjugated anti–mouse or anti–rabbit immunoglobulin added. The cells were incubated on ice for 20 minutes and washed twice with Perm/Wash buffer. In some experiments, cells were stained with cy-chrome–conjugated anti–human CD34 antibody and/or PI. In preadsorption studies, 1 μg monoclonal or polyclonal anti–survivin antibody was incubated with 20-fold molar excess of survivin/HIS for 30 minutes at 4°C before being added to fixed and permeabilized cells. For annexin-V staining, CD34+ cells and MO7e cells stimulated with or without growth factors were washed once with PBS and stained with FITC–annexin-V and PI according to the manufacturer's instructions. Stained cells were analyzed by means of a FACScan and ModFIT (for cell cycle) and CellQuest software (Becton Dickinson).
Cell-cycle fractionation with Hoechst-33342 and Pyronin-Y
Total CD34+ cells were stained with Hoechst-33342 (Hst) (Molecular Probes, Eugene, OR) and pyronin-Y (PY) (Polysciences, Warrington, PA), as described.32-34 Briefly, CD34+ cells were washed with Hst buffer, consisting of Hanks balanced salt solution containing 4 mM Hepes, 5.5 μM/L (1 mg/mL) glucose, 10% HI-FBS, and 100 μM verapamil. Cells were stained with 1 μg/mL Hst-33342 in Hst buffer for 45 minutes at 37°C, followed by staining with 3.3 μM PY in the same buffer for 45 minutes at 37°C. Cells were washed with Hst buffer and subjected to FACS on a FACStar Plus (Becton Dickinson) equipped with an argon laser providing the 488-nm excitation for PY and a krypton laser providing the 350-nm excitation. Sorting windows were constructed so that G0 cells were gated as Hstlow PYlow and G1 cells as HstlowPYhigh. Cells in G2/M were collected as HsthighPYhigh, and cells that showed intermediate staining with Hst and were PYhigh were collected as S-phase cells. Collected cells were immediately placed on ice for RT-PCR or fixed for intracellular staining. Viability and purity of sorted cells exceeded 98% and 90%, respectively.
Results
Expression of survivin in UCB and adult bone marrow CD34+ cells
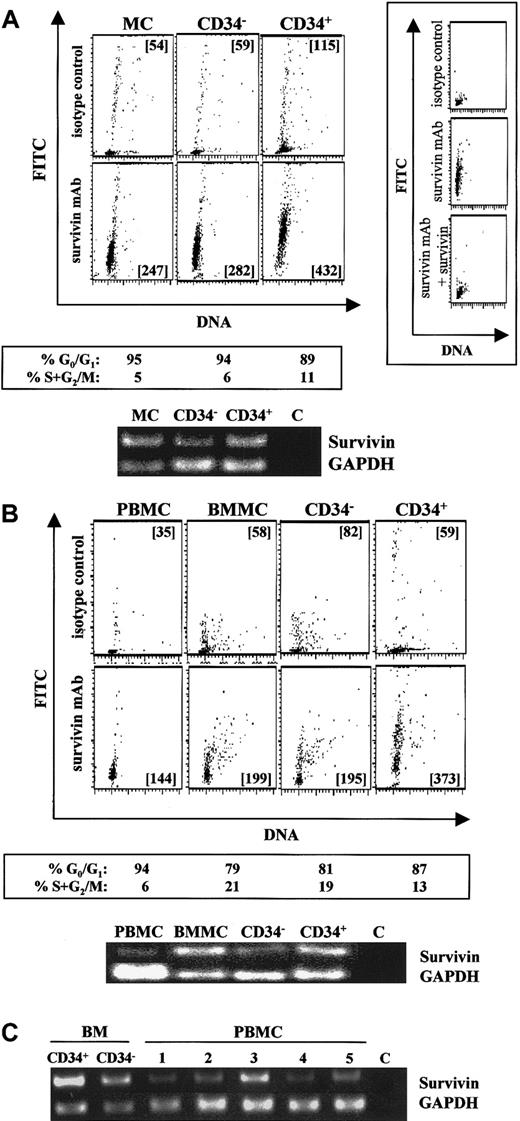
Intracellular staining using flow cytometry detected significant survivin protein in UCB mononuclear cells and freshly isolated CD34−(lin+) and CD34+ (purity exceeding 95%) cells (Figure 1A, lower panels; lanes 1-3), compared with individual isotype controls (Figure1A, upper panels). More than 92% of the survivin detected by flow cytometry was blocked by preadsorption of the antisurvivin monoclonal antibody with 20-fold molar excess survivin (Figure 1A, insert), confirming the specificity of the intracellular staining. Identical results were observed with the use of a polyclonal antisurvivin antibody, and 100% of the survivin detected with this antibody could be preadsorbed with excess survivin (data not shown). Although expressed in each of these cell populations, survivin protein was highest in freshly isolated CD34+ cells. Survivin mRNA in these same cell populations was detected by RT-PCR (Figure 1A), and like protein, survivin mRNA was highest in CD34+ cells. Simultaneous staining for DNA demonstrated that 90% of UCB mononuclear cells, CD34−(lin+) cells and CD34+cells were in G0/G1 of the cell cycle. Similarly, adult peripheral blood mononuclear cells, bone marrow mononuclear cells, isolated CD34+ cells, and CD34−(lin+) cells from the same donor all expressed survivin protein and mRNA (Figure 1B). Survivin mRNA and protein expression were higher in CD34+ cells (Figure 1B, panel 4 and lane 4) than in CD34−(lin+) cells (Figure 1B, panel 3 and lane 3) and were barely detectable in peripheral blood mononuclear cells (Figure 1B, panel 1 and lane 1). Cell-cycle analysis indicated that 87% of bone marrow CD34+ cells, 81% of CD34−(lin+) cells, and 94% of peripheral blood mononuclear cells were in G0/G1.
Survivin expression in normal hematopoietic cell populations.
(A) Survivin expression in UCB mononuclear cells (MCs), CD34−(lin+) cells (CD34−), and isolated CD34+ cells (CD34+). Cells were stained with PI (x-axis) and FITC–mouse IgG1 (upper panels) or FITC–anti-human survivin (lower panels) (y-axis). Mean channel fluorescence (MCF) of FITC signals is presented in each multivariate dot plot. Preadsorption of the antisurvivin monoclonal antibody (mAb) with excess human survivin/HIS for 30 minutes prior to addition to fixed and permeabilized cells reduced survivin detected by greater than 92% (panel A, right insert), confirming the specificity of the intracellular staining. Cell-cycle status of each cell population analyzed by ModFit software is shown below the corresponding plots. RT-PCR results (35 cycles) for survivin and GAPDH mRNAs from the same cell samples are presented. Lane 1: MC-UCB mononuclear cells. Lane 2: CD34−(lin+) cells. Lane 3: CD34+ cells. Lane 4: C, control sample without any RNA. Data are from a single UCB sample and representative of 5 experiments. (B) Survivin expression in adult peripheral blood mononuclear cells (PBMCs), adult bone marrow mononuclear cells (BMMCs), CD34−(lin+) cells (CD34−), and isolated CD34+. Peripheral blood and bone marrow are from the same donor. The upper panels represent isotype controls, and the lower panels represent survivin protein. Cell-cycle status of each cell population analyzed by Modifit software is shown below the corresponding multivariate dot plot. RT-PCR results (35 cycles) for survivin and GAPDH mRNAs from the same cell samples are presented. Lane 1: PBMCs; Lane 2: BMMCs; Lane 3: CD34−(lin+) cells; Lane 4: CD34+ cells; Lane 5: C, control sample without any RNA. Data are from a single donor and representative of 2 identical experiments with different donors. (C) Survivin mRNA in adult bone marrow CD34+ cells, CD34− cells, and PBMCs from 5 healthy donors. RT-PCR (35 cycles) for survivin and GAPDH mRNAs are shown. C represents control sample without RNA.
Survivin expression in normal hematopoietic cell populations.
(A) Survivin expression in UCB mononuclear cells (MCs), CD34−(lin+) cells (CD34−), and isolated CD34+ cells (CD34+). Cells were stained with PI (x-axis) and FITC–mouse IgG1 (upper panels) or FITC–anti-human survivin (lower panels) (y-axis). Mean channel fluorescence (MCF) of FITC signals is presented in each multivariate dot plot. Preadsorption of the antisurvivin monoclonal antibody (mAb) with excess human survivin/HIS for 30 minutes prior to addition to fixed and permeabilized cells reduced survivin detected by greater than 92% (panel A, right insert), confirming the specificity of the intracellular staining. Cell-cycle status of each cell population analyzed by ModFit software is shown below the corresponding plots. RT-PCR results (35 cycles) for survivin and GAPDH mRNAs from the same cell samples are presented. Lane 1: MC-UCB mononuclear cells. Lane 2: CD34−(lin+) cells. Lane 3: CD34+ cells. Lane 4: C, control sample without any RNA. Data are from a single UCB sample and representative of 5 experiments. (B) Survivin expression in adult peripheral blood mononuclear cells (PBMCs), adult bone marrow mononuclear cells (BMMCs), CD34−(lin+) cells (CD34−), and isolated CD34+. Peripheral blood and bone marrow are from the same donor. The upper panels represent isotype controls, and the lower panels represent survivin protein. Cell-cycle status of each cell population analyzed by Modifit software is shown below the corresponding multivariate dot plot. RT-PCR results (35 cycles) for survivin and GAPDH mRNAs from the same cell samples are presented. Lane 1: PBMCs; Lane 2: BMMCs; Lane 3: CD34−(lin+) cells; Lane 4: CD34+ cells; Lane 5: C, control sample without any RNA. Data are from a single donor and representative of 2 identical experiments with different donors. (C) Survivin mRNA in adult bone marrow CD34+ cells, CD34− cells, and PBMCs from 5 healthy donors. RT-PCR (35 cycles) for survivin and GAPDH mRNAs are shown. C represents control sample without RNA.
To further investigate the degree of survivin expression in peripheral blood, survivin protein and mRNA levels were quantitated in mononuclear and neutrophil cell fractions from 5 additional healthy donors. Mononuclear cells from all 5 blood samples demonstrated survivin protein expression by flow cytometry that was significantly greater than isotype control (data not shown), and survivin mRNA was detected in all samples (Figure 1C). No evidence of survivin protein expression was found in neutrophils from any of the peripheral blood samples.
Regulation of survivin expression by hematopoietic growth factors in CD34+ cells
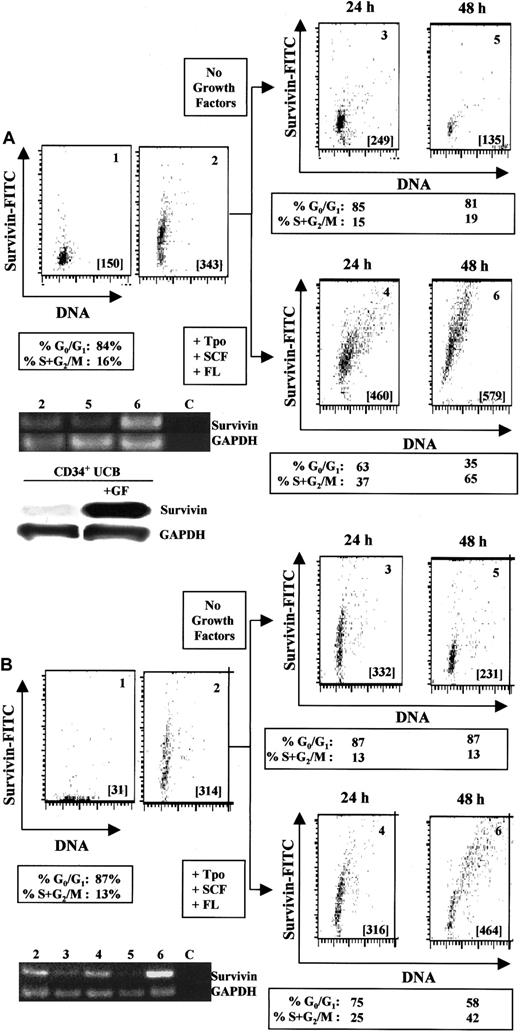
Incubation of UCB CD34+ cells with 100 ng/mL Tpo, SCF, and FL for 24 to 48 hours upregulates both survivin m-RNA (Figure2A, lane 6) and protein (Figure 2A, panels 4 and 6). In the absence of growth factors, survivin mRNA (Figure 2A, lane 5) and protein expression (Figure 2A, panels 3 and 5) gradually declined. Comparison by Western blot of survivin protein levels in cells cultured with or without Tpo, SCF, and FL for 48 hours also demonstrated growth factor–mediated upregulation of survivin protein (Figure 2A). Cell-cycle analysis of CD34+ cells cultured with Tpo, FL, and SCF indicated that 37% and 65% of CD34+ cells progressed to S+G2/M of the cell cycle after 24 and 48 hours, respectively, coincident with upregulation of survivin. The highest expression of survivin was observed in cells in G2/M. In the absence of growth factors, most of the CD34+ cells remained in G0/G1 (85% at 24 hours and 81% at 48 hours). Survivin upregulation in the presence of growth factors was not usually observed before 24 hours and was coincident with exit from G0/G1 and entry into the cell cycle, suggesting a link between survivin expression and exit of hematopoietic cells from quiescence. No upregulation of survivin was observed in CD34−(lin+) cells, which are composed mainly of differentiating cells with little proliferative capacity, with up to 48 hours of culture with growth factors (data not shown).
Regulation of survivin in CD34+ cells by hematopoietic growth factors.
(A) Multivariate flow dot plots for survivin expression (panel 2) versus isotype control (panel 1) in freshly isolated UCB CD34+ cells (purity in excess of 95%). Identical results were observed with the use of either an antisurvivin mAb (shown) or a polyclonal antisurvivin antibody. Cell-cycle analysis indicated that 84% of the CD34+ cells were in G0/G1 and 16% in S+G2/M. Panels 3 and 5 represent survivin expression in CD34+ cells cultured without growth factors for 24 and 48 hours, respectively. Panels 4 and 6 represent survivin expression in CD34+ cells cultured with Tpo, SCF, and FL for 24 and 48 hours. Cell-cycle distribution of each cell population is presented below the appropriate panels. All cells were counterstained with anti-CD34 mAb and gated to include only CD34+ cells. Cell samples from each population were analyzed for survivin and GAPDH by RT-PCR (35 cycles). Lane 2: freshly harvested UCB CD34+ cells. Lane 5: CD34+ cells cultured without growth factors for 48 hours. Lane 6: CD34+cells cultured in the presence of Tpo, SCF, and FL for 48 hours. Lane C: control sample without RNA template. Data are from 1 UCB sample and representative of 5 experiments. For Western blot analysis, UCB CD34+ cells were cultured with or without Tpo, SCF, and FL for 48 hours. Lysates from 1 × 106cells were separated on 10% SDS-PAGE gels and probed with anti–human polyclonal antibody. (B) Multivariate flow dot plots for survivin expression (panel 2) versus isotype control (panel 1) in freshly isolated adult bone marrow CD34+ cells (purity in excess of 97%). Identical results were observed with the use of either an antisurvivin mAb (shown) or a polyclonal antisurvivin antibody. Cell-cycle analysis indicated that 87% of the CD34+ cells were in G0/G1 and 13% in S+G2/M. Panels 3 and 5 represent survivin expression in CD34+ cells cultured in the absence of growth factors for 24 and 48 hours. Panels 4 and 6 represent survivin expression in CD34+ cells cultured in the presence of Tpo, SCF, and FL for 24 and 48 hours. Cell-cycle distribution of each cell population is presented below the appropriate panels. All cells were counterstained with anti-CD34 mAb and gated to include only CD34+ cells. Cell samples from each population were analyzed for survivin and GAPDH mRNA by RT-PCR (35 cycles). Lane 2: freshly isolated bone marrow CD34+ cells. Lanes 3 and 5: CD34+ cells cultured without growth factors for 24 and 48 hours, respectively. Lanes 4 and 6: CD34+ cells cultured in the presence of Tpo, SCF, and FL for 24 and 48 hours, respectively. Lane C represents control sample without any RNA template. Data are from 1 of 2 identical experiments.
Regulation of survivin in CD34+ cells by hematopoietic growth factors.
(A) Multivariate flow dot plots for survivin expression (panel 2) versus isotype control (panel 1) in freshly isolated UCB CD34+ cells (purity in excess of 95%). Identical results were observed with the use of either an antisurvivin mAb (shown) or a polyclonal antisurvivin antibody. Cell-cycle analysis indicated that 84% of the CD34+ cells were in G0/G1 and 16% in S+G2/M. Panels 3 and 5 represent survivin expression in CD34+ cells cultured without growth factors for 24 and 48 hours, respectively. Panels 4 and 6 represent survivin expression in CD34+ cells cultured with Tpo, SCF, and FL for 24 and 48 hours. Cell-cycle distribution of each cell population is presented below the appropriate panels. All cells were counterstained with anti-CD34 mAb and gated to include only CD34+ cells. Cell samples from each population were analyzed for survivin and GAPDH by RT-PCR (35 cycles). Lane 2: freshly harvested UCB CD34+ cells. Lane 5: CD34+ cells cultured without growth factors for 48 hours. Lane 6: CD34+cells cultured in the presence of Tpo, SCF, and FL for 48 hours. Lane C: control sample without RNA template. Data are from 1 UCB sample and representative of 5 experiments. For Western blot analysis, UCB CD34+ cells were cultured with or without Tpo, SCF, and FL for 48 hours. Lysates from 1 × 106cells were separated on 10% SDS-PAGE gels and probed with anti–human polyclonal antibody. (B) Multivariate flow dot plots for survivin expression (panel 2) versus isotype control (panel 1) in freshly isolated adult bone marrow CD34+ cells (purity in excess of 97%). Identical results were observed with the use of either an antisurvivin mAb (shown) or a polyclonal antisurvivin antibody. Cell-cycle analysis indicated that 87% of the CD34+ cells were in G0/G1 and 13% in S+G2/M. Panels 3 and 5 represent survivin expression in CD34+ cells cultured in the absence of growth factors for 24 and 48 hours. Panels 4 and 6 represent survivin expression in CD34+ cells cultured in the presence of Tpo, SCF, and FL for 24 and 48 hours. Cell-cycle distribution of each cell population is presented below the appropriate panels. All cells were counterstained with anti-CD34 mAb and gated to include only CD34+ cells. Cell samples from each population were analyzed for survivin and GAPDH mRNA by RT-PCR (35 cycles). Lane 2: freshly isolated bone marrow CD34+ cells. Lanes 3 and 5: CD34+ cells cultured without growth factors for 24 and 48 hours, respectively. Lanes 4 and 6: CD34+ cells cultured in the presence of Tpo, SCF, and FL for 24 and 48 hours, respectively. Lane C represents control sample without any RNA template. Data are from 1 of 2 identical experiments.
Similar results were obtained with the use of adult bone marrow CD34+ cells (Figure 2B). Culture of CD34+ cells with Tpo, SCF, and FL for 24 and 48 hours resulted in upregulation of survivin mRNA (Figure 2B, lane 6) and protein (Figure 2B, panel 6) expression. In the absence of growth factors, survivin mRNA (Figure 2B, lanes 3 and 5) and protein expression (Figure 2B, panels 3 and 5) decreased over 48 hours. Survivin mRNA and protein expression correlated with cell-cycle progression, with the highest expression of survivin being observed in CD34+ cells in G2/M.
Survivin expression in T lymphocytes
The murine survivin homolog TIAP is absent in resting T cells, but induced within 12 hours after activation.26 To determine if survivin expression and cytokine regulation observed in hematopoietic cells occurred in adult human T lymphocytes, peripheral blood T cells were isolated and survivin expression and cell cycle analyzed by multivariate flow cytometry before and after stimulation. In freshly isolated T lymphocytes, 97% of cells were in G0/G1 and expressed a low level of survivin protein (MCF = 189). Survivin expression was higher in cells in S+G2/M phase (MCF = 406) than in cells in G0/G1 (MCF = 175). Ninety percent of the cells progressed to S+G2/M and expressed high survivin levels (MCF = 445) after 72 hours' stimulation with anti-CD3, anti-CD28, and rhIL-2, compared with cells maintained without stimulation (MCF = 123) where 98% of the cells remained in G0/G1. Survivin mRNA expression in these cultures mirrored protein expression (data not shown).
Survivin expression and apoptosis in UCB CD34+cells
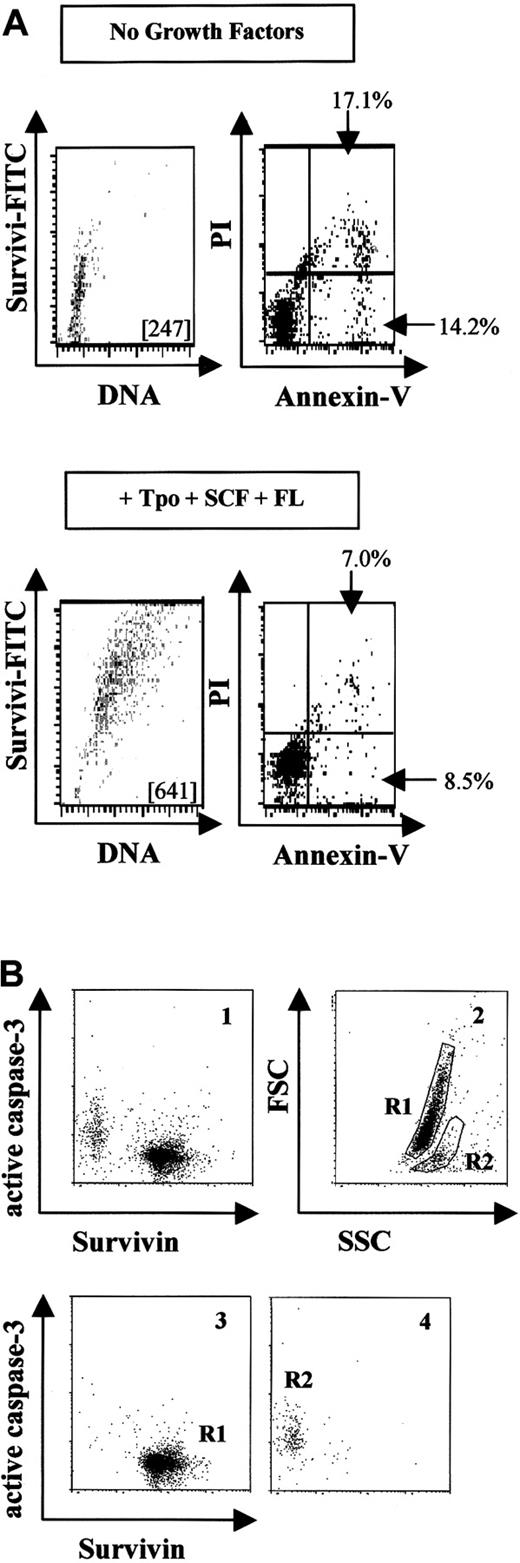
Since survivin is an antiapoptotic protein in cancer cells, we investigated survivin expression and apoptosis using annexin-V and PI in UCB CD34+ cells incubated with or without growth factors. Freshly isolated CD34+ cells demonstrated the characteristic expression of survivin (MCF = 287), with 9.8% of the cells being annexin-V positive. Without growth factors, survivin protein levels remained unchanged or decreased slightly after 48 hours (Figure 3A), and 31% of cells were apoptotic. In contrast, CD34+ cells cultured with Tpo, SCF, and FL expressed approximately 3-fold more survivin than cells cultured without growth factors, and only 15% of cells were apoptotic. Survivin expression was significantly decreased in CD34−(lin+) cells after 48 hours in culture, regardless of growth-factor addition. By annexin-V staining, 57% (with Tpo, SCF, and FL) and 83% (without growth factors) of these cells were apoptotic at 48 hours (data not shown). Analysis of hypodiploid DNA content and survivin expression in CD34+ cells cultured in the absence or presence of Tpo, FL, and SCF for 48 hours demonstrated an inverse correlation between apoptosis and survivin expression similar to that observed with annexin-V staining (Table1).
Survivin expression and apoptosis in UCB CD34+ cells.
(A) UCB CD34+ cells were cultured without (upper panels) or with (lower panels) Tpo, SCF, and FL for 48 hours, and survivin expression and apoptosis were quantitated by means of annexin-V/PI. The MCF of FITC-survivin is shown in each dot plot. The percentage of early apoptotic cells (annexin-VhighPIlow) and apoptotic and dying cells (annexin-VhighPIhigh) is shown in each blot. Data are representative of 3 experiments. (B) UCB CD34+ cells were cultured in the absence of growth factors in 10% FBS for 48 hours and then stained with FITC–anti-human survivin mAb and PE–anti-human active caspase-3 antibody (panel 1). Panel 2 shows forward and side scatter, representing R1 and R2 as viable and apoptotic cells, respectively. Panels 3 and 4 show survivin and caspase-3 counterstaining in the gated R1 and R2 cells from panel 2. Cells in R1 appeared as survivinhigh/active caspase-3low while cells in R2 appeared as survivinlow/active caspase-3high. Data are from 1 of 3 experiments. Identical results were observed with the use of polyclonal anti–human survivin antibody.
Survivin expression and apoptosis in UCB CD34+ cells.
(A) UCB CD34+ cells were cultured without (upper panels) or with (lower panels) Tpo, SCF, and FL for 48 hours, and survivin expression and apoptosis were quantitated by means of annexin-V/PI. The MCF of FITC-survivin is shown in each dot plot. The percentage of early apoptotic cells (annexin-VhighPIlow) and apoptotic and dying cells (annexin-VhighPIhigh) is shown in each blot. Data are representative of 3 experiments. (B) UCB CD34+ cells were cultured in the absence of growth factors in 10% FBS for 48 hours and then stained with FITC–anti-human survivin mAb and PE–anti-human active caspase-3 antibody (panel 1). Panel 2 shows forward and side scatter, representing R1 and R2 as viable and apoptotic cells, respectively. Panels 3 and 4 show survivin and caspase-3 counterstaining in the gated R1 and R2 cells from panel 2. Cells in R1 appeared as survivinhigh/active caspase-3low while cells in R2 appeared as survivinlow/active caspase-3high. Data are from 1 of 3 experiments. Identical results were observed with the use of polyclonal anti–human survivin antibody.
The effects of hematopoietic growth factors on hypodiploid DNA content and survivin protein expression in umbilical cord blood CD34+ cells
Umbilical cord blood CD34+ cells were cultured in the absence or presence of 100 ng/mL each of Tpo, FL, and SCF for 48 hours. Data are from 1 of 2 identical experiments.
Tpo indicates thrombopoietin; FL, Flt3 ligand; SCF, stem cell factor.
Hypodiploid DNA content was determined by flow cytometry on cells stained with 0.5 μg/mL propidium iodide at 48 hours.
Survivin expression was determined by intracellular staining with 5 μg/mL polyclonal antisurvivin and flow cytometry at 48 hours. Data are presented as mean channel fluorescence.
Since caspase-3 is activated when UCB CD34+ cells are deprived of growth factors35 and caspase-3 activation is inhibited by survivin in cancer cells,24 the relationship between survivin expression and caspase-3 activation in growth factor–starved UCB CD34+ cells was examined. Cells were cultured in the absence of growth factors in the presence of 10% HI-FBS for 48 hours, and survivin and active caspase-3 were examined by multivariate flow cytometry (Figure 3B). Two distinct cell populations appeared: survivinhigh/active caspase-3low and survivinlow/active caspase-3high (Figure 3B, panel 1). When gated for viable cells (R1) and apoptotic cells (R2) by forward and side scatter (Figure 3B, panel 2), viable cells (R1) corresponded exclusively to the survivinhigh/active caspase-3low population (Figure 3B, panel 3), whereas apoptotic cells (R2) were survivinlow/active caspase-3high (Figure 3B, panel 4). Similar findings were observed in cells starved for 24 hours (data not shown). Culture of UCB CD34+ cells with 50 μM ZVAD-fmk for 48 hours in the absence of growth factors resulted in a 33.8% increase in cell viability and a 34.7% reduction in active caspase-3, with only a 5.9% reduction in survivin expression compared with CD34+ cells cultured without caspase inhibitor.
Survivin expression in UCB CD34+ cells sorted by cell cycle
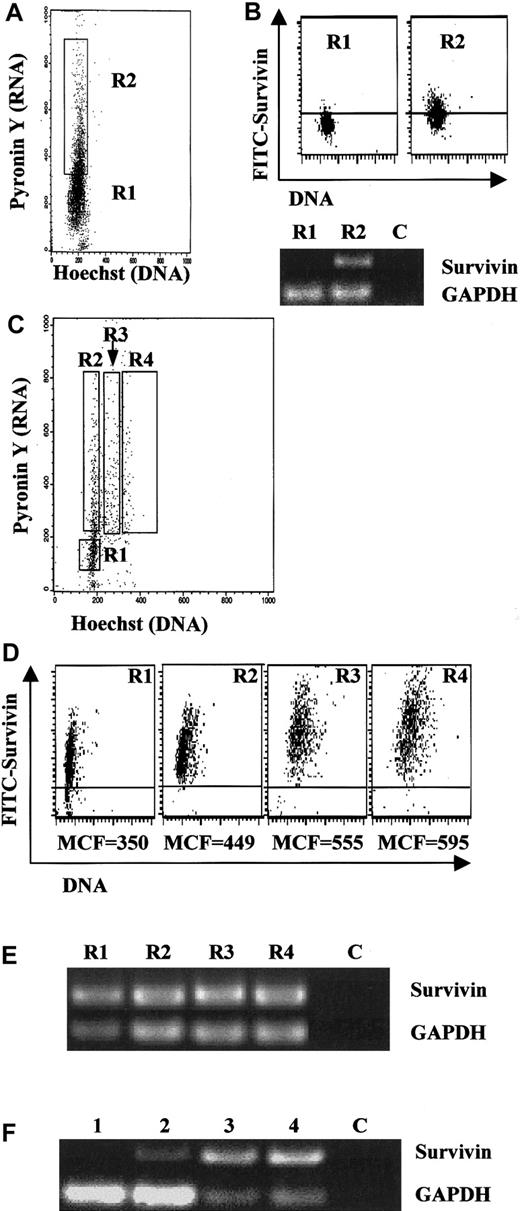
In transformed and neoplastic cells, survivin is expressed primarily during G2/M phase.23 In order to investigate survivin expression in CD34+ cells relative to cell cycle, UCB CD34+ cells were stained with Hst-33342 and PY and FACS-sorted into G0 and G1 populations. The gating criteria are shown in Figure4A. The population of HstlowPYlow cells (R1 gate) represents G0 CD34+ cells and the HstlowPYhigh population (R2) represents cells in G1. Analysis of survivin mRNA by RT-PCR indicated that little or no survivin was expressed in fresh UCB G0CD34+ cells (Figure 4B). In contrast, low levels of survivin protein were detected by intracellular flow cytometry, with 8% of the cells staining positive (Figure 4B). Survivin mRNA and protein were detected in G1 CD34+ cells, with more than 50% of the cells staining positive for survivin by flow analysis (Figure 4B).
Survivin expression in UCB CD34+ cells sorted by cell cycle.
(A) Fresh UCB CD34+ cells were sorted by Hst and PY staining as described in “Materials and methods” into G0 (R1) and G1 (R2) cell populations. Data are representative of 3 UCB samples. (B) Cells sorted in panel A were subjected to RT-PCR (40 cycles) and intracellular staining for survivin. Identical results were observed with the use of either an antisurvivin mAb (shown) or a polyclonal antisurvivin antibody. Upper panel: intracellular staining for survivin in fresh G0CD34+ (R1) and G1 CD34+ (R2) following cell sorting by Hst/PY. The area below the horizontal bar in each dot blot represents isotype control. The MCF of FITC-survivin is shown below each dot blot. Lower panel: 50 ng total RNA was used for reverse transcription and one tenth of the cDNA was used for PCR. Lane C was loaded with RT-PCR sample run without any RNA template. Data are representative of 3 UCB samples. (C) UCB CD34+ cells cultured with Tpo, SCF, and FL for 48 hours were stained with Hst and PY and sorted into gates R1 through R4 representing G0, G1, S, and G2/M populations as described in “Materials and methods.” All cells were counterstained with anti-CD34 mAb and gated to include only CD34+ cells. Data are from 1 of 3 identical experiments. (D) Cells in each gate in panel C were analyzed by multivariate flow cytometry with PI and FITC-antisurvivin mAb. The area below the horizontal bar in each dot blot represents isotype control. MCF of FITC-survivin is shown below each dot blot. Data are from 1 of 3 experiments with identical results. Identical results were observed with the use of FITC-antisurvivin polyclonal antibody. (E) RT-PCR (40 cycles) for survivin and GAPDH mRNA in sorted cell populations from panel C. Lane C was loaded with RT-PCR sample run without any RNA template. (F) Semiquantitative RT-PCR for survivin and GAPDH mRNA in UCB CD34+ cells in G0 and G1 before and after growth-factor stimulation. Gates were set exactly as in Figures 4A and 4C. Lane 1: freshly isolated G0 CD34+ cells. Lane 2: freshly isolated G1 CD34+ cells. Lane 3: G0 CD34+ cells after 48 hours' stimulation with Tpo, SCF, and FL. Lane 4: G1 CD34+ cells after 48 hours' stimulation with Tpo, SCF, and FL. Lane C was loaded with RT-PCR sample without any RNA template.
Survivin expression in UCB CD34+ cells sorted by cell cycle.
(A) Fresh UCB CD34+ cells were sorted by Hst and PY staining as described in “Materials and methods” into G0 (R1) and G1 (R2) cell populations. Data are representative of 3 UCB samples. (B) Cells sorted in panel A were subjected to RT-PCR (40 cycles) and intracellular staining for survivin. Identical results were observed with the use of either an antisurvivin mAb (shown) or a polyclonal antisurvivin antibody. Upper panel: intracellular staining for survivin in fresh G0CD34+ (R1) and G1 CD34+ (R2) following cell sorting by Hst/PY. The area below the horizontal bar in each dot blot represents isotype control. The MCF of FITC-survivin is shown below each dot blot. Lower panel: 50 ng total RNA was used for reverse transcription and one tenth of the cDNA was used for PCR. Lane C was loaded with RT-PCR sample run without any RNA template. Data are representative of 3 UCB samples. (C) UCB CD34+ cells cultured with Tpo, SCF, and FL for 48 hours were stained with Hst and PY and sorted into gates R1 through R4 representing G0, G1, S, and G2/M populations as described in “Materials and methods.” All cells were counterstained with anti-CD34 mAb and gated to include only CD34+ cells. Data are from 1 of 3 identical experiments. (D) Cells in each gate in panel C were analyzed by multivariate flow cytometry with PI and FITC-antisurvivin mAb. The area below the horizontal bar in each dot blot represents isotype control. MCF of FITC-survivin is shown below each dot blot. Data are from 1 of 3 experiments with identical results. Identical results were observed with the use of FITC-antisurvivin polyclonal antibody. (E) RT-PCR (40 cycles) for survivin and GAPDH mRNA in sorted cell populations from panel C. Lane C was loaded with RT-PCR sample run without any RNA template. (F) Semiquantitative RT-PCR for survivin and GAPDH mRNA in UCB CD34+ cells in G0 and G1 before and after growth-factor stimulation. Gates were set exactly as in Figures 4A and 4C. Lane 1: freshly isolated G0 CD34+ cells. Lane 2: freshly isolated G1 CD34+ cells. Lane 3: G0 CD34+ cells after 48 hours' stimulation with Tpo, SCF, and FL. Lane 4: G1 CD34+ cells after 48 hours' stimulation with Tpo, SCF, and FL. Lane C was loaded with RT-PCR sample without any RNA template.
Survivin expression and cell cycle were also examined in freshly isolated UCB CD34+ cells cultured with Tpo, SCF, and FL. After 48 hours' culture, CD34+ cells were gated and sorted into 4 populations, HstlowPYlow (R1), HstlowPYhigh (R2), Hstintermediate PYhigh (R3), and HsthighPYhigh (R4), representing cells in G0, G1, S, and G2/M of the cell cycle (Figure 4C), respectively. Each population was analyzed for survivin mRNA by RT-PCR (Figure 4E) and for protein by intracellular flow cytometry (Figure 4D). Survivin protein expression was observed in approximately 85% of G0 cells and 100% of cells in G1, S, and G2/M. Survivin protein expression increased moderately, with cell-cycle progression being lowest in G0 cells and highest (1.7-fold increase) in cells in G2/M. Survivin mRNA was detected in all cell populations.
To further investigate growth-factor upregulation of survivin relative to cell cycle, UCB CD34+ cells were sorted into G0 and G1 populations, and survivin mRNA was determined by semiquantitative RT-PCR before and after culture with Tpo, SCF, and FL for 48 hours. Survivin mRNA was detected in freshly isolated G1 (Figure 4F, lane 2) but not in G0cells (Figure 4F, lane 1), as previously observed (Figure 4B). After stimulation with Tpo, SCF, and FL, survivin mRNA was upregulated in both G0 and G1 CD34+ cells (Figure4F, lanes 3 and 4, respectively).
Growth-factor regulation of survivin, cell cycle, and apoptosis in MO7e cells
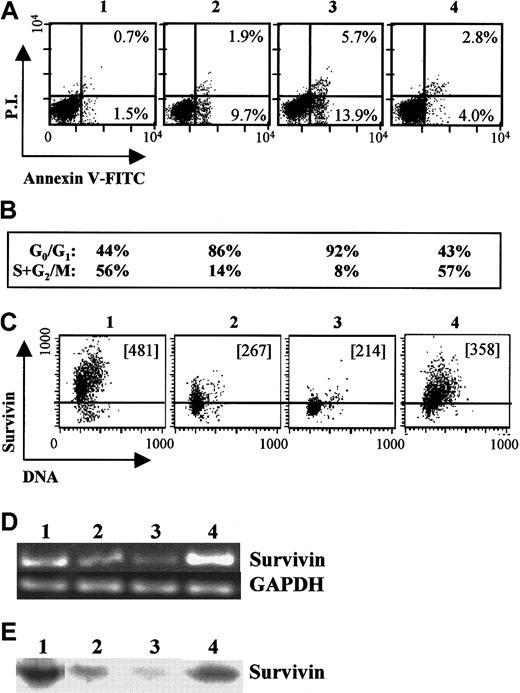
We examined growth-factor regulation of survivin, cell cycle, and apoptosis in GM-CSF–dependent MO7e cells (Figure5). MO7e cells growing in 10% HI-FBS and GM-CSF express significant survivin mRNA and protein, as demonstrated by RT-PCR (Figure 5D, lane 1), Western blotting (Figure 5E, lane 1), and intracellular flow cytometry (Figure 5C, panel 1). After 24 hours of growth-factor withdrawal, survivin mRNA (Figure 5D, lane 2) and protein levels (Figure 5E, lane 2, and 5C, panel 2) declined significantly, concomitant with cell-cycle arrest at G0/G1 (86% of cells in G0/G1 versus 44% of cells in log phase) (Figure 5B, column 2). An increase in apoptotic cells was also observed, with 12% of cells apoptotic after 24 hours of GM-CSF and FBS starvation versus 2% of cells growing in the presence of these growth factors (Figure 5A, panel 2). Factor withdrawal for an additional 24 hours resulted in further reduction of survivin mRNA (Figure 5D, lane 3) and protein levels (Figure 5E, lane 3, and 5C, panel 3); an increase in apoptotic cells to 20% (Figure 5A, panel 3); and continued cell-cycle arrest (Figure 5B, panel 3). In contrast, re-addition of GM-CSF and SCF after 24 hours of growth-factor and serum starvation significantly enhanced survivin mRNA (Figure 5D, lane 4) and protein expression (Figure 5E, lane 4, and 5C, panel 4); blocked apoptosis compared with factor-starved cells (Figure 5A, panel 4); and stimulated cell-cycle progression (Figure 5B, panel 4). Survivin expression levels increased as cells progressed through the cell cycle and were maximal during G2/M.
Survivin expression in factor-dependent MO7e cells.
Log phase MO7e cells growing in 20% FBS and 20 ng/mL GM-CSF (panel, lane 1) were washed and factor-starved for 24 hours in the absence of GM-CSF and FBS. Starved cells (panel, lane 2) were suspended in RPMI plus 10% FBS and cultured in the absence (panel, lane 3) or presence of GM-CSF and SCF (panel, lane 4) for an additional 24 hours. (A) Cells were harvested and stained with annexin-V and PI to examine apoptosis of each cell group. The percentage of early apoptotic cells (annexinhighPIlow) and apoptotic/dead cells (annexinhighPIhigh) are shown for each cell population. (B) Cell-cycle analysis for G0/G1 and S+G2/M for the corresponding cell populations. (C) Survivin expression and DNA staining for each cell population. The MCF is included in each dot plot. The area below the horizontal bar in each dot blot represents isotype control. (D) RT-PCR (35 cycles) for survivin and GAPDH mRNA for the corresponding cell populations. Negative controls indicated no amplification (not shown). (E) Western blots for survivin protein in lysates from each of the corresponding cell populations.
Survivin expression in factor-dependent MO7e cells.
Log phase MO7e cells growing in 20% FBS and 20 ng/mL GM-CSF (panel, lane 1) were washed and factor-starved for 24 hours in the absence of GM-CSF and FBS. Starved cells (panel, lane 2) were suspended in RPMI plus 10% FBS and cultured in the absence (panel, lane 3) or presence of GM-CSF and SCF (panel, lane 4) for an additional 24 hours. (A) Cells were harvested and stained with annexin-V and PI to examine apoptosis of each cell group. The percentage of early apoptotic cells (annexinhighPIlow) and apoptotic/dead cells (annexinhighPIhigh) are shown for each cell population. (B) Cell-cycle analysis for G0/G1 and S+G2/M for the corresponding cell populations. (C) Survivin expression and DNA staining for each cell population. The MCF is included in each dot plot. The area below the horizontal bar in each dot blot represents isotype control. (D) RT-PCR (35 cycles) for survivin and GAPDH mRNA for the corresponding cell populations. Negative controls indicated no amplification (not shown). (E) Western blots for survivin protein in lysates from each of the corresponding cell populations.
Analysis of survivin expression in UCB CD34+ cells and leukemic cells
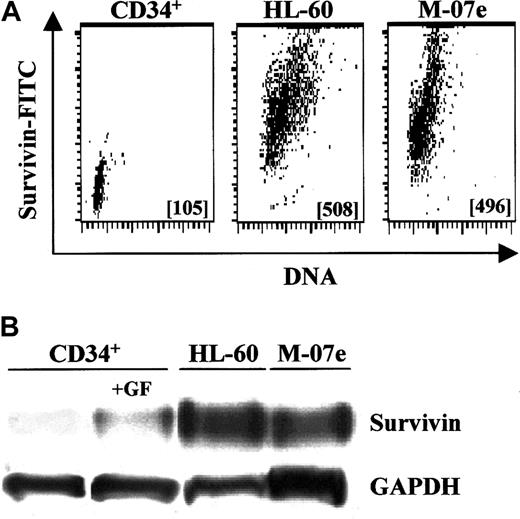
Survivin expression in primary CD34+ cells was compared with factor-independent HL-60 promyelocytic leukemia cells and GM-CSF–dependent MO7e cells. With the use of intracellular staining and flow cytometry, exponentially growing HL-60 and MO7e cells expressed approximately 5-fold more survivin than freshly isolated CD34+ cells (Figure 6A), while Western analysis indicated that these leukemic cells expressed up to 8-fold (range, 2.5- to 8.3-fold) more survivin (data not shown). By Western analysis, exponentially growing HL-60 and MO7e cells expressed 2.7- and 2.3-fold more survivin, respectively, than CD34+cells cultured with Tpo, SCF, and FL for 48 hours (Figure 6B).
Relative survivin expression in UCB CD34+cells and leukemic cells.
(A) Multivariate intracellular flow cytometry plots of survivin expression in fresh UCB CD34+ cells, and exponentially growing HL-60 and MO7e leukemia cells. Survivin MCF is shown in each data plot. Data are from 1 of 2 experiments with identical results. (B) Western blot analysis of survivin levels in freshly isolated UCB CD34+ cells; CD34+ cells stimulated with 100 ng/mL each of Tpo, FL, and SCF for 48 hours; and exponentially growing HL-60 and MO7e leukemia cells. Cell lysates from 1 × 106cells were loaded in each lane and blotted with polyclonal anti–human survivin. Data are from 1 of 3 experiments.
Relative survivin expression in UCB CD34+cells and leukemic cells.
(A) Multivariate intracellular flow cytometry plots of survivin expression in fresh UCB CD34+ cells, and exponentially growing HL-60 and MO7e leukemia cells. Survivin MCF is shown in each data plot. Data are from 1 of 2 experiments with identical results. (B) Western blot analysis of survivin levels in freshly isolated UCB CD34+ cells; CD34+ cells stimulated with 100 ng/mL each of Tpo, FL, and SCF for 48 hours; and exponentially growing HL-60 and MO7e leukemia cells. Cell lysates from 1 × 106cells were loaded in each lane and blotted with polyclonal anti–human survivin. Data are from 1 of 3 experiments.
Discussion
Multivariate intracellular flow cytometry, cell-cycle separation and analysis, Western blots, and RT-PCR indicate that survivin is expressed in normal hematopoietic cells, particularly CD34+cells enriched for early and late stem and progenitor cells. In addition, survivin mRNA and protein expression in CD34+cells and factor-dependent leukemic cells was regulated by hematopoietic growth factors. Expression of survivin was highest in G2/M phase of the cell cycle, but in contrast to cancer cells, which express survivin preferentially during G2/M phase, CD34+ cells express survivin in all phases of the cell cycle.
Survivin mRNA and protein are expressed during G1 in freshly isolated CD34+ cells when these cells are analyzed by means of Hst-33342 and PY staining. CD34+ cells in G0 did not express survivin mRNA, but low levels of survivin protein could be detected by intracellular flow cytometry. The detection of survivin protein in freshly isolated G0CD34+ cells in the absence of mRNA might represent survivin protein carried over through mitosis, differences in protein versus message stability, or minor contamination of G0 cells with G1 cells. In this regard, survivin expression does not completely disappear in CD34+ cells even after 48 hours without growth factors. After sorting CD34+ cells on the basis of cell cycle, significant increases in both survivin mRNA and protein were observed in G0 and G1 cells after growth-factor stimulation, suggesting that survivin was specifically upregulated in CD34+ cells by hematopoietic growth factors coincident with exit from quiescence. These results are in contrast to survivin expression in a G2/M–specific manner in HeLa cells or immortalized NIH3T3 cells.23 26
Survivin upregulation in CD34+ hematopoietic cells is not as dramatic as in cancer cells or 3T3 cells. In HeLa cells, survivin mRNA and protein are undetectable in G0/G1cells and increase 6-fold during S-phase and 40-fold during G2/M.23 Survivin mRNA or protein in CD34+ cells routinely increases 3.3 ± 1.1-fold after cytokine stimulation. This difference in degree of survivin upregulation probably relates to differences between growth-regulated and transformed cells. In this regard, survivin promoter activity is 10-fold higher in transformed cells than in normal cells,36 suggesting deregulation of survivin transcription in cancer cells.37 Our observations concerning the magnitude of survivin upregulation in CD34+ cells is similar to the 2.2-fold increase in survivin protein observed in human primary endothelial cells after stimulation with angiopoietin-1,28 although these cells expressed survivin only during G2/M. In contrast, factor-independent HL-60 cells express 6.2 ± 2.8-fold and up to 8-fold more survivin than freshly isolated UCB CD34+ cells by multivariate flow cytometry and Western analysis, respectively. HL-60 cells express approximately 3-fold more survivin than growth-factor–stimulated CD34+ cells. Despite retention of a normal cytokine regulatory phenotype,31 MO7e cells also expressed more robust levels of survivin than CD34+ cells (5.8 ± 1.5-fold by flow cytometry; 2.4- to 8.5-fold by Western analysis) and expressed approximately 2-fold more survivin than growth-factor–stimulated CD34+ cells by Western analysis. These data agree with the findings that human myeloid leukemia cells express approximately 10-fold higher survivin levels than adult marrow CD34+ cells.38
Currently, reported data favor the hypothesis that survivin overcomes a normal apoptotic block in G2/M of the cell cycle,7,23 probably by downregulating the Cdk4/cyclin D complex; activating the Cdk2/cyclin E complex; and releasing p21, which blocks caspase-3 activation thereby suppressing cell death signaling.24 39 In cancer cells, control of this apoptosis checkpoint has been lost, either directly by loss of regulation of survivin expression or via some other pathway that enhances survivin production. In CD34+ cells, the decision to enter mitosis rather than initiate apoptosis may require only modest levels of survivin, perhaps proportional to the degree of growth-factor stimulation.
Survivin is expressed in all fetal tissue and normal thymus, but not in most adult tissues.2 In hematopoietic cells, survivin expression is highest in CD34+ cells that are enriched for stem and progenitor cells, intermediate in CD34−(lin+) cells that are primarily late-stage immature cells and differentiated cells, and lowest in differentiated nondividing peripheral blood mononuclear cells. These and other data presented here suggest an inverse correlation between survivin expression and hematopoietic cell differentiation. To further address the issue of whether survivin expression is developmentally regulated or occurs only in proliferating cells of the hematopoietic system, we examined survivin expression in adult peripheral blood T lymphocytes. Survivin is expressed in freshly isolated T cells, primarily in the small fraction of cells that are in S+G2/M. However, as cells in G0/G1enter cell cycle following stimulation, survivin expression increases. This is consistent with data on mouse TIAP, which is rapidly upregulated upon T-cell activation.26 The fact that survivin expression is growth-factor regulated in both primitive hematopoietic cells and adult somatic T cells suggests that survivin is expressed during proliferation, at least in the lymphohematopoietic system. Whether cell proliferation leads to survivin expression or cells proliferate because of induction of survivin expression remains to be determined.
In normal CD34+ cells and MO7e cells, survivin upregulation was associated with inhibition of apoptosis and decreased survivin expression correlated with increased apoptosis, as shown by annexin-V staining or analysis of hypodiploid DNA content. These observations suggest an antiapoptotic effector function of survivin in hematopoietic cells, similar to that observed in other cell systems.8,23,26 Activation of caspase-3 is associated with growth-factor withdrawal–induced apoptosis in UCB CD34+cells,35 and suppression of casapase-3 activation by survivin is well documented, at least in cancer cells.7-9,20,23,25,26 Our multivariate flow cytometry results for survivin and active caspase-3 expression in UCB CD34+ cells undergoing growth-factor withdrawal–induced apoptosis demonstrate an inverse correlation between survivin and active caspase-3, ie, survivinhigh/active caspase-3low in viable cells and survivinlow/active caspase-3high in apoptotic cells. Survivin upregulation by hematopoietic growth factor is consistent with caspase-3 inactivation and strongly suggests an antiapoptotic role for survivin in normal adult CD34+cells. Inhibition of caspase activity without an effect on survivin levels in CD34+ cells by the caspase inhibitor ZVAD-fmk resulted in an antiapoptotic phenotype equivalent to growth-factor addition and its sequela of survivin upregulation and is consistent with caspases' being downstream of survivin.8 24
In summary, we have demonstrated that survivin expression may be involved in the normal regulation of adult hematopoietic cell proliferation and survival and suggest that survivin-targeted therapy for cancer cells or tumor angiogenesis may also disrupt normal hematopoiesis. We do not know if disruption of survivin production or action in primary normal hematopoietic cells will have a direct consequence on hematopoietic cell survival or proliferation. However, the tight association between survivin expression, cell cycle, and apoptosis in normal CD34+ cells and GM-CSF–dependent MO7e cells strongly suggests that this is the case. In support of this hypothesis, phosphothioate oligonucleotide survivin antisense therapy in HL-60 cells blocks survivin gene translation, initiates apoptosis measured by DNA fragmentation, and reduces cell proliferation and total viable cell number by greater than 85% within 3 days (data not shown). These results in hematopoietic cells are consistent with survivin antisense therapy in cancer cell models.19,20,22 In addition, culture of CD34+ cells with early- (Tpo, SCF, FL) or late-acting (G-CSF, GM-CSF, M-CSF) growth-factor cocktails for 7 weeks demonstrates a positive correlation between survivin mRNA levels and continued production of granulocyte-macrophage colony-forming units, a function of more primitive hematopoietic cells, but not total nucleated differentiated cell production (data not shown). Finally, in UCB CD34+ cells, growth-factor withdrawal results in activation of caspase-3 and apoptosis,35 a result consistent with our demonstration of downregulation of survivin and activation of caspase-3 in UCB CD34+ cells.
We thank Dr Hal Broxmeyer and Charlie Mantel for helpful discussions; Dr Eddie Srour and Susan Rice for their help with cell-cycle gating and sorting; and Hui-min Bian and Jonathan Pelus for technical support.
Supported by the Walther Oncology Center, Indiana University School of Medicine, and the Walther Cancer Institute, Indianapolis, IN.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Louis M. Pelus, Walther Oncology Center, Indiana University School of Medicine, 1044 West Walnut St, Indianapolis, IN 46202; e-mail: lpelus@iupui.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal