Genetic polymorphisms of the low-affinity receptors of the Fc domain of IgG (FcγR) have been proposed to be associated with an array of hematologic, autoimmune, and other diseases. Most studies have addressed the R/H131polymorphism of the FcγRIIa isoform1 and theV/F158 polymorphism of the FcγRIIIaisoform.2 For the majority of the associations, only single studies have been performed. But for conditions such as heparin-induced thrombocytopenia, systemic lupus erythematosus (SLE) and SLE-related nephritis, and idiopathic thrombocytopenic purpura, data have been generated from multiple teams of investigators. Information from different teams has often been controversial or even conflicting. We believe that there is a need to standardize and synthesize the accumulating data across these various studies.
Lehrnbecher et al3 made an effort to accumulate in a systematic fashion the epidemiologic data on genetic associations involving low-affinity FcγR receptor polymorphisms. Unfortunately, the synthesis of the data was problematic. Typically, when there was more than one study addressing the same association, the number of subjects in the patient and control arms were summed for the various genotypes and statistical tests were performed in the resulting contingency tables with the summed data. This approach is methodologically inappropriate. The synthesis of data across diverse studies needs to take into account the within study variance, and when there is detectable statistical heterogeneity in the measure of association between the studies, it would be prudent to also take into account an estimate of the variance between the studies.4Such a formal meta-analytic approach may yield very different results compared to simple summation.
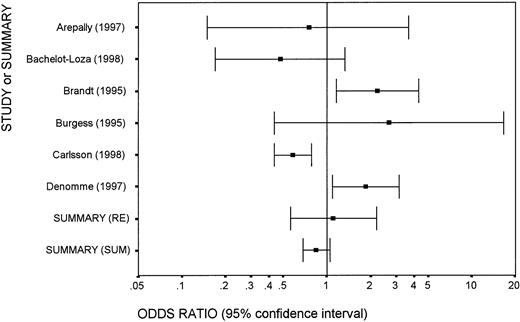
As an example, the figure shows a meta-analysis for the association of the FcγRIIa H/H131 genotype with heparin-induced thrombocytopenia, analyzing the same databases as in the Lehrnbecher et al paper, which had suggested a possible negative association ofH/H131 with heparin-induced thrombocytopenia (P = .13). To simply sum up the data from 6 studies,H/H131 is found in 151 of the 626 patients (24.1%) versus in 359 of the 1313 controls (27.3%). The crude odds ratio is 0.84 (95% CI, 0.68-1.05). In truth, here is a situation in which there is highly statistically significant heterogeneity between the 6 studies (P < .001 for heterogeneity based on the Q statistic).4 As shown in Figure1, some studies suggest a strongly positive association, while others suggest a negative association. Simple summation is misleading. By random effects calculations,5 the summary odds ratio is 1.11 (95% CI, 0.56-2.19) and the P value for the association is .77, which suggests absolutely no consistent effect.
Meta-analysis of 6 studies addressing the association of FcγRIIa R131H polymorphism with heparin-induced thrombocytopenia.
Each study is shown as a point-estimate odds ratio, with lines extending to the 95% confidence interval of the odds ratio. The summary effect is obtained with random effects (RE) calculations, and for comparison we also show a summary effect based on simply adding up the numbers across studies (SUM). The numerical data are presented in Lehrnbecher et al.3
Meta-analysis of 6 studies addressing the association of FcγRIIa R131H polymorphism with heparin-induced thrombocytopenia.
Each study is shown as a point-estimate odds ratio, with lines extending to the 95% confidence interval of the odds ratio. The summary effect is obtained with random effects (RE) calculations, and for comparison we also show a summary effect based on simply adding up the numbers across studies (SUM). The numerical data are presented in Lehrnbecher et al.3
The table shows the summary odds ratios by fixed and random effects models,5 as well as the estimated P value for the heterogeneity Q statistic for 5 associations where multiple studies have been performed. In the presence of statistically significant heterogeneity, the fixed effects estimates are also not appropriate. Thus, while the original report suggested a trend for an association of FcγRIIa R/R131genotype with idiopathic thrombocytopenic purpura (P = .089) and the fixed effects result is also fairly similar, the 2 available studies are very heterogeneous and the pooled estimate by random effects has very wide confidence intervals.
Meta-analysis estimates for the association of polymorphisms of the low-affinity Fcγ receptors with various disease outcomes
| Outcome (no. of studies) . | FcγRIIa: HH vs HR +RR . | FcγRIIa: RR vs HR +HH . | ||||
|---|---|---|---|---|---|---|
| Heterogeneity P* . | ORr (95% CI) . | ORf (95% CI) . | Heterogeneity P* . | ORr (95% CI) . | ORf (95% CI) . | |
| SLE (12) | .009 | 0.61 (0.44-0.85) | 0.70 (0.57-0.85) | .810 | 1.37 (1.13-1.67) | 1.38 (1.14-1.67) |
| HIT (6) | <.001 | 1.11 (0.56-2.19) | 0.86 (0.69-1.08) | .023 | 0.81 (0.49-1.34) | 1.02 (0.81-1.29) |
| ITP (2) | .067 | 0.81 (0.19-3.50) | 0.92 (0.45-1.87) | .011 | 1.62 (0.24-11.2) | 1.71 (0.87-3.38) |
| SLE nephritis (7) | .156 | 0.71 (0.44-1.14) | 0.68 (0.47-0.98) | .617 | 1.35 (0.97-1.88) | 1.36 (0.98-1.88) |
| FcγRIIIa: FF vs VF+VV | FcγRIIIa: VV vs VF+ FF | |||||
| SLE (2) | .493 | 2.49 (1.67-3.71) | 2.48 (1.66-3.70) | .549 | 0.67 (0.38-1.19) | 0.67 (0.38-1.18) |
| Outcome (no. of studies) . | FcγRIIa: HH vs HR +RR . | FcγRIIa: RR vs HR +HH . | ||||
|---|---|---|---|---|---|---|
| Heterogeneity P* . | ORr (95% CI) . | ORf (95% CI) . | Heterogeneity P* . | ORr (95% CI) . | ORf (95% CI) . | |
| SLE (12) | .009 | 0.61 (0.44-0.85) | 0.70 (0.57-0.85) | .810 | 1.37 (1.13-1.67) | 1.38 (1.14-1.67) |
| HIT (6) | <.001 | 1.11 (0.56-2.19) | 0.86 (0.69-1.08) | .023 | 0.81 (0.49-1.34) | 1.02 (0.81-1.29) |
| ITP (2) | .067 | 0.81 (0.19-3.50) | 0.92 (0.45-1.87) | .011 | 1.62 (0.24-11.2) | 1.71 (0.87-3.38) |
| SLE nephritis (7) | .156 | 0.71 (0.44-1.14) | 0.68 (0.47-0.98) | .617 | 1.35 (0.97-1.88) | 1.36 (0.98-1.88) |
| FcγRIIIa: FF vs VF+VV | FcγRIIIa: VV vs VF+ FF | |||||
| SLE (2) | .493 | 2.49 (1.67-3.71) | 2.48 (1.66-3.70) | .549 | 0.67 (0.38-1.19) | 0.67 (0.38-1.18) |
Numerical data for the calculations are as presented by Lehrnbecher et al.3
ORf indicates fixed-effects odds ratio; ORf, random-effects odds ratio; SLE, systemic lupus erythematosus; HIT, heparin-induced thrombocytopenia; and ITP, idiopathic thrombocytopenic purpura.
P value for the Q statistic testing for between-study heterogeneity of the odds ratios; heterogeneity is typically considered statistically significant for P < .10.
There are some other serious biases that need to be considered in synthesizing information from genetic association studies. A key issue is that all the pertinent information needs to be retrieved and synthesized. Excluding studies because they do not give numerical data on their published reports, as was done by Lehrnbecher et al, may lead to skewed estimates. It may be more common not to report exact numbers when no significant association is seen than when a significant relationship is revealed. The summary data may then suggest a spurious association, simply because “negative” studies were excluded. A similar bias can arise, if negative studies are not published at all, a situation that is often more difficult to discern.6 Even in this situation, there are quantitative techniques in meta-analysis such as variance-related diagnostics7 and recursive cumulative meta-analysis8 that can give us an impression about the possible extent of bias and heterogeneity. Furthermore, bias may occur when only the early studies in a field (hypothesis-generating studies) are synthesized, since their results may differ from the subsequent hypothesis-validating studies. Most of these genetic effects may be of modest magnitude,9 as also suggested by the preliminary data in the table, but they may be important to prove or to refute in building our knowledge about the genetic background of these diseases.
For all these reasons, we believe that a careful meta-analysis is important to perform in this field, and we have undertaken the conduct of this project. The available information for some associations is already very extensive. For example, there were data for 12 studies in the original report of Lehrnbecher et al for the association ofFcγRIIa R/H131 with SLE and 7 studies for the association with SLE nephritis, separating different racial subgroups in each study. To date, our preliminary comprehensive search has identified 22 and 26 respective studies for these 2 associations. A large number of studies has also accumulated on the FcγRIIIa V/F158polymorphism. We are making an effort to communicate with experts in the field who could provide key missing data and potentially additional databases. All-inclusiveness would be important for ensuring that all pertinent information is assembled in the meta-analysis. We would like to take the opportunity to invite investigators who have worked on low-affinity Fcγ receptor gene polymorphisms to communicate with us and contribute additional data for this international effort. Establishing a mechanism for performing comprehensive meta-analyses would be important, as more polymorphisms with potential clinical significance are being identified in this rapidly changing field of research.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal