CEACAM1 on leukocytic, endothelial, and epithelial cells functions in homophilic adhesion, tumor suppression, regulating cell adhesion and proliferation, and in heterophilic adhesion as a receptor for E-selectin and Neisseria meningiditis, Neisseria gonorrhoeae, Haemophilus influenzae, and murine coronaviruses. The 8 transmembrane isoforms of human CEACAM1 possess an extracellular N-terminal IgV domain, followed by variable numbers of IgC2 domains. To establish which key amino acids contribute specifically to CEACAM1 homophilic adhesion, exposed amino acids in the N-terminal domain of a soluble form of CEACAM1 were subjected to mutagenesis. Analyses of mutant proteins with conformationally dependent antibodies indicated that most mutations did not substantially affect the structural integrity of CEACAM1. Nevertheless, decreased adhesion was observed for the single mutants V39A or D40A (single-letter amino acid codes) in the CC′ loop and for the triple mutants located in the GFCC′C" face of the N-terminal domain. Interestingly, whereas single mutations in R64 or D82 that are predicted to form a salt bridge between the base of the D and F β strands close to the critical V39 and D40 residues also abolish adhesion, an amino acid swap (R64D and D82R), which maintains the salt bridge was without significant effect. These studies indicate that the CC′ loop plays a crucial role in the homophilic adhesion of CEACAM1. They further predict that specific hydrophobic amino acid residues on the nonglycosylated GFCC′C" face of CEACAM1 N-terminal domain are not only involved in heterophilic interactions with Opa proteins and H influenzae, but are also critical for protein-protein interactions between 2 CEACAM1 molecules on opposing cells.
Introduction
The human carcinoembryonic antigen (CEA) family is composed of 29 genes tandemly arranged on chromosome 19q13.2. Based on nucleotide homologies, these genes are classified into 2 major subfamilies, the CEACAM and the pregnancy-specific glycoprotein (PSG) subgroups. The CEACAM-encoded proteins include CEA, the biliary glycoproteins (CEACAM1), nonspecific cross-reacting antigen (CEACAM6), and the CEA gene members, CEACAM3 (CGM1), CEACAM4 (CGM7), CEACAM7 (CGM2), and CEACAM8 (CGM6).1,2 Protein structural analyses indicate that CEACAM subgroup members belong to the immunoglobulin (Ig) superfamily of adhesion molecules. The complexity of the CEACAM subgroup is increased by differential splicing and posttranslational modifications of some of its members. This is exemplified by the human CEACAM1 isoforms, where at least 8 transmembrane variants are generated by differential splicing of a single gene.1 2 These 8 isoforms possess an extracellular N-terminal IgV-set domain, followed by no (CEACAM1-1L and -1S), 2 (A1, B for CEACAM-3L and -3S), or 3 (A1, B, A2 for CEACAM-4L and -4S) IgC2-set domains, or with the A2 domain replaced by a serine-threonine–rich non-Ig sequence (Y, Z for CEACAM1-3AL and -3AS). Alternative splicing of the cytoplasmic exons generates CEACAM1 variants with either long (L) or short (S) cytoplasmic tails.
The CEACAM1 molecules are recognized by CD66a monoclonal antibodies (Mabs)3 and are expressed widely, occurring on monocytes, granulocytes and their precursors, activated T cells,4 and CD16−CD56+ natural killer cells, B cells, and the epithelium and endothelium of a variety of tissues.1,2Both apical membrane staining of simple epithelia with CD66/CEACAM antibodies and localization of CEACAM1 to sites of cell-cell contact (eg, between hepatocytes, stratified epithelia, junctional epithelium that forms a transition zone between gingival epithelium and teeth, and between pericytes and endothelial cells of blood vessel walls) have been demonstrated. These spatiotemporal expression patterns, the structural relationship of the CEACAM1 molecules to the Ig superfamily, and the presence of specific signal transduction motifs in their cytoplasmic tails provided the first clues to their diverse functions as homophilic and heterophilic adhesion molecules and as regulators of signal transduction. Homophilic adhesion, which has been confirmed by studies in vitro for the human CEACAM1-4L, -4S, -3L, -1L, and -4AL isoforms5-7 and for rodent CEACAM1 molecules,1,2 is thought to be important in the embryonic organization of the intestinal epithelium and of hepatocytes in the liver, in placental trophoblasts, during muscle and tooth development, during vascularization of the central nervous system,1,2in neutrophil activation and adhesion during inflammatory responses,8 in lymphoregulation and immunosurveillance,4 in angiogenesis,9 and in the negative regulation of cell proliferation.1,2Heterophilic adhesion of CEACAM1 to other CEACAM family members, E-selectin, to fimbral proteins of Escherichia coli andSalmonella, to Opa proteins of Neisseria meningiditisand Neisseria gonorrhoeae, to Haemophilus influenzae, and to murine coronaviruses has also been reported.10-18 This heterophilic adhesion mediatesNeisseria, Haemophilus, or coronavirus transmission; facilitates bacterial colonization of the gut and bacterial phagocytosis by neutrophils; and is involved in the initial tethering of granulocytes to E-selectin on the endothelium prior to their transendothelial migration during inflammatory responses.
Because the N-terminal IgV set domain of CEACAM1 has been implicated in mediating homophilic adhesion,7,19-23 we have determined the key amino acid residues (single-letter amino acid codes used throughout) involved in such interactions using site-directed mutagenesis. The basic structure of the IgV N-terminal domain of CEACAM1 is a predicted tertiary fold of a stacked pair of β-pleated sheets. There are 9 component β strands, with strands A, B, E, and D lying in one sheet and strands C, C′, C", F, and G being antiparallel in the other. When homology can be shown with nuclear magnetic resonance (NMR) or crystallographically well characterized proteins like CD2 and CD58,24-27 many properties of 3-dimensional structure may be predicted. Thus, selection of amino acids for mutagenesis was based on a 3-dimensional model for the N-domain of CEACAM1 and the identification of exposed amino acids from sequence alignments of the N-terminal amino acid sequences of human CEACAM1 with rat and human CD225 and human CD58.25-27 From these studies, our results suggest that residues on the GFCC′C" face of the N-terminal domain, particularly those within the CC′ loop, play a crucial role in such homophilic interactions.
Materials and methods
Mabs
The rat CD66/CEACAM MAb, YTH71.3.2 (rIgG2a isotype),3 and the mouse 34B1, 5F4, and 26H7 MAbs (all mIgG1 isotypes)4 were produced as described. The CD66/CEACAM MAb, clone 85A12, was obtained from Oxoid (Basingstoke, United Kingdom). All remaining mouse CD66/CEACAM MAbs were obtained from the Vth and VIth Leukocyte Differentiation Antigen Workshop.28 29 The CD14 (Tük 2; mIgG2a isotype) and CD31 (JC/70A; mIgG1 isotype) MAbs were purchased from Dakopatts (Glostrup, Denmark) and the anti-NCAM (BCA8; mIgG1 isotype) from R & D Systems (Abingdon, United Kingdom). The anti-MUC 18 MAb, 1B8, was kindly provided by Professor I. Hart, ICRF, London, United Kingdom. The isotype-matched mouse IgG1, IgG2a, and IgG2b irrelevant negative control MAbs were purchased from Dakopatts. YTH76.9 (Serotec, Oxford, United Kingdom) to HLA class 1 molecules was used as the rat IgG2a-negative control.
Stable transfectants
Chinese hamster ovary (CHO) cells stably expressing CEACAM1-4L and -4S and CEACAM1-1S were generated as described,4,5,30as were stable HeLa transfectants expressing CEA, CEACAM3, CEACAM6, and CEACAM8.31-33 Nontransfected CHO cells or CHO cells stably transfected with pSV2Neo (CHO-Neo) were used as negative controls.
Flow cytometric analysis and quantification
For quantitating the levels of CEACAM molecules on transfectants, flow cytometry was performed on a FACScan or FACSCalibur flow cytometer (Becton Dickinson, Sunnyvale, CA) using the LysisII or Cellquest software for data processing. Cells (1-2 × 105) were labeled with CD66/CEACAM-specific MAbs or irrelevant negative control MAbs of the same isotype (50 μg/mL), phycoerythrin-linked goat (Southern Biotechnology Associates, Birmingham, AL), or fluorescein isothiocyanate–conjugated rabbit F(ab)2 (Dakopatts) antimouse IgG (1:50) and then propidium iodide (5 μg/mL).5 Absolute quantification of the number of expressed CEACAM molecules was carried out using Quantum simply cellular microbeads (Sigma Chemical, St Louis, MO), using the MAbs COL1, 9A6, 80H3, T84.66, and D14HD11 to quantify CEACAM3, CEACAM6, CEA, and CEACAM1, respectively, and revealed CEA on HeLa 20 000 sites/cell, CEACAM1-4L on CHO 211 000 sites/cell, CEACAM1-4S on CHO 39 000 sites/cell, CEACAM1-1S on CHO 441 000 sites/cell, CEACAM8 on HeLa 18 000 sites/cell, CEACAM6 on HeLa 106 000 sites/cell, and CEACAM3 on HeLa 34 000 sites/cell.
Solublerecombinant chimeric proteins
Epitope mapping of CD66/CEACAM MAbs
The reactivity of the MAbs with the soluble domain deletion constructs of CEACAM1 or with negative control constructs, NCAM(D1-7)-Fc or MUC18(D1-5)-Fc, was determined in triplicate and repeated at least twice using alkaline phosphatase–based enzyme-linked immunosorbent assays (ELISAs).7
Identification of the conformational-dependent and -independent CD66/CEACAM MAbs
Because conformational-dependent MAbs provide useful and sensitive probes for analyzing structural alterations in mutant proteins, the capacities of CD66/CEACAM MAbs to recognize native as opposed to denatured forms of soluble recombinant CEACAM1 constructs were determined. Proteins were denatured by boiling for 5 minutes. in 0.1% (wt/vol) sodium dodecyl sulfate (SDS), and 0.2% (vol/vol) β-mercaptoethanol. Samples of either untreated (native) or denatured CEACAM1-4-Fc and NCAM(D1-7)-Fc recombinant proteins (100 and 10 ng/100 μL) were analyzed for CD66 MAb antibody binding after slot blot transfer to immobilon-polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA), and incubation with CD66/CEACAM or isotype-matched negative MAbs and sheep F(ab)2 antimouse IgG–horseradish peroxidase. The membranes were developed using the chemiluminescence kit (ECL; Amersham International, Little Chalfont, Bucks, United Kingdom).
Site-directed mutagenesis of soluble recombinant CEACAM1-3-Fc complementary DNAs
The initial site-directed mutagenesis of the N-terminal domain of CEACAM1-3-Fc complementary DNA (cDNA) used a set of sense and antisense primers containing the appropriate single, double, or triple mutations (Table 1) as detailed35 and illustrated in Figure1. With CEACAM1-3-Fc as template, the first polymerase chain reaction (PCR) used either a sense primer (primer 1: 5′ GAGAACCCACTGCTTAACTGG 3′) specific to the πH3M vector plus an antisense primer that contained the appropriate mismatched base(s) (Table 1) or a sense primer containing appropriate mismatched base(s) (Table 1) plus an antisense primer (primer 2: 5′ CTGATCCGGAGAATTCCTTACCTGT AGTGACTATGATCGTCTT GATGT 3′) to the end of the B domain, to create 2 PCR products that overlapped within the region spanned by the mutagenized sense and antisense primers. The reaction was carried out at 94°C for 10 minutes, followed by 25 cycles of 94°C for 1 minute, 47°C for 1 minute, 72°C for 2 minutes, with a final extension period of 72°C for 10 minutes. Aliquots (5 μL) of each PCR product were annealed at 94°C for 1 minute and cooled to 37°C at 1°C for 1 minute before the addition of 40 μL PCR mix containing primers 1 and 2 only. PCR was carried out at 72°C for 2 minutes, followed by 30 cycles of 94°C for 1 minute, 50°C for 1 minute, and 72°C for 1 minute with a 5-minute extension step at 72°C. The PCR products were digested with HindIII and EcoR1 and cloned into the pIg vector.7Using the Quickchange Mutagenesis Kit (Stratagene Europe, Amsterdam, The Netherlands), site-directed mutagenesis was carried out to generate CEACAM1-3-Fc mutants R64A, D82A, R38A, D40A, R64D, and D82R (Figure 1and Table 1). After confirming mutated clones by automatic sequencing, each cDNA mutant was transfected into Cos-1 cells and the soluble proteins isolated on protein A-Sepharose as above.7
CEACAM1 primers for site-directed mutagenesis
| Name . | Sequence 5′…3′ . |
|---|---|
| BGPAMPs | CAUCAUCAUCAUAAGCTTATGGGGCACCTC |
| NTMas | GCCATTTTCTTGGGGCAGCTCCGGGTATAC |
| NTMs | GTATACCCGGAGCTGCCCCAAGAAAATGGC |
| Trans-cyt-as | CUACUACUACUAAGACTATGAAGTTGGTTG |
| MutS32s | CTTTTTGGCTACGCCTGGTACAAAGG |
| MutS32as | CCTTTGTACCAGGCGTAGCCAAAAAG |
| MutY34s | TGGCTACAGCTGGGCCAAAGGGGAAAG |
| MutY34as | CTTTCCCCTTTGGCCAGCTGTAGCCA |
| MutV39s | GGAAAGAGCGGATGGCAA |
| MutV39as | TTGCCATCCGCTCTTTCC |
| MutQ44s | GGATGGCAACCGTGCAATTGTAGGATATGC |
| MutQ44as | GCATATCCTACAATTGCACGGTTGCCATCC |
| MutT87s | GGATTCTACGCCCTACAAGTC |
| MutT87as | GACTTGTAGGGCGTAGAATCC |
| MutQ89s | GGATTCTACACCCTAGCAGTCATAAAGTCAG |
| MutQ89as | CTGACTTTATGACTGCTAGGGCGTAGAATCC |
| MutI91s | CACCCTACAAGTCGCAAAGTCAGATCTTG |
| MutI91as | CAAGATCTGACTTTGCGACTTGTAGGGTG |
| MutV96s | GATCTTGTGAATGCAGAAGCAACTG |
| MutV96as | CAGTTGCTTCTGCATTCACAAGATC |
| MutV96E98s | GTCAGATCTTGCGAATGCAGAAGCAACTG |
| MutV96E98as | CAGTTGCTTCTGCATTCGCAAGATCTGAC |
| MutS32Y34V39s | GCAACTTTTTGGCTACGCCTGGGCCAAAGGGGAAAGAGCG |
| MutS32Y34V39as | CGCTCTTTCCCCTTTGGCCCAGGCGTAGCCAAAAAGTTGC |
| MutT87Q89I91s | CACAGGATTCTACGCCCTAGCAGTCGCAAAGTCAG |
| MutT87Q89I91as | CTGACTTTGCGACTGCTAGGGCGTAGAATCCTGTG |
| MutL18L20s | GAGGGGAAGGAGGTTGCTCTCGCTGTCCACAATCTGCCCC |
| MutL18L20as | GGGGCAGATTGTGGACAGCGAGAGCAACCTCCTTCCCCTC |
| MutS72L74s | CAATATACCCCAATGCAGCCCTGGCGATCCAGAACGTCACCC |
| MutS72L74as | GGGTGACGTTCTGGATCGCCAGGGCTGCATTGGGGTATATTG |
| IIH3Ms | GAGAACCCACTGCTTAACTGG |
| BGPNA1B1as | CTGATCCGGAGAATTCCTTACCTGTAGTGACTATGATCGTCTTGACTGT |
| MutR38As | GGTACAAAGGGGAAGCAGTGGATGGCAACCG |
| MutR38Aas | CGGTTGCCATCCACTGCTTCCCCTTTGTACC |
| MutD40As | GGGGAAAGAGTGGCCGGCAACCGTCAA |
| MutD40Aas | TTGACGGTTGCCGGCCACTCTTTCCCC |
| MutR64Ds | CCGCAAACAGCGGTGACGAGACAATATACCC |
| MutR64Das | GGGTATATTGTCTC GTCACCGCTGTTTGCGG |
| MutD82As | GAACGTCACCCAGAATGCCACAGGATTCTACAC |
| MutD82Aas | GTGTAGAATCCTGTGGCATTCTGGGTGACGTTC |
| MutD82Rs | GAACGTCACCCAGAATCGCACAGGATTCTACAC |
| MutD82Ras | GTGTAGAATCCTGTGCGATTCTGGGTGACGTTC |
| Name . | Sequence 5′…3′ . |
|---|---|
| BGPAMPs | CAUCAUCAUCAUAAGCTTATGGGGCACCTC |
| NTMas | GCCATTTTCTTGGGGCAGCTCCGGGTATAC |
| NTMs | GTATACCCGGAGCTGCCCCAAGAAAATGGC |
| Trans-cyt-as | CUACUACUACUAAGACTATGAAGTTGGTTG |
| MutS32s | CTTTTTGGCTACGCCTGGTACAAAGG |
| MutS32as | CCTTTGTACCAGGCGTAGCCAAAAAG |
| MutY34s | TGGCTACAGCTGGGCCAAAGGGGAAAG |
| MutY34as | CTTTCCCCTTTGGCCAGCTGTAGCCA |
| MutV39s | GGAAAGAGCGGATGGCAA |
| MutV39as | TTGCCATCCGCTCTTTCC |
| MutQ44s | GGATGGCAACCGTGCAATTGTAGGATATGC |
| MutQ44as | GCATATCCTACAATTGCACGGTTGCCATCC |
| MutT87s | GGATTCTACGCCCTACAAGTC |
| MutT87as | GACTTGTAGGGCGTAGAATCC |
| MutQ89s | GGATTCTACACCCTAGCAGTCATAAAGTCAG |
| MutQ89as | CTGACTTTATGACTGCTAGGGCGTAGAATCC |
| MutI91s | CACCCTACAAGTCGCAAAGTCAGATCTTG |
| MutI91as | CAAGATCTGACTTTGCGACTTGTAGGGTG |
| MutV96s | GATCTTGTGAATGCAGAAGCAACTG |
| MutV96as | CAGTTGCTTCTGCATTCACAAGATC |
| MutV96E98s | GTCAGATCTTGCGAATGCAGAAGCAACTG |
| MutV96E98as | CAGTTGCTTCTGCATTCGCAAGATCTGAC |
| MutS32Y34V39s | GCAACTTTTTGGCTACGCCTGGGCCAAAGGGGAAAGAGCG |
| MutS32Y34V39as | CGCTCTTTCCCCTTTGGCCCAGGCGTAGCCAAAAAGTTGC |
| MutT87Q89I91s | CACAGGATTCTACGCCCTAGCAGTCGCAAAGTCAG |
| MutT87Q89I91as | CTGACTTTGCGACTGCTAGGGCGTAGAATCCTGTG |
| MutL18L20s | GAGGGGAAGGAGGTTGCTCTCGCTGTCCACAATCTGCCCC |
| MutL18L20as | GGGGCAGATTGTGGACAGCGAGAGCAACCTCCTTCCCCTC |
| MutS72L74s | CAATATACCCCAATGCAGCCCTGGCGATCCAGAACGTCACCC |
| MutS72L74as | GGGTGACGTTCTGGATCGCCAGGGCTGCATTGGGGTATATTG |
| IIH3Ms | GAGAACCCACTGCTTAACTGG |
| BGPNA1B1as | CTGATCCGGAGAATTCCTTACCTGTAGTGACTATGATCGTCTTGACTGT |
| MutR38As | GGTACAAAGGGGAAGCAGTGGATGGCAACCG |
| MutR38Aas | CGGTTGCCATCCACTGCTTCCCCTTTGTACC |
| MutD40As | GGGGAAAGAGTGGCCGGCAACCGTCAA |
| MutD40Aas | TTGACGGTTGCCGGCCACTCTTTCCCC |
| MutR64Ds | CCGCAAACAGCGGTGACGAGACAATATACCC |
| MutR64Das | GGGTATATTGTCTC GTCACCGCTGTTTGCGG |
| MutD82As | GAACGTCACCCAGAATGCCACAGGATTCTACAC |
| MutD82Aas | GTGTAGAATCCTGTGGCATTCTGGGTGACGTTC |
| MutD82Rs | GAACGTCACCCAGAATCGCACAGGATTCTACAC |
| MutD82Ras | GTGTAGAATCCTGTGCGATTCTGGGTGACGTTC |
s indicates forward primer; as, reverse primer.
Amino acid alignment of the N-terminal CEACAM1 IgV domain.
(A) Alignment of the N-domain of CEACAM1 with other members of the CEA family. Interface 1 and 2: # symbols indicate the position of mutated amino acids in CEACAM1 that affect YTH71.3.2 Mab or Opa protein binding, respectively. Peptide sequences that regulate the expression of CD11/CD18 and L-selectin levels on neutrophils are both underlined in pink and the residues are shown in green or pink. Interface 3 and 4: # symbols indicate the position of mutated amino acids in CEA or CEACAM8, respectively, that affect Opa protein binding in reference 52. The amino acid sequence that is critical for mouse CEACAM1 binding to murine corona viruses is indicated in brown and underlined. Amino acids in CEACAM1 conserved in other family members are indicated in bold type (100% identity); gaps are denoted by dashes (–). Strand indicates secondary structure elements and β strand positions are underlined and marked under the sequence and lettered A to G. (B) Alignment of the N-domain of CEACAM1 with human (H) and rat (R) CD2 and human CD58. Interface 5: Red # symbols represent the position of mutants that abrogate CEACAM1 homophilic interactions. Blue # symbols represent the position of mutants that abrogate anti-CEACAM Mab binding. For the CEACAM1 [H] sequence, residues in red are those mutated in this manuscript. Interface 6 and 7: # symbols indicate known amino acid residues at the interface in human CD2 and CD58, respectively. These are based on x-ray crystallographic coordinates for human and rat CD2 and human CD58.25-27 Mutated human CEACAM1 residues are indicated by arrows, with each amino acid being substituted with alanine (A) either as a single, double, or triple mutant or with arginine (R) for D82 or aspartic acid (D) for R64 as indicated in Figures 3, 6, and 7 and Table 1.
Amino acid alignment of the N-terminal CEACAM1 IgV domain.
(A) Alignment of the N-domain of CEACAM1 with other members of the CEA family. Interface 1 and 2: # symbols indicate the position of mutated amino acids in CEACAM1 that affect YTH71.3.2 Mab or Opa protein binding, respectively. Peptide sequences that regulate the expression of CD11/CD18 and L-selectin levels on neutrophils are both underlined in pink and the residues are shown in green or pink. Interface 3 and 4: # symbols indicate the position of mutated amino acids in CEA or CEACAM8, respectively, that affect Opa protein binding in reference 52. The amino acid sequence that is critical for mouse CEACAM1 binding to murine corona viruses is indicated in brown and underlined. Amino acids in CEACAM1 conserved in other family members are indicated in bold type (100% identity); gaps are denoted by dashes (–). Strand indicates secondary structure elements and β strand positions are underlined and marked under the sequence and lettered A to G. (B) Alignment of the N-domain of CEACAM1 with human (H) and rat (R) CD2 and human CD58. Interface 5: Red # symbols represent the position of mutants that abrogate CEACAM1 homophilic interactions. Blue # symbols represent the position of mutants that abrogate anti-CEACAM Mab binding. For the CEACAM1 [H] sequence, residues in red are those mutated in this manuscript. Interface 6 and 7: # symbols indicate known amino acid residues at the interface in human CD2 and CD58, respectively. These are based on x-ray crystallographic coordinates for human and rat CD2 and human CD58.25-27 Mutated human CEACAM1 residues are indicated by arrows, with each amino acid being substituted with alanine (A) either as a single, double, or triple mutant or with arginine (R) for D82 or aspartic acid (D) for R64 as indicated in Figures 3, 6, and 7 and Table 1.
Analysis of the conformational integrity of mutated CEACAM1-3-Fc proteins
To determine if any decreases in CHO-CEACAM1-4L adhesion to the mutant CEACAM1-3-Fc proteins were due to a modification in the region of contact between the 2 molecules or to a more drastic change in their tertiary structure, conformational analysis was performed for all the mutated proteins, using conformational-dependent and -independent CD66/CEACAM MAbs as detailed in the epitope mapping section.7
In vitro adhesion assays
Stable CHO CEACAM1-4L, CEACAM1-4S, CEACAM1-1S, or CHO-Neo transfectants were labeled with 1 μg/mL 2′,7′-bis-(2-carboxyethyl)-5-(and -6)-carboxy fluoresceinacetoxymethylester (BCECF-am; Molecular Probes, Eugene, OR) for 30 minutes at 37°C, prior to washing in phosphate-buffered saline (PBS)-0.2% (wt/vol) bovine serum albumin (BSA) or Puck saline (Gibco BRL, Paisley, Scotland)-0.2% (wt/vol) BSA and 5 to 10 × 104 added to Immulon 3 microtiter plates (Dynex Technologies, Chantilly, VA) precoated with 1 μg/well purified goat antihuman Fc antibody (Sigma Chemical) and 1 μg/well of the appropriate soluble recombinant protein for 60 minutes at 37°C. BCECF-am fluorescence in each well was read on the Cytofluor II plate reader (Perseptive Biosystems, Hertford, United Kingdom) at an excitation wavelength of 485/20 nm, a gain of 70 and an emission wavelength of 530/30 nm. The plates were washed in PBS-0.2% (wt/vol) BSA and the percentage of cells adhering to the constructs estimated from the subsequent fluorescence determinations. Adhesion assays were carried out with 4 to 6 replicates on 2 to 7 independent occasions.
Molecular modeling of the human CEACAM1 N-terminal domain
The sequence from the N-terminal domain of CEACAM1 was run against a database of sequences from x-ray and NMR structures, using the program BLASTP.36 This identified the best templates for model building. Five Ig variable domains, human CD4,37Bence-Jones VL dimer REI,38 rat CD2,39 human CD2,40 and human CD58,40 were then superimposed and used as a sequence and structural template. The superimposition was entirely automatic and not biased to any particular region. It was performed using Multisup (Dr P.A. Bates, ICRF, London, United Kingdom), a program based on a pairwise superposition alogorithm.41 The CEACAM1 sequence, plus other members of the family, were then automatically aligned to the structural template using a local program Mseq (Dr P.A. Bates, ICRF, London, United Kingdom). Due to the low identity between the CEACAM family and the templates, average 10%, sections of the alignment were manually adjusted to conserve buried hydrophobics and known features of the variable Ig fold. Secondary structure selection, loop building, and side chain replacements were done automatically using 3D-jigsaw,42 (Dr P.A. Bates, ICRF, London, United Kingdom) a program that optimizes fragment and side chain conformations.
Results
The human CEACAM-4L and CEACAM-4S isoforms both mediate homophilic adhesion
The CEACAM1-4L and CEACAM1-4S isoforms share identical extracellular domains and transmembrane sequences, but their cytoplasmic tails are composed of 73 or 9 amino acids, respectively. Previous studies indicate that rat CEACAM1 and human CEACAM1-4L, CEACAM1-3L, CEACAM1-4S, and CEACAM1-1L transfectants adhere homophilically.5,6,22,23,30 43-45 We have examined the ability of human CEACAM1-4L, CEACAM1-4S, and CEACAM1-1S transfectants to adhere directly to immobilized recombinant proteins carrying the entire extracellular domain CEACAM1-4L/S. Our results show that, despite the higher level of CEACAM1-1S expression on CHO-CEACAM1-1S transfectants, only the CEACAM1-4L and CEACAM1-4S transfectants were able to bind significantly (30%-62% of the input cells added) to immobilized CEACAM1-4-Fc molecules (Figure2). Low to negligible levels of adhesion were observed when CHO-CEACAM1-1S cells, possessing the N-terminal, transmembrane, and short cytoplasmic domains only, were examined for their ability to adhere to this same CEACAM1-4-Fc protein (Figure 2). This suggested that the N terminal domain in such transfectants was not as readily accessible for binding to recombinant soluble CEACAM1-4-Fc proteins or that, without the IgC2 domains or long cytoplasmic tail, the avidity of adhesion was decreased and could not be maintained in this type of receptor/ligand binding assay.
CHO-CEACAM1-4L transfectants adhere preferentially to immobilized recombinant human CEACAM1-3-Fc and CEACAM1-4-Fc domain deletion variants.
The 96-well Immulon 3 plates were coated with goat antihuman Fc prior to the addition of the CEACAM1-1-Fc, CEACAM1-3-Fc, and CEACAM1-4-Fc soluble proteins. CD31(D1-3)-Fc, MUC 18(D1-5)-Fc CD14-Fc, and NCAM(D1-7)-Fc were used as negative control constructs. CHO transfectants were labeled with the fluorescent tag, BCECF-am, and allowed to adhere to these soluble constructs for 60 minutes at 37°C. The total fluorescence of each well was then determined using the Cytofluor II fluorescence plate reader. The plates were then washed and the number of cells adhering determined by fluorescence estimations in the Cytofluor II as a percentage of the total cells added per well.
CHO-CEACAM1-4L transfectants adhere preferentially to immobilized recombinant human CEACAM1-3-Fc and CEACAM1-4-Fc domain deletion variants.
The 96-well Immulon 3 plates were coated with goat antihuman Fc prior to the addition of the CEACAM1-1-Fc, CEACAM1-3-Fc, and CEACAM1-4-Fc soluble proteins. CD31(D1-3)-Fc, MUC 18(D1-5)-Fc CD14-Fc, and NCAM(D1-7)-Fc were used as negative control constructs. CHO transfectants were labeled with the fluorescent tag, BCECF-am, and allowed to adhere to these soluble constructs for 60 minutes at 37°C. The total fluorescence of each well was then determined using the Cytofluor II fluorescence plate reader. The plates were then washed and the number of cells adhering determined by fluorescence estimations in the Cytofluor II as a percentage of the total cells added per well.
The N-terminal domain alone does not mediate strong homophilic interactions
To investigate whether CHO-CEACAM1-4L transfectants were able to adhere to different extracellular domains of CEACAM1-4L/S in the absence of a cellular background, we constructed soluble recombinant domain deletion variants of CEACAM1-4L/S containing the N-terminal domain (CEACAM1-1-Fc) or the N-terminal domain linked to the A1B (CEACAM1-3-Fc) or A1BA2 (CEACAM1-4-Fc) domains, which mimicked the extracellular domains of the CEACAM1-1L/S, CEACAM1-3L/S, and CEACAM1-4L/S isoforms, respectively. Figure 2 shows the mean ± SD of 7 independent experiments in which immobilized CEACAM1-1-Fc bound weakly to CHO-CEACAM1-4L transfectants, in contrast to the much stronger adhesion to immobilized CEACAM1-3-Fc and CEACAM1-4-Fc proteins. An average of 26% ± 9% of the CHO-CEACAM1-4L cells adhered to the CEACAM1-1-Fc construct compared to 52% ± 7% and 56% ± 5% to the CEACAM1-3-Fc and CEACAM1-4-Fc proteins, respectively. For the CHO-CEACAM1-4S transfectants, slightly higher levels of adhesion were obtained in the presence as opposed to the absence of the A2 domain. However, binding of these cells to the CEACAM1-1-Fc protein was similar to that observed using irrelevant CD31(D1-3)-Fc and CD14-Fc controls (Figure 2). For the CHO-CEACAM1-1S transfectants, essentially no binding to the CEACAM1-1-Fc– or CEACAM1-3-Fc–immobilized proteins above background was detected, whereas adhesion to the CEACAM1-4-Fc construct was low to negligible (Figure 3). Because the CEA, CEACAM3, CEACAM6, and CEACAM8 molecules have been shown previously30 46-48 to interact heterophilically when expressed in cell lines, we examined the adhesion of HeLa-CEA, -CEACAM3L, and -CEACAM6 to immobilized recombinant forms of CEACAM1-4-Fc (Figure 2). Only weak adhesion (1.7- to 2.1-fold higher than for CD14-Fc) occurred despite the fact that these molecules share 89% to 91% amino acid identity in their N-terminal domains (Figure1A). Taken together, our studies show that the A2 domain and long cytoplasmic tail are not essential for this homophilic interaction, although they appear to stabilize or increase avidity of binding.
Molecular model of the N-terminal IgV set domain of human CEACAM1/BGP.
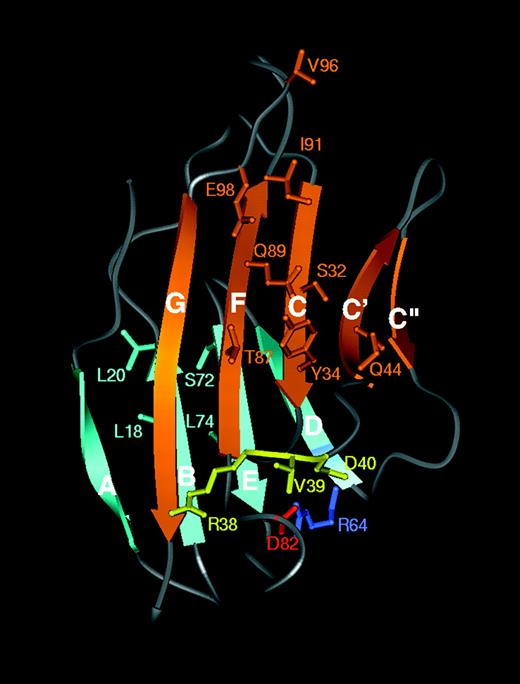
Ribbon diagram of the N-terminal domain of CEACAM1 showing the predicted homophilic interface. The β strands are labeled A to G according to convention. The GFCC′C" face is in gold and the ABED face is in cyan. The mutated amino acids analyzed in the adhesion assays are indicated on the model according to the one-letter amino acid code. Particularly noticeable are the amino acids in CC′ and FG loops, which protrude from the IgV domain. The conserved intradomain salt bridge R64 (light blue) and D82 (red) can be clearly seen to link the base of the D and F strands.
Molecular model of the N-terminal IgV set domain of human CEACAM1/BGP.
Ribbon diagram of the N-terminal domain of CEACAM1 showing the predicted homophilic interface. The β strands are labeled A to G according to convention. The GFCC′C" face is in gold and the ABED face is in cyan. The mutated amino acids analyzed in the adhesion assays are indicated on the model according to the one-letter amino acid code. Particularly noticeable are the amino acids in CC′ and FG loops, which protrude from the IgV domain. The conserved intradomain salt bridge R64 (light blue) and D82 (red) can be clearly seen to link the base of the D and F strands.
Definition of key amino acid residues on the CFG face of the CEACAM1 N-terminal domain involved in homophilic adhesion
It has been demonstrated that (1) rat CEACAM1 lacking the N-terminal domain but containing the A1B1A2 domains failed to mediate adhesion,20,21 and (2) mutagenesis of a single amino acid within the GPAYSGRET N-domain sequence of rat CEACAM1 and corresponding to R64 in human CEACAM1 prevented aggregation of Sf9 transfectants.19 This mutation would be predicted to destabilize the tertiary structure of the N-terminal domain of rat CEACAM1 by preventing intrastrand salt bridge formation between the base of β strands D and F at residues R64 and D82.49 To determine the importance of these and other amino acid residues from the N-terminal domain of human CEACAM1 in homophilic adhesion and because high levels of homophilic adhesion had been achieved between the CEACAM1-3-Fc construct and the CHO-CEACAM1-4L transfectants, we subjected specific amino acid residues within the N-terminal domain of CEACAM1-3-Fc to site-directed mutagenesis. The primary amino acid sequences of this N-terminal domain of CEACAM1 were aligned with those of other human CEACAM family members (Figure 1A) and with human and rat CD2 and human CD58 (Figure 1B). This alignment was based on x-ray crystallographic coordinates for CD2 and CD58.25 From the molecular model for the N-domain of CEACAM1 (Figure 3), we also predicted which amino acid residues in the N-terminal domain of CEACAM1 contributed to the different β strands of the Ig structure. Figure 3shows the predicted arrangement of 9 β strands arranged on 2 faces, GFCC′C" and ABED. Initially, 9 specific mutations in residues predicted to be solvent accessible and exposed at the surface of CEACAM1 were targeted onto the GFCC′C" face and 4 on the opposite ABED face. Two mutations at residues R38 and D40 adjacent to V39 on the GFCC′C" face were also made. The amino acid residues selected were all substituted with alanine (A) residues. The single salt bridge across the GFCC′C" interface is predicted to involve residues R43 and E98. To disrupt this, the E98A mutation was made. In addition, residues R64 and D82 are predicted to form an intrafold salt bridge between the GFCC′C" and ABED faces, but this does not occur on the GFCC′C" face. These were mutated individually to alanine residues (R64A, D82A) or to R64D (aspartic acid), or D82R (arginine) to disrupt this salt bridge. To regain function, the salt bridge within the R64D mutant was restored but in the opposite amino acid orientation by introducing a second mutation at D82R (Figure 1B).
Identification of CEACAM1 N-domain reactive Mabs
To determine which CD66/CEACAM MAbs recognize the CEACAM family members, we analyzed a set of MAbs on CEACAM1-4L, CEACAM1-4S, CEACAM1-1S, CEA, CEACAM3L, CEACAM6, and CEACAM8 CHO or HeLa transfectants and on different CEACAM1 soluble isoforms. Two of these MAbs, 26H7 and 5F4, appeared to be specific for CEACAM1. The other MAbs reacted variably with different CEACAM molecules (Table2). The 26H7, 5F4, 12-140-4, 4/3/17, COL-4, YG-C28F2, D14HD11, 34B1, B18.7.7, D11-AD11, HEA 81, CLB-gran-10, F34-187, T84.1, B6.2, and B1.1 MAbs recognize the N-terminal domain of CEACAM1. The F36-54, YG-C94G7, 12-140-5 and TET-2 MAbs react with both the CEACAM1-3-Fc and CEACAM1-4-Fc constructs, but not the CEACAM1-1-Fc protein (Table 2).
Epitope mapping of CD66/CEACAM Mabs
| Mab . | δ N (CEACAM6 1-59 aa)* . | N (CEACAM6 1-122 aa)* . | CEA CEACAM1,3,6,8* . | CEACAM1 N-domain binding . | CEACAM1-1-Fc . | CEACAM1-3-Fc . | CEACAM1-4-Fc . | NCAM(D1-5)-Fc/ Muc 18-Fc† . |
|---|---|---|---|---|---|---|---|---|
| 26H7 | − | − | CEACAM1 | + | 1.498 ± 0.127 | 1.672 ± 0.068 | 1.659 ± 0.167 | 0.083 ± 0.003 |
| 5F4 | − | − | CEACAM1 | + | 1.774 ± 0.024 | 1.933 ± 0.090 | 1.931 ± 0.044 | 0.082 ± 0.001 |
| TEC-11 | − | − | CEACAM1 | − | 0.205 ± 0.005 | 0.188 ± 0.004 | 1.298 ± 0.121 | 0.191 ± 0.004† |
| 12-140-4‡ | nd | nd | CEACAM1 CEA | + | 1.560 ± 0.087 | 1.628 ± 0.253 | 1.731 ± 0.085 | 0.257 ± 0.018 |
| 4/3/17‡,2-153 | − | + | CEACAM1 CEA | + | 2.118 ± 0.106 | 2.178 ± 0.155 | 2.444 ± 0.101 | 0.416 ± 0.066† |
| b 7.8.5 | − | − | CEA | − | nd | 0.268 ± 0.019 | 0.267 ± 0.034 | 0.252 ± 0.007 |
| YG-C35D6 | − | − | CEACAM6 CEA | − | 0.360 ± 0.033 | 0.339 ± 0.040 | 0.356 ± 0.030 | 0.284 ± 0.010 |
| COL-4‡ | nd | nd | CEACAM1,3 CEA | + | 2.396 ± 0.60 | 2.319 ± 0.023 | 2.421 ± 0.122 | 0.310 ± 0.006 |
| F36-54 | − | − | CEACAM1,6 CEA | − | 0.187 ± 0.011 | 0.834 ± 0.031 | 0.999 ± 0.279 | 0.178 ± 0.016 |
| 34B1 | nd | nd | CEACAM1,3,6 CEA | + | 1.364 ± 0.117 | 1.507 ± 0.050 | 1.460 ± 0.089 | 0.800 ± 0.005 |
| YG-C28F2 | nd | nd | CEACAM1,3,6 CEA | + | 1.952 ± 0.275 | 1.872 ± 0.140 | 2.035 ± 0.164 | 0.271 ± 0.004 |
| D14HD11‡ | + | + | CEACAM1,3,6 CEA | + | 2.248 ± 0.168 | 2.175 ± 0.144 | 2.294 ± 0.163 | 0.221 ± 0.008 |
| b18.7.7‡ | + | + | CEACAM1,3,6 CEA | + | 1.795 ± 0.277 | 2.073 ± 0.346 | 1.822 ± 0.147 | 0.201 ± 0.058 |
| D11-AD11‡ | + | + | CEACAM1,3,6 CEA | + | 2.248 ± 0.168 | 2.175 ± 0.144 | 2.294 ± 0.163 | 0.221 ± 0.008 |
| HEA 81‡ | + | + | CEACAM1,3,6 CEA | + | 0.505 ± 0.120 | 0.577 ± 0.083 | 0.379 ± 0.062 | 0.206 ± 0.005† |
| B1.12-153 | − | + | CEACAM1,3,6 CEA | + | 0.870 ± 0.046 | 0.890 ± 0.126 | 1.051 ± 0.125 | 0.285 ± 0.002† |
| CLB-gran-102-155 | − | + | CEACAM1,3,6 CEA | + | 1.946 ± 0.389 | 2.071 ± 0.055 | 1.944 ± 0.086 | 0.206 ± 0.017 |
| F34-187‡,2-153 | − | + | CEACAM1,3,6 CEA | + | 2.019 ± 0.284 | 2.189 ± 0.148 | 2.135 ± 0.263 | 0.205 ± 0.006 |
| T84.1‡,2-155 | − | + | CEACAM1,3,6 CEA | + | 1.572 ± 0.138 | 1.701 ± 0.238 | 1.488 ± 0.317 | 0.235 ± 0.025 |
| B6.2 | nd | nd | CEACAM1,3,6 CEA | + | 1.913 ± 0.469 | 1.972 ± 0.335 | 2.325 ± 0.246 | 0.365 ± 0.039 |
| B1.13 | nd | nd | CEACAM1,3,6 CEA | + | 1.660 ± 0.050 | 1.698 ± 0.213 | 1.808 ± 0.194 | 0.253 ± 0.018 |
| YG-C94G7 | − | − | CEACAM1,6,8 CEA | − | 0.342 ± 0.037 | 1.469 ± 0.227 | 1.458 ± 0.062 | 0.279 ± 0.023 |
| 12-140-5 | − | − | CEACAM1,6,8 CEA | − | 0.444 ± 0.055 | 2.432 ± 0.033 | 2.357 ± 0.062 | 0.475 ± 0.051 |
| TET-2 | − | − | CEACAM1,6,8 CEA | − | 0.327 ± 0.023 | 2.041 ± 0.061 | 2.198 ± 0.262 | 0.289 ± 0.023 |
| mIgG1 | − | − | − | − | 0.147 ± 0.003 | 0.147 ± 0.003 | 0.150 ± 0.008 | 0.145 ± 0.005† |
| mIgG2a | − | − | − | − | 0.142 ± 0.002 | 0.135 ± 0.002 | 0.200 ± 0.010 | 0.206 ± 0.017† |
| mIgG2b | − | − | − | − | 0.199 ± 0.010 | 0.220 ± 0.019 | 0.144 ± 0.006 | 0.145 ± 0.011† |
| mIgG1 | − | − | − | − | 0.196 ± 0.016 | 0.223 ± 0.037 | 0.206 ± 0.008 | 0.184 ± 0.020 |
| mIgG2a | − | − | − | − | 0.188 ± 0.029 | 0.196 ± 0.004 | 0.201 ± 0.057 | 0.170 ± 0.009 |
| mIgG2b | − | − | − | − | 0.265 ± 0.111 | 0.156 ± 0.014 | 0.168 ± 0.012 | 0.166 ± 0.021 |
| mIgM | − | − | − | − | 0.171 ± 0.026 | 0.193 ± 0.026 | 0.201 ± 0.049 | 0.178 ± 0.028 |
| Mab . | δ N (CEACAM6 1-59 aa)* . | N (CEACAM6 1-122 aa)* . | CEA CEACAM1,3,6,8* . | CEACAM1 N-domain binding . | CEACAM1-1-Fc . | CEACAM1-3-Fc . | CEACAM1-4-Fc . | NCAM(D1-5)-Fc/ Muc 18-Fc† . |
|---|---|---|---|---|---|---|---|---|
| 26H7 | − | − | CEACAM1 | + | 1.498 ± 0.127 | 1.672 ± 0.068 | 1.659 ± 0.167 | 0.083 ± 0.003 |
| 5F4 | − | − | CEACAM1 | + | 1.774 ± 0.024 | 1.933 ± 0.090 | 1.931 ± 0.044 | 0.082 ± 0.001 |
| TEC-11 | − | − | CEACAM1 | − | 0.205 ± 0.005 | 0.188 ± 0.004 | 1.298 ± 0.121 | 0.191 ± 0.004† |
| 12-140-4‡ | nd | nd | CEACAM1 CEA | + | 1.560 ± 0.087 | 1.628 ± 0.253 | 1.731 ± 0.085 | 0.257 ± 0.018 |
| 4/3/17‡,2-153 | − | + | CEACAM1 CEA | + | 2.118 ± 0.106 | 2.178 ± 0.155 | 2.444 ± 0.101 | 0.416 ± 0.066† |
| b 7.8.5 | − | − | CEA | − | nd | 0.268 ± 0.019 | 0.267 ± 0.034 | 0.252 ± 0.007 |
| YG-C35D6 | − | − | CEACAM6 CEA | − | 0.360 ± 0.033 | 0.339 ± 0.040 | 0.356 ± 0.030 | 0.284 ± 0.010 |
| COL-4‡ | nd | nd | CEACAM1,3 CEA | + | 2.396 ± 0.60 | 2.319 ± 0.023 | 2.421 ± 0.122 | 0.310 ± 0.006 |
| F36-54 | − | − | CEACAM1,6 CEA | − | 0.187 ± 0.011 | 0.834 ± 0.031 | 0.999 ± 0.279 | 0.178 ± 0.016 |
| 34B1 | nd | nd | CEACAM1,3,6 CEA | + | 1.364 ± 0.117 | 1.507 ± 0.050 | 1.460 ± 0.089 | 0.800 ± 0.005 |
| YG-C28F2 | nd | nd | CEACAM1,3,6 CEA | + | 1.952 ± 0.275 | 1.872 ± 0.140 | 2.035 ± 0.164 | 0.271 ± 0.004 |
| D14HD11‡ | + | + | CEACAM1,3,6 CEA | + | 2.248 ± 0.168 | 2.175 ± 0.144 | 2.294 ± 0.163 | 0.221 ± 0.008 |
| b18.7.7‡ | + | + | CEACAM1,3,6 CEA | + | 1.795 ± 0.277 | 2.073 ± 0.346 | 1.822 ± 0.147 | 0.201 ± 0.058 |
| D11-AD11‡ | + | + | CEACAM1,3,6 CEA | + | 2.248 ± 0.168 | 2.175 ± 0.144 | 2.294 ± 0.163 | 0.221 ± 0.008 |
| HEA 81‡ | + | + | CEACAM1,3,6 CEA | + | 0.505 ± 0.120 | 0.577 ± 0.083 | 0.379 ± 0.062 | 0.206 ± 0.005† |
| B1.12-153 | − | + | CEACAM1,3,6 CEA | + | 0.870 ± 0.046 | 0.890 ± 0.126 | 1.051 ± 0.125 | 0.285 ± 0.002† |
| CLB-gran-102-155 | − | + | CEACAM1,3,6 CEA | + | 1.946 ± 0.389 | 2.071 ± 0.055 | 1.944 ± 0.086 | 0.206 ± 0.017 |
| F34-187‡,2-153 | − | + | CEACAM1,3,6 CEA | + | 2.019 ± 0.284 | 2.189 ± 0.148 | 2.135 ± 0.263 | 0.205 ± 0.006 |
| T84.1‡,2-155 | − | + | CEACAM1,3,6 CEA | + | 1.572 ± 0.138 | 1.701 ± 0.238 | 1.488 ± 0.317 | 0.235 ± 0.025 |
| B6.2 | nd | nd | CEACAM1,3,6 CEA | + | 1.913 ± 0.469 | 1.972 ± 0.335 | 2.325 ± 0.246 | 0.365 ± 0.039 |
| B1.13 | nd | nd | CEACAM1,3,6 CEA | + | 1.660 ± 0.050 | 1.698 ± 0.213 | 1.808 ± 0.194 | 0.253 ± 0.018 |
| YG-C94G7 | − | − | CEACAM1,6,8 CEA | − | 0.342 ± 0.037 | 1.469 ± 0.227 | 1.458 ± 0.062 | 0.279 ± 0.023 |
| 12-140-5 | − | − | CEACAM1,6,8 CEA | − | 0.444 ± 0.055 | 2.432 ± 0.033 | 2.357 ± 0.062 | 0.475 ± 0.051 |
| TET-2 | − | − | CEACAM1,6,8 CEA | − | 0.327 ± 0.023 | 2.041 ± 0.061 | 2.198 ± 0.262 | 0.289 ± 0.023 |
| mIgG1 | − | − | − | − | 0.147 ± 0.003 | 0.147 ± 0.003 | 0.150 ± 0.008 | 0.145 ± 0.005† |
| mIgG2a | − | − | − | − | 0.142 ± 0.002 | 0.135 ± 0.002 | 0.200 ± 0.010 | 0.206 ± 0.017† |
| mIgG2b | − | − | − | − | 0.199 ± 0.010 | 0.220 ± 0.019 | 0.144 ± 0.006 | 0.145 ± 0.011† |
| mIgG1 | − | − | − | − | 0.196 ± 0.016 | 0.223 ± 0.037 | 0.206 ± 0.008 | 0.184 ± 0.020 |
| mIgG2a | − | − | − | − | 0.188 ± 0.029 | 0.196 ± 0.004 | 0.201 ± 0.057 | 0.170 ± 0.009 |
| mIgG2b | − | − | − | − | 0.265 ± 0.111 | 0.156 ± 0.014 | 0.168 ± 0.012 | 0.166 ± 0.021 |
| mIgM | − | − | − | − | 0.171 ± 0.026 | 0.193 ± 0.026 | 0.201 ± 0.049 | 0.178 ± 0.028 |
+ indicates positive; −, nonreactive or negative; nd, not determined.
Summary from the Boston and Osaka Leucocyte Culture Conferences or after testing by flow cytometry on HeLa-CEA, CEACAM3L, CEACAM6, CEACAM8, and CHO-CEACAM1-4L, -4S and -1S.
Muc 18-Fc used in place of NCAM-Fc as a negative control; N domain covers 1-180 aa of CEACAM1-4L; δ N domain of CEACAM6 covers 1-59 aa; N domain of CEACAM6 covers 1-122 aa.
Mabs to the N-terminal domain that cross-block the binding of 125I-labeled 12-140-4 to CEACAM1 or CEA transfectants indicating similar or closely linked epitopes; other N-domain Mabs were not analyzed in cross-blocking experiments.
Does not cross-block CLB-gran/10.
Cross-blocks CLB-gran/10.
Identification of conformationally dependent CD66/CEACAM MAbs
Our results classify the CD66/CEACAM MAbs analyzed into 3 groups. Only one MAb, F34-187, reacted equally well with the native and denatured forms of the CEACAM1-4-Fc protein. Eight MAbs, CLB-gran-10, T84.1, B18.7.7, D14HD11, HEA 81, B1.1, 34B1, and 4/3/17, reacted with the native form and, to a lesser extent, with the denatured protein. Nine MAbs, YG-C94G7, TET-2, 12-140-5, COL-4, 26H7, 5F4, B6.2, YG-C28F2, and 12-140-4, of which the latter 6 are N-terminal domain reactive, reacted preferentially with the native protein and were conformationally dependent (Figure 4).
The identification of conformational-dependent and -independent MAbs.
Samples (100 μL) of either untreated (native) or SDS/β-mercaptoethanol and boiled (denatured) CEACAM1-4-Fc soluble recombinant proteins at concentrations of 100 ng/100 μL (1) and 10 ng/100 μL (2) were slot blot transferred onto immobilon-PVDF membranes and their reactivities with the CD66/CEACAM MAbs determined as described in “Materials and methods.” NCAM(D1-7)-Fc was used as a negative control and did not bind the CD66/CEACAM MAbs (data not shown). This experiment was repeated at least twice for each MAb.
The identification of conformational-dependent and -independent MAbs.
Samples (100 μL) of either untreated (native) or SDS/β-mercaptoethanol and boiled (denatured) CEACAM1-4-Fc soluble recombinant proteins at concentrations of 100 ng/100 μL (1) and 10 ng/100 μL (2) were slot blot transferred onto immobilon-PVDF membranes and their reactivities with the CD66/CEACAM MAbs determined as described in “Materials and methods.” NCAM(D1-7)-Fc was used as a negative control and did not bind the CD66/CEACAM MAbs (data not shown). This experiment was repeated at least twice for each MAb.
Analysis of the conformational integrity of mutated CEACAM1-3-Fc soluble proteins and identification of sites for CD66/CEACAM1 MAb reactivity
Conformational analysis was carried out on the mutated CEACAM1-3-Fc constructs using these MAbs to establish whether any alterations in their adhesion to CHO-CEACAM1-4L transfectants might result from a major change in their tertiary structure, rather than in the region of contact between the 2 opposing molecules. The majority of MAbs tested reacted with the mutated and native CEACAM1 constucts in a manner similar to that observed for the conformational independent MAb, F34-187 (Figure 5A), suggesting that most single amino acid substitutions did not drastically affect the general tertiary CEACAM1-3-Fc structure. Three of the conformationally dependent MAbs showed major reductions in binding. These were (1) COL-4, which did not react with the I91A or T87AQ89AI91A mutants; (2) 12-140-4, which failed to bind to the Y34A or S32AY34AV39A mutants; and (3) 5F4, which did not bind the Y34A, S32AY34AV39A, or T87AQ89AI91A mutants and showed reduced adhesion to constructs carrying single mutations, T87A, Q89A, and I91A, in the F β strand and FG loop. Most N-domain–specific MAbs showed reduced binding to single R64 and D82 mutants (Figure 5B). This was restored in all cases after introduction of the second D82R mutation into the R64D mutant (R64D D82R; Figure 5B) and reformation of the salt bridge, suggesting that this salt bridge was required for the conformational integrity of CEACAM1.
Binding of conformational-dependent and -independent MAbs to mutated CEACAM1-3-Fc constructs.
MAb binding to (A) the initial CEACAM1-3-Fc mutants and (B) to mutants surrounding the V39 residue and involved in R64 to D82 intrafold salt bridge. Each mutated CEACAM1-3-Fc protein, the unmodified CEACAM1-4-Fc protein, or BSA at concentrations of 1 μg/100 μL were plated in triplicate onto 96-well Immulon 3 plates that had been precoated with goat antihuman Fc and their reactivity with the MAbs indicated on each graph assessed by ELISAs as described in “Materials and methods.” The binding of each MAb to the unmodified CEACAM1-4-Fc (A) or CEACAM1-3-Fc (B) constructs was normalized to 100% and the relative binding of these MAbs to each mutated constructed or to the BSA (Nil) negative control was calculated as a percentage of binding to the unmodified CEACAM1-4-Fc or CEACAM1-3-Fc, respectively. The graphs are from a representative experiment and show means ± SD of 3 to 6 replicate measurements. The reactivity of each construct was analyzed on at least 2 independent occasions and generated similar results.
Binding of conformational-dependent and -independent MAbs to mutated CEACAM1-3-Fc constructs.
MAb binding to (A) the initial CEACAM1-3-Fc mutants and (B) to mutants surrounding the V39 residue and involved in R64 to D82 intrafold salt bridge. Each mutated CEACAM1-3-Fc protein, the unmodified CEACAM1-4-Fc protein, or BSA at concentrations of 1 μg/100 μL were plated in triplicate onto 96-well Immulon 3 plates that had been precoated with goat antihuman Fc and their reactivity with the MAbs indicated on each graph assessed by ELISAs as described in “Materials and methods.” The binding of each MAb to the unmodified CEACAM1-4-Fc (A) or CEACAM1-3-Fc (B) constructs was normalized to 100% and the relative binding of these MAbs to each mutated constructed or to the BSA (Nil) negative control was calculated as a percentage of binding to the unmodified CEACAM1-4-Fc or CEACAM1-3-Fc, respectively. The graphs are from a representative experiment and show means ± SD of 3 to 6 replicate measurements. The reactivity of each construct was analyzed on at least 2 independent occasions and generated similar results.
The GFCC′C" face and CC′ loop of N terminal domain of CEACAM1 are crucial for mediating homophilic adhesion
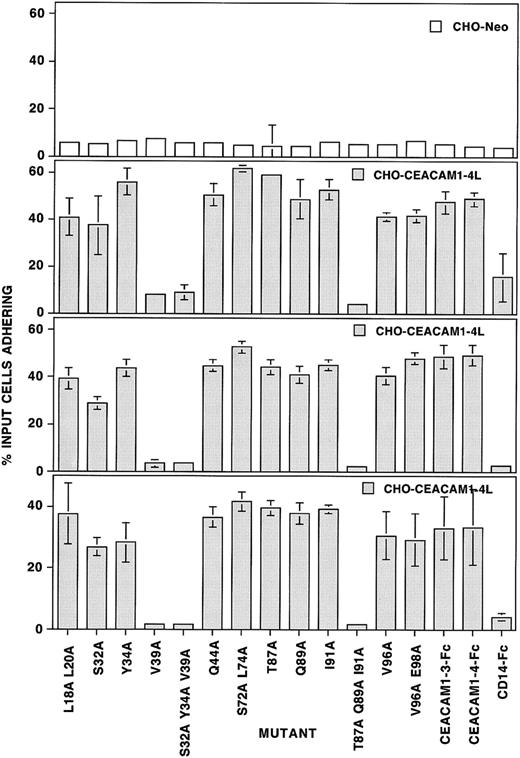
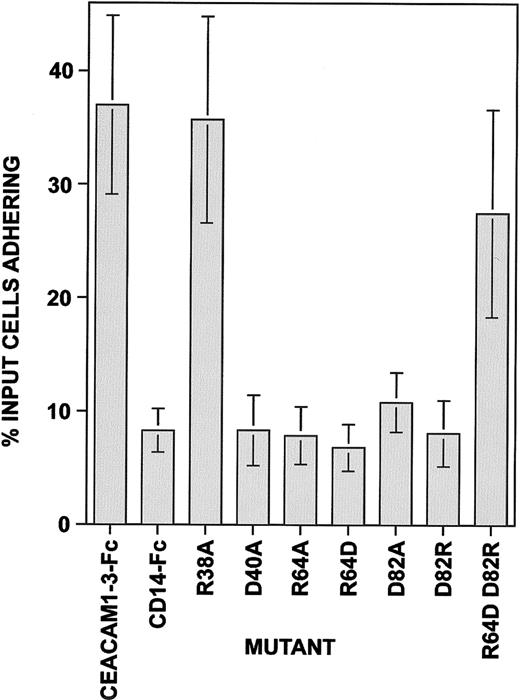
Our results show that mutation of valine 39 to alanine (V39A) on the CC′ loop abrogated the adhesion of CEACAM1-3-Fc to CHO-CEACAM1-4L transfectants (Figure 6). A slight but less significant decrease in adhesion of the transfectants to soluble proteins carrying a mutation at serine 32 (S32A) was also observed. The mutation at tyrosine 34 (Y34A) did not affect adhesion, despite the lack of reactivity of this mutant protein with the 5F4 and 12-140-4 MAbs (Figures 5 and 6). As with the V39 mutation, the triple mutation (S32A Y34A V39A) also ablated adhesion. Further mutation of single amino acids on either side of V39, in particular R38A and D40A, revealed that the latter but not the former mutation, abolished homophilic adhesion (Figure 7). These results indicate that residues V39 and D40 in the CC′ loop affect adhesion more significantly than those tested in the C β strand (S32A, Y34A). Mutation of residues T87, Q89, and I91 to alanine residues in the F β strand had no major effects on adhesion when used alone, again despite the fact that mutation of I91A abrogated 5F4 and COL-4 MAb binding (Figures 5 and 6). However, when all 3 mutations were present on the same molecule, adhesion was dramatically inhibited (Figure 6), even when the concentration of this mutant was doubled (data not shown). This may be due to a more severe change in the domain conformation with all the accessible polar residues of the F β strand substituted by neutral residues. Residues V96 and E98 are located in the FG loop of the N-terminal domain, with E98 predicted to form a salt bridge with R43 across the GFCC′C" interface. Substitution of V96 with alanine either alone or as a double mutant, V96E98, inhibited adhesion slightly, whereas the E98 mutation had no effect. Thus, the predicted salt bridge between residues R43 and E98 is not important for homophilic adhesion. The control ABED face mutants, L18L20 (B β strand) and S72L74 (E β strand) had no overall significant effect on binding. Alignment studies predicted a second intrafold salt bridge between residues R64 and D82 in the N-terminal domain of CEACAM1 at the base of the D and F β strands (Figures 1 and 3). Our results (Figure7) confirm studies by Sippel and coworkers19 that mutation of the equivalent R64 residue in rodent CEACAM1 inhibits homophilic adhesion and they further reveal that single amino acid mutations involving these salt bridge residues (R64A, R64D, D82A, or D82R) abrogate homophilic adhesion of CEACAM1. When this salt bridge is restored, albeit in the opposite amino acid orientation, the gain-of-function mutant is capable of binding to human CEACAM1 homophilically (Figure 7). These results strongly suggest that neither the R43 to E98 salt bridge at the GFCC′C′ interface nor the R64 to D82 intrafold salt bridge is involved directly in the binding site, but that the R64 to D82 salt bridge is required for fold stability and thus indirectly affects the binding site.
The GFCC′C" face of the CEACAM1 N-terminal domain contributes to homophilic adhesion.
The 96-well Immulon 3 plates were coated with goat antihuman Fc prior to the addition of the mutated CEACAM1-3-Fc proteins or of the unmodified CEACAM1-3-Fc and CEACAM1-4-Fc soluble constructs. CD14-Fc was used as the negative control. CHO-CEACAM1-4L transfectants were labeled with the fluorescent tag, BCECF-am, and allowed to adhere to these soluble constructs for 60 minutes at 37°C. The total fluorescence of each well was then determined using the Cytofluor II fluorescence plate reader. The plates were then washed and the number of cells adhering determined by fluorescence estimations in the Cytofluor II as a percentage of the total cells added per well. CHO-Neo cells were used as a negative control for CHO cell binding. Six replicates were used for each construct and the experiments performed on 3 separate occasions. The results show the mean ± SD of 6 replicates for one typical CHO-Neo experiment and for 3 CHO-CEACAM1-4L binding experiments.
The GFCC′C" face of the CEACAM1 N-terminal domain contributes to homophilic adhesion.
The 96-well Immulon 3 plates were coated with goat antihuman Fc prior to the addition of the mutated CEACAM1-3-Fc proteins or of the unmodified CEACAM1-3-Fc and CEACAM1-4-Fc soluble constructs. CD14-Fc was used as the negative control. CHO-CEACAM1-4L transfectants were labeled with the fluorescent tag, BCECF-am, and allowed to adhere to these soluble constructs for 60 minutes at 37°C. The total fluorescence of each well was then determined using the Cytofluor II fluorescence plate reader. The plates were then washed and the number of cells adhering determined by fluorescence estimations in the Cytofluor II as a percentage of the total cells added per well. CHO-Neo cells were used as a negative control for CHO cell binding. Six replicates were used for each construct and the experiments performed on 3 separate occasions. The results show the mean ± SD of 6 replicates for one typical CHO-Neo experiment and for 3 CHO-CEACAM1-4L binding experiments.
Residues in the CC′ loop are directly involved in homophilic adhesion.
The adhesion assay is described in the legend to Figure 6. Six replicates were used for each construct and the experiments performed on 6 separate occasions. The results show the mean ± SD of 6 experiments.
Residues in the CC′ loop are directly involved in homophilic adhesion.
The adhesion assay is described in the legend to Figure 6. Six replicates were used for each construct and the experiments performed on 6 separate occasions. The results show the mean ± SD of 6 experiments.
Discussion
Using an alanine-scanning mutagenesis approach, we have identified key amino acids on the N-terminal IgV domain of human CEACAM1 that are involved in homophilic adhesion. Most notably, the V39 and D40 residues on the CC′ loop play a critical role in this process. Residues closely associated with V39 and D40 and positioned in the molecular model in the lower region of the GFCC′C" face, particularly R38, Y34, Q44, and T87, as well as those at some distance away and positioned at the top of the GFCC′C" face, namely I91, V96 and E98, if mutated individually, and those on the ABED face did not abrogate adhesion.
The importance of the GFCC′C" face of the CEACAM1 N-terminal domain in regulating adhesive interactions and signaling is underscored in several other experimental systems. First, CEACAM1 peptides that activate neutrophils by up-regulating their expression of CD11/CD18 and down-regulating their expression of L-selectin, thereby enhancing adhesion of neutrophils to endothelium, span the first 48 amino acids of the N-terminal domain, an area that encompasses the CC′ loop sequence.50 Heterophilic interactions of the CEACAM1 N-terminal domain with murine corona viruses, H influenzaeand Opa proteins of N meningiditis and N gonorrheae also occur on the GFCC′C" face of CEACAM1.18,50-57 Amino acids between 34 to 52 in the CC′ loop region of the murine CEACAM1 N-terminal domain are crucial for murine corona virus interaction.51 Similarly, amino acids between residues 27 to 42 (particularly the triplet Q27L28F29) and S32, Y34, V39, Q44, Q89, and I91 on the GFCC′C" face of human CEACAM1 form differential adhesiotopes for the surrogate pathogen coreceptors described above.18,53,54 These adhesiotopes are predicted to either reside in a groove formed by homophilic interactions of CEACAM1 in cis that involve the V39 and D40 CC′ loop residues or are exposed after disruption of CEACAM1 cisdimerization by cytokine (eg, tumor necrosis factor-α) activation that precedes binding of CEACAM1 by pathogenic bacteria.58The CEACAM1 molecules also function as inhibitory coreceptors for activated human intestinal intraepithelial T cells (iIEL) in the gastrointestinal tract.4 Interestingly, the CEACAM1-specific Mab, 5F4, which inhibits the activation of peripheral blood T cells and the cytolytic function of iIEL, shows significantly reduced binding to the N-domain CEACAM1 mutants, Y34A, T87A, Q89A, or I91A on the GFCC′C" face. A less marked but significant decrease in binding to the Y34 mutant was also observed with the other functional Mabs, 34B1 and 26H7. Whether this is mediated by a heterophilic interaction involving residues distinct from those involved in homophilic interactions remains to be determined. Nevertheless, these studies demonstrate the importance of the GFCC′C" face in proinflammatory and immunoregulatory roles, for regulating a signaling function of CEACAM1, and for mediating interactions with pathogens. They further support the view54 that pathogenic bacteria use residues that are unlikely to undergo adaptive mutations because they are involved in the normal function of CEACAM1 receptors.
One might envisage that the molecular mechanisms designed to control CEACAM1 homophilic and heterophilic adhesion resemble, but are distinct from, the interactions observed for other adhesion receptor-ligand pairs, such as of CD2 with CD58, of ICAM-1 with ICAM-1, LFA-1, rhinoviruses or Plasmodium falciparum–infected erythrocytes, and of cadherins with cadherins. Such studies when taken together support the proposal that the GFCC′C" faces of the Ig family members may have evolved as a sticky patch to recognize a variety of protein-protein interactions.59 Superimposing 2 domains of CEACAM1 in a similar orientation to the packing geometry found for CD2-CD2 and CD2-CD58 indicates that the interface of CEACAM1 is much less hydrophilic than that found in CD2 interactions with either CD2 or CD58,60,61 with the CD2-CD58 interface being particularly dominated by charged residues. No less than 10 salt bridges and 5 hydrogen bonds have been identified at this interface.25Thus, a significant number of charged residues are engaged in a complex salt bridge network ensuring high specificity with interacting coreceptors. For CEACAM-1, assuming a similar packing arrangement, only one potential salt bridge involving residues R43 and E98 at the base of the C′ strand and the top of the G strand can be identified at the adhesive interface. Mutation of one of the contributing residues to this predicted salt bridge, E98, had no effect on and was not essential for homophilic adhesion.
A conserved intrastrand salt bridge is also predicted, but this does not lie on the GFCC′C" interface in our proposed molecular model. One of the contributing amino acids to this salt bridge is R64 at the base of the D β strand, whereas its partner is predicted to be D82 at the base of the F β strand. Mutation of either of these residues individually results in a loss of homophilic adhesion. This confirms the observation of Sippel and coworkers19 that mutation of the equivalent R64 residue in rodent CEACAM1 abolishes homophilic adhesion. However, re-formation of the salt bridge in an opposite amino acid orientation restored homophilic adhesion, suggesting that R64 and D82 are not critical for homophilic interactions, but are required for maintaining the stability and tertiary structure of the molecule. Interestingly, Taheri and colleagues62 reported recently that residues predicted to form a salt bridge in the N-terminal domain of the related CEA molecule did not significantly contribute to the adhesive interface. Thus, we would expect that both the homophilic and heterophilic interfaces of CEACAM1 are more hydrophobic than hydrophilic in nature and may therefore be expected to bind with greater affinity than interactions involving CD2 and CD58. The human CD2 and CD58 molecules on opposing cells are thought to pack face-to-face with the GFCC′C" faces of their N-terminal domains in a “hand-shaking” arrangement.25 61 This CD2-CD58 sensing mechanism generated by the hydrophilic nature of the interacting GFCC′C" faces maintains a high specificity and fast on-off rate to function in immune responses, with the affinity of the monomeric CD2-CD58 interaction being low (Kd of approximately 1 μM). This may not be true for CEACAM1 where a higher affinity may be needed.
Discriminating between cis and trans interactions is important for both heterophilic and homophilic adhesion molecules when expressed on the same and opposing cells, with cisinteractions being able to either block or enhance interactions intrans. Insight into the molecular mechanisms for CEACAM1 interactions may come from studies on molecules such as ICAM-1 or N-cadherin. The crystal structure of the 2 N-terminal domains of ICAM-1 has revealed that both domains function in integrin (LFA-1) binding, the first interacting with residues on LFA-1 and the second involved in orienting the recognition surface of the first domain so that the counterreceptors can achieve optimal contact in trans, while preventing recognition of cis counterreceptors on the same cell.61 Interestingly, the ICAM-1 binding sites for its cognate coreceptor LFA-1 and its surrogate pathogenic counterreceptors are almost all different.61 The position of amino acids on the GFCC′C" face of CEACAM1 used for homophilic, NeisserialOpa protein and H influenzae adhesion is reminiscent of but differs significantly from the critical amino acids on ICAM-1 that are required for interactions of its N-terminal domain incis and trans. In the case of N-cadherin, the functional N-terminal domain contains 7 β strands in an antiparallel orientation that resemble the Ig fold.63 Its crystal structure reveals 2 types of intermolecular interactions, one mediating intracellular and the other intercellular cadherin dimerization. Initially, dimerization occurs in cis so that the dimers are parallel to each other. This allows 2 monomeric halves of the dimer interface on opposing cells to interact with enhanced avidity in an antiparallel fashion to form an “adhesion zipper.” Such enhanced homophilic adhesion has been suggested for CEA49 and remains a possibility that should be investigated for CEACAM1.
Previous studies have shown that the N-terminal domain is crucial for homophilic interactions of rat CEACAM1.21 Although single deletions of the IgC2 domains, A1, B, and A2, did not affect adhesion, deletions of the N-domain alone or of the A1, B, and A2 domains together abrogated adhesion and deletions of both the B and A2 domains reduced adhesion substantially.21 The IgC2 domains as well as the N domain of CEACAM1 have been implicated in corona virus receptor activity,51 and H influenzaebinding.18 We have observed significantly reduced binding of CHO-CEACAM1-4L and -4S transfectants to immobilized human CEACAM1 N-domain constructs. The avidity of adhesion was also reduced if the A2 domain was removed from the CEACAM1-4 soluble molecule. Whereas previous studies have shown that cells expressing CEACAM1-4L and -4S are able to mediate homophilic adhesion,6,64-67our studies show that CEACAM1-4S in contrast to CEACAM1-4L transfectants bind less avidly to immobilized CEACAM1 molecules and suggest that the IgC2 and cytoplasmic domains of CEACAM1 may regulate the specificity and avidity of homophilic and heterophilic interactions or signaling functions. Several observations support this view. First, no transmembrane isoform of CEACAM1 containing N and A1 ectodomains only has been identified,2 suggesting that such an isoform does not have functional significance. Second, multiple splice variants of CEACAM1 lacking IgC2 domains and possessing long or short cytoplasmic tails exist,2 yet each CEACAM1 isoform will preferentially form homodimers incis,68,69 a process regulated by its interaction with calmodulin68,69 and by the relative levels of expression of the different isoforms on a single cell type.1 Third, cross-linking studies suggest that the ectodomains of specific CEACAM1 isoforms are closely associated,1 with the formation of homodimers appearing to require interactive sequences in the extracellular domain, cytoplasmic tail, and transmembrane region.1,44 70
It might be speculated that the expression levels, the conformation, and association of the different CEACAM1 isoforms in the cell membrane regulate the interaction of their N-terminal domains in cisor trans and thereby their ability to mediate homophilic or heterophilic functions. Homophilic interactions or dimerization of CEACAM1 molecules in cis may maintain the receptor in an inactive conformation for interacting with opposing cells, or place the N terminal GFCC′C" face in the correct orientation to increase the avidity of binding to homophilic or heterophilic counterreceptors on the same or opposing cells as has been described for ICAM-1 and the cadherins and predicted for CEA.49 Engagement of CEACAM1 homophilically on the same epithelial cell may prevent its interaction in trans and may deliver a negative signal inhibiting epithelial cell proliferation, a regulatory mechanism that is lost when CEACAM1 levels are decreased during epithelial tumor formation.1,2 Alternatively, the activation of CEACAM1 molecules on the surface of neutrophils during inflammation may control the presentation of sLex residues to E-selectin ligands on endothelial cells and regulate CD11/CD18 and L-selectin levels.11,49,71 On endothelial and epithelial cells, CEACAM1 activation, perhaps by inducing or inhibiting homophilic interactions or dimerization, may also regulate isoform concentrations on the cell surface, orient the molecules, or increase the avidity of adjacent residues on the GFCC′C" face of CEACAM1 forNeisserial Opa proteins or H influenzae.14-17 52-56
A high degree of sequence similarity exists between the N-domains of CEACAM-1 and CEA. A model of CEA49 has suggested that the Ig domains of CEA dimerize and subsequently align in parallel on the surface of the cell, making residues in the N-terminal domain of CEA accessible for homophilic adhesion in trans. However, subsequent biochemical studies23 have suggested that high affinity homophilic binding involves domains 1 and 6 of CEA. Nevertheless, this does not preclude a first phase of lower affinity binding between just the N-terminal domains of CEA as the 2 cell surface membranes approach each other. The subtle sequence variations on the GFCC′C" faces of CEA and CEACAM-1 are thus a key feature for further study and for x-ray crystallographic analyses.
Supported by the National Blood Service, Medical Research Council and Leukaemia Research Fund, United Kingdom, and by European Union Biotech and INTAS/RFBR grants (to S.M.W., A.M.T., G.-Q.Z., R.D., and Y.Z.); by a postdoctoral grant of the Fundacao para a Ciencia e Tecnologia, Portugal (to A.M.T.); by the Deutsche Krebshilfe Project number 70-2028 (to F.G.); and by National Institutes of Health grant DK51362 (to R.S.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Suzanne M. Watt, Stem Cell Laboratory, National Blood Service, The John Radcliffe Hospital, Headington, Oxford, OX3 9DS, United Kingdom; e-mail: swatt@molbiol.ox.ac.uk.

![Fig. 1. Amino acid alignment of the N-terminal CEACAM1 IgV domain. / (A) Alignment of the N-domain of CEACAM1 with other members of the CEA family. Interface 1 and 2: # symbols indicate the position of mutated amino acids in CEACAM1 that affect YTH71.3.2 Mab or Opa protein binding, respectively. Peptide sequences that regulate the expression of CD11/CD18 and L-selectin levels on neutrophils are both underlined in pink and the residues are shown in green or pink. Interface 3 and 4: # symbols indicate the position of mutated amino acids in CEA or CEACAM8, respectively, that affect Opa protein binding in reference 52. The amino acid sequence that is critical for mouse CEACAM1 binding to murine corona viruses is indicated in brown and underlined. Amino acids in CEACAM1 conserved in other family members are indicated in bold type (100% identity); gaps are denoted by dashes (–). Strand indicates secondary structure elements and β strand positions are underlined and marked under the sequence and lettered A to G. (B) Alignment of the N-domain of CEACAM1 with human (H) and rat (R) CD2 and human CD58. Interface 5: Red # symbols represent the position of mutants that abrogate CEACAM1 homophilic interactions. Blue # symbols represent the position of mutants that abrogate anti-CEACAM Mab binding. For the CEACAM1 [H] sequence, residues in red are those mutated in this manuscript. Interface 6 and 7: # symbols indicate known amino acid residues at the interface in human CD2 and CD58, respectively. These are based on x-ray crystallographic coordinates for human and rat CD2 and human CD58.25-27 Mutated human CEACAM1 residues are indicated by arrows, with each amino acid being substituted with alanine (A) either as a single, double, or triple mutant or with arginine (R) for D82 or aspartic acid (D) for R64 as indicated in Figures 3, 6, and 7 and Table 1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/5/10.1182_blood.v98.5.1469/5/m_h81711478001.jpeg?Expires=1765963907&Signature=319k6IFsHT~zwsnJ11bv60z5Z7jkuj~KC5ufyYInUZGuZeV9j0~vm02Bq1JWyLju-j-KpzlIfi7FDK9sXSJ2DHUKkHWFgNMhOhGGpfHndZgl5qeK47fpbFyysFXc-PoNpboJW~8LF-vRLyE~wXHzOO-vLQiyXrg6EvSM3ab2tx-3OYXONSrRxbb6VfH7hOPcfsi0r6UlwpraSux74Ixpcw4acwyKZK1NSju4HQJZx9tOLBVb39GZMUjDmTc-RDXuhPnFJI0g-Nks4l-cwbutsBUiBS80HslLYf~54eku3gAlgwKiMpLofk3hMPZ5RlVn-~wswBFUWeq3CyRLSWWtlw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal