In the inflammatory response, leukocyte rolling before adhesion and transmigration through the blood vessel wall is mediated by specific cell surface adhesion receptors. Neutrophil rolling involves the interaction of P-selectin expressed on activated endothelium and its counter-receptor on neutrophils, P-selectin glycoprotein ligand-1 (PSGL-1). Here, it is reported that P-selectin binding to neutrophils is lost under conditions that cause the release of proteinases from neutrophil primary granules. Treatment of neutrophils with the purified neutrophil granule proteinases, cathepsin G and elastase, rapidly abolished their capacity to bind P-selectin. This inactivation corresponded to loss of the N-terminal domain of PSGL-1, as assessed by Western blot analysis. A loss of intact PSGL-1 protein from the surfaces of neutrophils after the induction of degranulation was also detected by Western blot analysis. Cathepsin G initially cleaved near the PSGL-1 N-terminus, whereas neutrophil elastase predominantly cleaved at a more C-terminal site within the protein mucin core. Consistent with this, cathepsin G cleaved a synthetic peptide based on the PSGL-1 N-terminus between Tyr-7/Leu-8. Under conditions producing neutrophil degranulation in incubations containing mixtures of platelets and neutrophils, the loss of PSGL-1, but not P-selectin, from platelet-neutrophil lysates was detected. Cathepsin G- or neutrophil elastase-mediated PSGL-1 proteolysis may constitute a potential autocrine mechanism for down-regulation of neutrophil adhesion to P-selectin.
Introduction
Recruitment of neutrophils to inflammatory sites is initiated by rolling of circulating cells on activated endothelium before firmer adhesion and subsequent transmigration.1-4Neutrophil rolling is mediated predominantly by expression of adhesive receptors of the selectin family on activated endothelial cells.5,6 P-selectin is stored on Weibel-Palade body membranes and rapidly expressed on the surface of activated endothelial cells; E-selectin expression is induced in response to inflammatory stimuli. The counter-receptor for P-selectin on neutrophils is P-selectin glycoprotein ligand-1 (PSGL-1), an approximately 220-kd glycoprotein homodimer. The mature protein consists of an extracellular N-terminal anionic sequence containing 1 to 3 sulfated tyrosine residues (Gln-1–Glu-20), a sialomucin domain, a transmembrane domain, and a cytoplasmic tail.7,8 Several lines of evidence have shown that the sulfated N-terminal sequence of PSGL-1 is critical for P-selectin binding. We found that a cobra venom metalloproteinase, mocarhagin, abolished P-selectin binding to neutrophils by selectively cleaving P-selectin between Tyr-10 and Asp-11.9 Further, P-selectin binding to recombinant PSGL-1 was dependent on coexpression with tyrosylsulfate sulfotransferase, whereas blocking sulfation of the recombinant receptor inhibited ligand binding.10-12
After initial rolling, tight adhesion of arrested neutrophils is regulated by other adhesion receptors, in particular members of the integrin family on neutrophils such as αLβ2 (LFA-1) and αMβ2 (Mac-1) binding to counter-receptors of the immunoglobulin superfamily, ICAM-1 and ICAM-2, expressed on endothelial cells.2-4During the transition stage, as neutrophils progress from rolling to stable adhesion, an important question is how the selectin-dependent interaction is down-regulated. One mechanism that appears to down-regulate P-selectin interaction is that neutrophil activation results in the surface redistribution of PSGL-1 from the tips of microvilli to the uropod of the activated neutrophil.13,14Another potential mechanism that could regulate PSGL-1 expression under certain conditions is proteolytic shedding. In this regard, Davenpeck et al15 have recently provided evidence for PSGL-1 shedding in response to treatment of neutrophils with platelet-activating factor (10−9-10−6 M) or phorbol 12-myristate acetate (PMA). Shedding was EDTA-sensitive but unaffected by other metalloproteinase inhibitors, suggesting the loss of PSGL-1 was either due to a metalloproteinase distinct from that involved in shedding of L-selectin or some other EDTA-sensitive mechanism.
Two other proteinases, cathepsin G and neutrophil elastase, are known to be released from primary granules after neutrophil activation. These are strong potential candidates for targeting PSGL-1 given that both have been implicated in regulating leukocyte function. Cathepsin G has been reported to regulate neutrophil–endothelial cell adhesion,16 whereas inhibitors of cathepsin G and elastase have been shown to attenuate neutrophil infiltration of inflamed microvessels in vivo using intravital microscopy.17Elastase has recently been shown to be released from adherent neutrophils under flow conditions in the presence ofN-formyl-methionyl-leucyl-phenylalanine (fMLP), complement fragment C5a, and interleukin-8 (IL-8)18 and from neutrophils adherent to endothelial cells in the presence of IL-1 or tumor necrosis factor.19 It has previously been shown that elastase treatment of neutrophils eliminates their capacity to bind P-selectin, presumably because of proteolysis of PSGL-1.20Interestingly, like mocarhagin, cathepsin G has been reported to cleave the platelet von Willebrand factor receptor, glycoprotein (GP) Ibα of the GP Ib-IX-V complex, within a sulfated tyrosine sequence that is analogous to that in PSGL-1.21-23 This suggests that cathepsin G may also cleave at or near the mocarhagin cleavage site in the PSGL-1–sulfated sequence.
In this study, we demonstrate that neutrophil secretion is accompanied by a diminished capacity of the cells to support the binding of P-selectin. Further, we show that purified cathepsin G and neutrophil elastase both potently inhibit P-selectin binding to PSGL-1, with cleavage of PSGL-1 and loss of the N-terminal–sulfated sequence required for P-selectin binding. These results imply that neutrophil elastase- or cathepsin G-mediated proteolysis of PSGL-1 may constitute an autocrine mechanism for the down-regulation of PSGL-1–dependent neutrophil adhesion.
Materials and methods
Materials
Aprotinin, benzamidine, soybean trypsin inhibitor, diisopropyl fluorophosphate (DFP), cytochalasin B, fMLP, platelet-activating factor (PAF), lipopolysaccharide, and pepstatin were purchased from Sigma (St Louis, MO). Leupeptin and thrombin-related agonist peptide (TRAP) were purchased from Auspep (Parkville, Australia), methyl-α-D-mannopyranoside and anti–PSGL-1 monoclonal antibody PL-1 were purchased from Beckman Coulter (Fullerton, CA), and concanavalin-A–Sepharose CL-6B, Ficoll-Hypaque, and jacalin-Sepharose were purchased from Pharmacia, (Uppsala, Sweden). Elastase inhibitor (Boc-Ala-Ala-Ala-NHO-Bz) and chymostatin were obtained from Calbiochem (La Jolla, CA). Purified human cathepsin G and neutrophil elastase were the kind gift of Dr Philip Hogg (Sydney, Australia), lysates from the myeloid leukemic KG1a cell line were from Dr Paul Simmons (Adelaide, Australia), and the anti–PSGL-1 monoclonal antibody PL-2 was from Dr Rodger McEver (Oklahoma City, OK). Rabbit IgG against a synthetic peptide based on the N-terminal sequence of PSGL-1 (Gln-1–Glu-15) was raised and affinity purified as described elsewhere.9Monoclonal antibody AK4 against human platelet P-selectin was raised in our laboratory,24 and platelet membrane P-selectin was purified from recently expired human platelet concentrates (a gift of the Red Cross Blood Bank, Victoria, Australia) as previously described.24 25
Binding of P-selectin to resting or degranulated neutrophils
Human neutrophils were isolated from venous blood using 25 U/mL heparin as an anticoagulant according to the method of Bignold and Ferrante.26 Measurement of binding of purified P-selectin was performed as previously described.24,25 Briefly, saturating amounts of detergent-free sodium iodide I 125-labeled P-selectin (1 μg/mL, final concentration)25 were incubated with neutrophils (2 × 106/mL, final concentration) in a 1:1 mixture of TSCa buffer (0.02 M Tris, 0.15 M sodium chloride, 1 mM calcium chloride, pH 7.4) and RPMI medium containing 1% (vol/vol) fetal bovine serum (RPMI/FBS) for 20 minutes at 22°C. Duplicate 50-μL aliquots were then layered onto 200-μL cushions of 17% (wt/vol) sucrose in a 1:1 mixture of TSCa buffer and RPMI/FBS and pelleted at 8750g for 2 minutes, and radioactivity associated with the pellet was measured in a gamma counter after aspiration of the supernatant. Nonspecific binding was assessed by including either a 50-fold excess of unlabeled P-selectin, 10 mM EDTA, or rabbit anti–P-selectin Fab fragments (50 μg/mL, final concentration) in parallel assays. To test the effect of proteinases on P-selectin binding, neutrophils were treated with cathepsin G or neutrophil elastase (0.5-20 μg/mL, final concentration) for 30 minutes at 22°C, then washed (8750g, 2 minutes) with a 1:1 mixture of TSCa buffer and RPMI and resuspended to the original volume in washing buffer before the addition of 125I-labeled P-selectin. Time course experiments were similarly performed and examined the effect of pretreatment of neutrophils with 20 μg/mL cathepsin G or neutrophil elastase for 5 to 180 minutes.
To examine the effect of neutrophil granule secretion on P-selectin binding and levels of neutrophil PSGL-1, neutrophils were stimulated with different agonists reported to induce degranulation in neutrophils.15,20,27 Neutrophils (2 × 107cells/mL) were treated with either PAF (0.1-10 μM), lipopolysaccharide (LPS) (100 μg/mL), or cytochalasin B (5 μg/mL) and fMLP (0.1 μM) for 10 to 60 minutes. In some incubations, neutrophils were treated with 0.1 mM elastase inhibitor, chymostatin, or 1 mM EDTA, either alone or in combination before the addition of degranulating agents. After centrifugation at 150g for 1 minute, neutrophils either were resuspended in RPMI/FBS and binding of125I-labeled P-selectin was assessed as described above or neutrophils were lysed by the addition of an equal volume of lysis buffer (0.01 M Tris/HCl pH 7.5 containing 0.15 M sodium chloride, 5 mM EDTA, 0.1% Triton X-100, 10 μg/mL each of soybean trypsin inhibitor, leupeptin, and pepstatin, and 1 mM NaF, sodium orthovanadate, and 5 mM phenylmethylsulfonyl fluoride) on ice for 30 minutes. Cell lysates were centrifuged at 150g for 20 minutes, and 50-μL aliquots of the supernatant were mixed with an equal volume of sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) reducing sample buffer, electrophoresed on 7.5% SDS–polyacrylamide gels, electrotransferred to nitrocellulose, and immunoblotted with anti–PSGL-1 monoclonal antibodies, either PL-1 against the N-terminal portion or PL-2 against a region of the ectodomain of PSGL-1.28 Some Western blots were probed with a rabbit polyclonal antibody directed against the N-terminal region of PSGL-1. All blots were probed with horseradish peroxidase-labeled anti–rabbit or anti–mouse IgG secondary antibodies (Silenus, Hawthorn, Australia), and visualized using the enhanced chemiluminescence (ECL) detection method (Amersham, Buckinghamshire, United Kingdom).
The effect of neutrophil granule secretion and platelet activation on levels of PSGL-1 and P-selectin in lysates of platelet-neutrophil suspensions was also assessed. Washed human platelets (prepared as previously described29) and neutrophils were resuspended separately in Tyrode buffer, pH 7.4, and mixed together in a ratio of approximately 50:1 in Tyrode buffer alone or containing combinations of 5 μg/mL cytochalasin B, 0.1 μM fMLP, and 62.5 μM TRAP. Similar experiments were performed using 0.1 mg/mL LPS or 10 μM PAF as agonists. Some incubations were preincubated with the proteinase inhibitor cocktail described above, which included 0.1 mM elastase inhibitor and chymostatin but not Triton X-100. These concentrations of inhibitor have been used elsewhere to inhibit elastase30and cathepsin G.31 After incubation at 22°C for 15 minutes, platelet-neutrophil suspensions were pelleted by centrifugation and treated with the proteinase inhibitor cocktail containing 1% Triton X-100 for 2 hours on ice. Lysates were centrifuged at 10 000g, and aliquots of supernatant were eluted on 7.5% SDS–polyacrylamide gels, transferred to membranes, and subjected to Western blot analysis using monoclonal antibodies against either P-selectin (AK4) or PSGL-1 (PL-1) and detected using ECL.
Digestion of surface-labeled neutrophils or PSGL-1–enriched fractions of KG1a cells
For assessing potential proteinase substrates on the membrane surfaces, neutrophils were surface-labeled using Na125I–lactoperoxidase or 3H-borohydride as previously described,32 33 resuspended in serum-free RPMI medium (2 × 107 cells/mL), and treated with cathepsin G or neutrophil elastase (20 μg/mL, final concentration) at 22°C. At various times, the cells were then centrifuged at 150g for 10 minutes, and the supernatant was quenched with soybean trypsin inhibitor before the addition of SDS-PAGE sample buffer. Pellets were washed once in RPMI medium, lysed with 1% (wt/vol) Triton X-100 at 4°C for 1 hour in the presence of DFP (0.5 mM), aprotinin (10 μg/mL), pepstatin (1 μM), leupeptin (100 μg/mL), and benzamidine (10 mM), and centrifuged at 8750g for 3 minutes. Supernatants were mixed with 1:1 SDS-PAGE sample buffer and electrophoresed on 5% to 20% SDS–polyacrylamide gels.
The KG1a leukemic myeloid cell line was readily available to our laboratory and was shown to be a convenient source of PSGL-1 protein. PSGL-1–enriched samples were prepared from KG1a cell lysates by affinity purification on a column of concanavalin-A–Sepharose CL-6B (1.5 × 25 cm), equilibrated with ETSTa buffer (10 mM Tris/HCl pH 7.4 containing 0.15 M sodium chloride, 1 mM EDTA, 2 mM calcium chloride, 0.1% (vol/vol) Triton X-100 and 0.02% (wt/vol) sodium azide). Bound protein was eluted at 25 mL/h with 0.2 M methyl-α-D-mannopyranoside in ETSTa buffer, dialyzed against TS buffer (0.01 M Tris/HCl pH 7.4, 0.15 M sodium chloride), and loaded onto a 1 × 10-cm jacalin–Sepharose column equilibrated with TS buffer containing 0.1% (vol/vol) Triton X-100 at 20 mL/h. Proteins were eluted with 0.01 M sodium phosphate buffer, pH 7.4, containing 0.15 M sodium chloride (PBS), 1% (vol/vol) octyl glucoside, and 0.8 M D-galactose. Peak fractions containing protein as assessed by SDS-PAGE and Coomassie blue staining were pooled and dialyzed against PBS buffer containing 1% (vol/vol) octyl glucoside and 0.02% (wt/vol) sodium azide. For digestion, the purified mucin fraction from the KG1a lysate was made 1 mM in calcium chloride and treated with 10 μg/mL (final concentration) of cathepsin G or neutrophil elastase at 22°C. At various times (0-30 minutes), samples were added to an equal volume of SDS-PAGE sample buffer, electrophoresed on 7.5% SDS–polyacrylamide gels, electrotransferred to nitrocellulose, and immunoblotted with either rabbit anti–N-terminal PSGL-1 peptide IgG or the monoclonal antibody PL-2. In the same manner, purified unlabeled P-selectin or125I-labeled P-selectin was digested with various amounts of cathepsin G or elastase for 0 to 90 minutes and electrophoresed on 7.5% SDS–polyacrylamide gels. Gels containing radioactive material were dried and subjected to autoradiography. Gels containing nonradioactive samples were electrotransferred to nitrocellulose and immunoblotted with anti–P-selectin polyclonal antibody or the anti–P-selectin monoclonal antibody AK4. All nitrocellulose blots were probed with the appropriate peroxidase-labeled secondary antibody, and bands were visualized using the ECL method.
Cathepsin G digestion of a synthetic peptide based on PSGL-1 for N-terminal amino acid sequencing
A synthetic peptide based on residues Thr-3–Glu-17 at the N-terminus of PSGL-1 was dissolved at 0.3 mg/mL in 50 mM HEPES buffer, pH 7.4, containing 0.15 M sodium chloride and 1 mM calcium chloride and was digested with cathepsin G (20 μg/mL, final concentration) for 4 hours at 22°C. Samples (100 μL) were added to 400 μL of 1% (vol/vol) trifluoroacetic acid (TFA) in methanol. Cathepsin G was added to some aliquots of the peptide after the addition of TFA. All samples were microfuged at 8750g for 5 minutes, and the supernatants were vacuum dried in a Savant Speedivac (Farmingham, NY). Samples were analyzed by reverse-phase high-performance liquid chromatography (HPLC) on a Waters 510 HPLC (Hartford, CT) system using a linear gradient of 0% to 70% (vol/vol) acetonitrile in 0.08% (vol/vol) TFA eluted at 1 mL/min over 30 minutes. Absorbance was monitored at 215 nm, and peptide peaks were collected manually. Amino acid sequences of peptide fractions were analyzed on an Applied Biosystems model 470A protein sequencer equipped with an on-line model 120A phenylthiohydantoin analyzer.16 Polybrene was used as the carrier.
Results
Neutrophil degranulation causes reduction of P-selectin binding and loss of PSGL-1 from the neutrophil surface
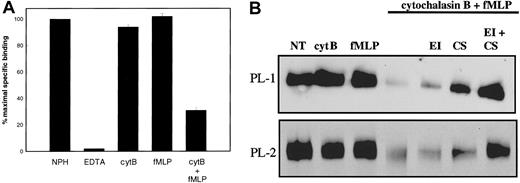
Initially, we investigated the role of secretion in regulating PSGL-1 function by stimulating neutrophils to release their primary granule contents and analyzing the effect on P-selectin binding. Unstimulated neutrophils supported P-selectin binding in the presence of Ca++ but not EDTA (Figure1A). Treatment of neutrophils with cytochalasin B or fMLP alone had little effect on levels of P-selectin binding to neutrophils. In contrast, neutrophils degranulated by treatment with cytochalasin B and fMLP27 showed a marked decrease in levels of 125I-labeled P-selectin binding (Figure 1A).
Effect of neutrophil granule secretion on P-selectin binding.
(A) Specific binding of 125I-labeled P-selectin (1 μg/mL, final concentration) to unstimulated neutrophils (2 × 107/mL, final concentration) at 22°C was measured in the presence of 1 mM Ca++ (NPH) or 1 mM EDTA (EDTA). Neutrophils were stimulated for 15 minutes with 5 μg/mL cytochalasin B (cytB), 30 minutes with 0.1 μM fMLP, or sequentially with 5 μg/mL cytochalasin B then 0.1 μM fMLP (cytB+fMLP), washed, and resuspended at 2 × 107/mL before measuring binding of125I-labeled P-selectin in the presence of 1 mM Ca++. Results shown represent the mean and standard deviation of 3 separate experiments performed with different donors. (B) Neutrophils were treated as in panel A. Some samples were treated with elastase inhibitor (EI) or chymostatin (CS) alone or in combination before the addition of cytochalasin B and fMLP. All samples were lysed in buffer containing Triton X-100. Neutrophil lysates were electrophoresed on SDS–polyacrylamide gels, transferred to nitrocellulose, and probed with either PL-1 (upper panel) or PL-2 (lower panel). Data are representative of at least 3 experiments with different donors.
Effect of neutrophil granule secretion on P-selectin binding.
(A) Specific binding of 125I-labeled P-selectin (1 μg/mL, final concentration) to unstimulated neutrophils (2 × 107/mL, final concentration) at 22°C was measured in the presence of 1 mM Ca++ (NPH) or 1 mM EDTA (EDTA). Neutrophils were stimulated for 15 minutes with 5 μg/mL cytochalasin B (cytB), 30 minutes with 0.1 μM fMLP, or sequentially with 5 μg/mL cytochalasin B then 0.1 μM fMLP (cytB+fMLP), washed, and resuspended at 2 × 107/mL before measuring binding of125I-labeled P-selectin in the presence of 1 mM Ca++. Results shown represent the mean and standard deviation of 3 separate experiments performed with different donors. (B) Neutrophils were treated as in panel A. Some samples were treated with elastase inhibitor (EI) or chymostatin (CS) alone or in combination before the addition of cytochalasin B and fMLP. All samples were lysed in buffer containing Triton X-100. Neutrophil lysates were electrophoresed on SDS–polyacrylamide gels, transferred to nitrocellulose, and probed with either PL-1 (upper panel) or PL-2 (lower panel). Data are representative of at least 3 experiments with different donors.
Proteolysis of PSGL-1 on neutrophils treated with cytochalasin B and fMLP was directly confirmed by immunoblotting. Neutrophils isolated from healthy donors were treated with either cytochalasin B or fMLP alone or in sequential combination, or in the presence of inhibitors of neutrophil elastase and cathepsin G, either alone or in combination. Cells were pelleted by centrifugation, the medium was removed, and cells were lysed in the presence of proteinase inhibitors. Aliquots of cell lysates were electrophoresed on 7.5% (wt/vol) SDS–polyacrylamide gels and transferred onto nitrocellulose. Levels of PSGL-1 that remained associated with the neutrophil surface were estimated by Western blotting with either PL-1 directed against the N-terminal region of PSGL-1 or PL-2 directed against an epitope within residues 147-194 of the mucin protein core.34 Figure 1B shows a loss of PL-1 reactivity (upper panel) and PL-2 reactivity (lower panel) in sample lanes containing neutrophils treated with both cytochalasin B and fMLP (lane 4), whereas treatment of neutrophils with either reagent alone (lanes 2 and 3) did not greatly alter levels of PL-1 or PL-2 reactivity compared with untreated neutrophils (lane 1). Inclusion of either an elastase inhibitor (lane 5) or the cathepsin G inhibitor, chymostatin (lane 6), substantially reduced the loss of the PL-1 and PL-2 epitopes, whereas inclusion of both inhibitors together protected against the loss of either epitope. These data suggest that the induction of neutrophil degranulation by treatment with cytochalasin B and fMLP resulted in the removal of the PL-1 and PL-2 epitopes from neutrophil PSGL-1. Roles for neutrophil elastase and neutrophil cathepsin G in the observed proteolysis are indicated; separate preincubation of neutrophils with inhibitors of elastase or cathepsin G partially blocked the loss of PSGL-1 reactivity to monoclonal antibodies. Inclusion of both inhibitors completely protected against the loss of PSGL-1 from the neutrophil cell surface, as determined by a comparison of band density.
Effect of physiological neutrophil agonists on PSGL-1
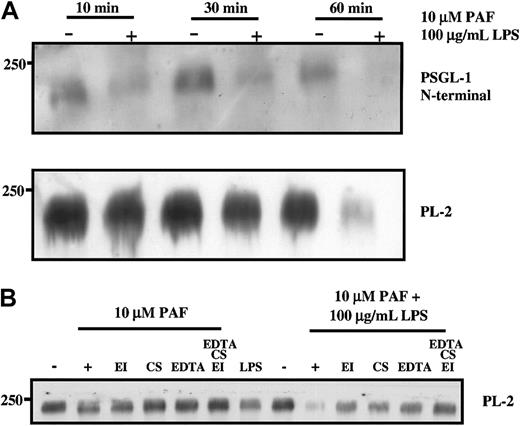
The loss of PSGL-1 from the surface of neutrophils was also observed after treatment of neutrophils with other agents reported to induce degranulation, including the combination of PAF and LPS as would occur in sepsis35 (Figure 2) and PMA (data not shown). The N-terminal portion of PSGL-1 was cleaved from the surface of neutrophils within 10 minutes of treatment with a combination of PAF (10 μM) and LPS (100 μg/mL) (Figure 2A), and the more C-terminal PL-2 epitope was lost after 1-hour incubation at 37°C. Loss of more than 50% of PSGL-1 protein from neutrophils isolated from 3 different donors was consistently observed with either PAF or LPS alone after 1-hour incubation (Figure 2B); however, the extent of loss varied between donors. The loss could be partially prevented by the inclusion of either elastase inhibitor or chymostatin or EDTA and could be completely prevented by including a combination of the inhibitors (Figure 2B). In contrast to the report of Davenpeck et al,15 we did not observe that lower concentrations of PAF (less than or equal to 1 μM) resulted in loss of PSGL-1 even after a 1-hour incubation. Because nanomolar PAF caused platelet aggregation, the reason for the discrepancy between our results and theirs is unclear, though we did consistently observe loss of PSGL-1 with higher concentrations of PAF (10 μM). In agreement with their studies, we did confirm that 10 nM PMA resulted in significant loss of PSGL-1 (data not shown). Using PAF and LPS in combination resulted in almost complete loss of PSGL-1 from the surface of the neutrophils, which was only partially protected by individual inhibitors but completely protected by the combination of inhibitors. Taken together, these results suggest that physiologically relevant agonists such as LPS and PAF are able to induce degranulation in neutrophils sufficient to cause a loss of the binding portion of PSGL-1 after 10 minutes. The results are further consistent with PSGL-1 shedding under these conditions, involving elastase, cathepsin G, and potentially metalloproteinases.
Neutrophil granule secretion induced by PAF or LPS results in loss of PSGL-1 from the cell surface.
Purified neutrophils were treated with 10 μM PAF or 100 μg/mL LPS for the indicated times (A) or 60 minutes (B) at 37°C. Some incubations included elastase inhibitor or chymostatin or EDTA. Cells were pelleted and lysed in buffer containing proteinase inhibitors and Triton X-100 as described in “Materials and methods.” Neutrophil lysates were electrophoresed on SDS–polyacrylamide gels, transferred to nitrocellulose, and probed with either polyclonal anti–PSGL-1 N-terminal antibody or PL-2 as indicated. Data are representative of at least 3 experiments with different donors.
Neutrophil granule secretion induced by PAF or LPS results in loss of PSGL-1 from the cell surface.
Purified neutrophils were treated with 10 μM PAF or 100 μg/mL LPS for the indicated times (A) or 60 minutes (B) at 37°C. Some incubations included elastase inhibitor or chymostatin or EDTA. Cells were pelleted and lysed in buffer containing proteinase inhibitors and Triton X-100 as described in “Materials and methods.” Neutrophil lysates were electrophoresed on SDS–polyacrylamide gels, transferred to nitrocellulose, and probed with either polyclonal anti–PSGL-1 N-terminal antibody or PL-2 as indicated. Data are representative of at least 3 experiments with different donors.
Treatment of neutrophils with elastase or cathepsin G results in loss of P-selectin binding
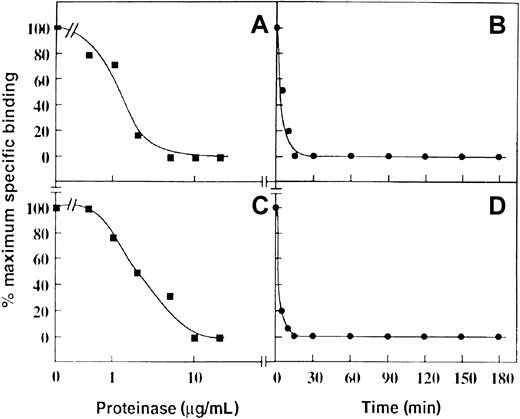
To test whether this mechanism could account for a loss of P-selectin binding, purified proteinases known to be present in neutrophil primary granules were incubated with neutrophils, and the ability of these cells to bind 125I-labeled P-selectin was assessed in a binding assay. Treatment of neutrophils with increasing amounts of purified cathepsin G or neutrophil elastase for 30 minutes at 22°C resulted in the loss of 125I–P-selectin binding to neutrophils with apparent IC50 values of approximately 1 μg/mL (Figure 3A) and 1-2 μg/mL (Figure 3C), respectively. These IC50 values are in the physiological range expected on neutrophil degranulation. In this regard, 106 neutrophils/mL maximally degranulated would release approximately 1.6 μg/mL cathepsin G36 and 2.5 μg/mL elastase.35 Equivalent data were obtained when the cells were washed before P-selectin binding (not shown) was measured, suggesting inactivation was irreversible, and consistent with proteolysis of PSGL-1 rather than any effect on P-selectin. At concentrations of 20 μg/mL of either proteinase, there was complete loss of 125I-labeled P-selectin binding after approximately 10 minutes for cathepsin G (Figure 3B) and 15 minutes for neutrophil elastase (Figure 3D).
Effect of cathepsin G and neutrophil elastase on binding of P-selectin to neutrophils.
Specific binding of 125I-labeled P-selectin (1 μg/mL, final concentration) to neutrophils (2 × 107/mL, final concentration) pretreated with cathepsin G (A and B) or neutrophil elastase (C and D). For the dose-response curves (A and C), the digestion time was 30 minutes at 22°C. For the time course (B and D), the final concentration of the enzymes was 20 μg/mL. Data are representative of 3 separate experiments.
Effect of cathepsin G and neutrophil elastase on binding of P-selectin to neutrophils.
Specific binding of 125I-labeled P-selectin (1 μg/mL, final concentration) to neutrophils (2 × 107/mL, final concentration) pretreated with cathepsin G (A and B) or neutrophil elastase (C and D). For the dose-response curves (A and C), the digestion time was 30 minutes at 22°C. For the time course (B and D), the final concentration of the enzymes was 20 μg/mL. Data are representative of 3 separate experiments.
Purified PSGL-1 is a substrate for elastase and cathepsin G
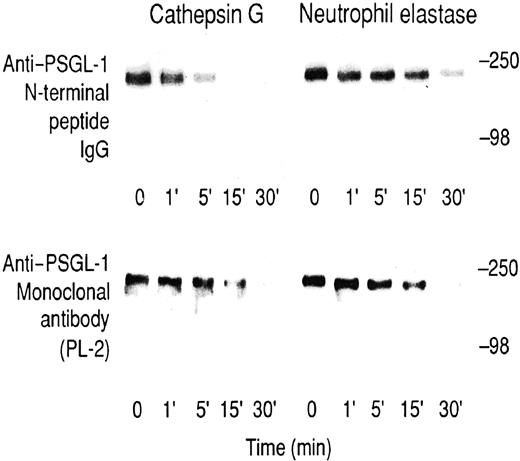
To further analyze potential cathepsin G or neutrophil elastase substrates, we initially digested neutrophils that had been surface-labeled with 125I–lactoperoxidase or3H-borohydride. Supernatants of cells treated with cathepsin G over a 60-minute time course and analyzed on SDS–polyacrylamide gels were indistinguishable from untreated neutrophils (not shown). After neutrophil elastase treatment, several unidentified labeled bands were released after 60 minutes (not shown), suggesting neutrophil elastase is less selective in its effect on neutrophils than cathepsin G. These studies failed to identify a substrate cleaved on a time scale corresponding to the observed loss of P-selectin binding. Therefore, a PSGL-1–enriched fraction was purified from lysates of KG1a cells. This sample contained PSGL-1 since it was Western blotted by an anti–PSGL-1 monoclonal antibody, PL-2, and a rabbit anti–N-terminal peptide IgG raised against the PSGL-1 sequence, Gln-1–Glu-15. The epitope for the latter antibody has previously been shown to be lost after cleavage of PSGL-1 with mocarhagin, which cleaves between Tyr-10 and Asp-11.9 Consistent with published results,9 mocarhagin treatment resulted in a time-dependent loss of the N-terminal peptide IgG epitope, which disappeared within 5 minutes, whereas there was no loss of the PL-2 epitope (data not shown). Cathepsin G digestion showed a partial loss of the epitope for the anti–N-terminal peptide IgG at 5 minutes and complete loss after 15 minutes (Figure 4, top left panel), a time scale comparable to the loss of P-selectin binding to cathepsin G–treated neutrophils (compare with Figure 3B). In contrast, the PL-2 epitope showed only partial loss at 15 minutes and complete loss after 30 minutes (Figure 4, lower left panel), suggesting slower secondary cleavage(s) was also occurring. For neutrophil elastase digestion of PSGL-1, the epitopes for the anti–N-terminal peptide IgG antibody and PL-2 were lost with a similar time course (Figure 4, right panels). Under the conditions described, we did not observe the appearance of any lower molecular weight PL-1 or PL-2 immunoreactive bands after digestion of any of the samples. Together, these results suggest that though cathepsin G preferentially cleaves near the N-terminus of PSGL-1, neutrophil elastase cleaves PSGL-1 downstream of the PL-2 epitope.
Digestion of KG1a lysates.
Time course for the digestion of KG1a lysates with cathepsin G (left panels) or neutrophil elastase (right panels) at a final concentration of 20 μg/mL. Upper panels were immunoblotted with rabbit anti–PSGL-1 N-terminal peptide IgG, and lower panels were immunoblotted with the anti–PSGL-1 monoclonal antibody PL-2. Blots were visualized using the ECL method. Prestained molecular weight markers are myosin (250 kd) and bovine serum albumin (98 kd).
Digestion of KG1a lysates.
Time course for the digestion of KG1a lysates with cathepsin G (left panels) or neutrophil elastase (right panels) at a final concentration of 20 μg/mL. Upper panels were immunoblotted with rabbit anti–PSGL-1 N-terminal peptide IgG, and lower panels were immunoblotted with the anti–PSGL-1 monoclonal antibody PL-2. Blots were visualized using the ECL method. Prestained molecular weight markers are myosin (250 kd) and bovine serum albumin (98 kd).
We also investigated whether purified P-selectin was a substrate for cathepsin G and elastase. Purified P-selectin was sensitive to digestion with low concentrations of cathepsin G (2.0 μg/mL) and elastase (0.2 μg/mL) and with cleavage essentially complete at 90 minutes (data not shown). Figure 5B shows aliquots of P-selectin (1 μg/mL) that were treated with high concentrations of either cathepsin G or neutrophil elastase (20 μg/mL) for 90 minutes, electrophoresed under nonreducing conditions, transferred to nitrocellulose, and probed with the anti–P-selectin monoclonal antibody AK4. Both cathepsin G and elastase used P-selectin as a substrate, and treatment of P-selectin with cathepsin G generated a 97-kd fragment recognized by AK4.
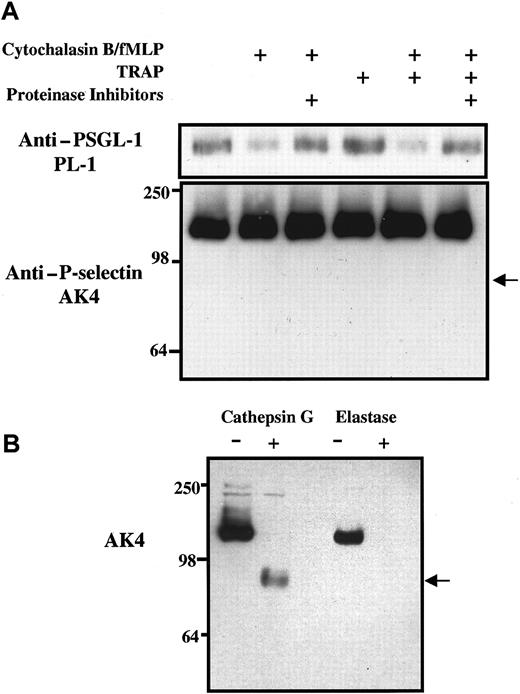
Effect of neutrophil granule secretion and platelet activation on levels of PSGL-1 and P-selectin in lysates of platelet-neutrophil suspensions.
(A) Purified and washed human platelets and neutrophils were resuspended in Tyrode buffer, pH 7.4, and were mixed together in a ratio of approximately 50:1 in Tyrode buffer alone or containing 5 μg/mL cytochalasin B, 0.1 μM fMLP, or 62.5 μM TRAP as shown. Some incubations were preincubated with a proteinase inhibitor cocktail that included 0.1 mM elastase inhibitor and chymostatin. After incubation at 22°C for 15 minutes, platelet-neutrophil suspensions were pelleted and treated with the proteinase inhibitor cocktail containing 1% Triton X-100. Aliquots of lysates were eluted on 7.5% SDS–polyacrylamide gels, transferred to membranes, and subjected to Western blot analysis using monoclonal antibodies against either P-selectin (AK4) or PSGL-1 (PL-1). The arrow indicates the expected position of the P-selectin fragment recognized by AK4 if P-selectin had been digested by cathepsin G. Data are representative of at least 3 experiments with different donors. (B) Purified125I-labeled P-selectin was treated for 1 hour with purified human elastase or cathepsin G, eluted on 7.5% SDS–polyacrylamide gels, transferred to membranes, and subjected to Western blot analysis using monoclonal antibody against P-selectin (AK4).
Effect of neutrophil granule secretion and platelet activation on levels of PSGL-1 and P-selectin in lysates of platelet-neutrophil suspensions.
(A) Purified and washed human platelets and neutrophils were resuspended in Tyrode buffer, pH 7.4, and were mixed together in a ratio of approximately 50:1 in Tyrode buffer alone or containing 5 μg/mL cytochalasin B, 0.1 μM fMLP, or 62.5 μM TRAP as shown. Some incubations were preincubated with a proteinase inhibitor cocktail that included 0.1 mM elastase inhibitor and chymostatin. After incubation at 22°C for 15 minutes, platelet-neutrophil suspensions were pelleted and treated with the proteinase inhibitor cocktail containing 1% Triton X-100. Aliquots of lysates were eluted on 7.5% SDS–polyacrylamide gels, transferred to membranes, and subjected to Western blot analysis using monoclonal antibodies against either P-selectin (AK4) or PSGL-1 (PL-1). The arrow indicates the expected position of the P-selectin fragment recognized by AK4 if P-selectin had been digested by cathepsin G. Data are representative of at least 3 experiments with different donors. (B) Purified125I-labeled P-selectin was treated for 1 hour with purified human elastase or cathepsin G, eluted on 7.5% SDS–polyacrylamide gels, transferred to membranes, and subjected to Western blot analysis using monoclonal antibody against P-selectin (AK4).
Relative loss of PSGL-1 and P-selectin in neutrophil-platelet suspensions
To assess the susceptibility of P-selectin and PSGL-1 to proteolysis under conditions of neutrophil degranulation and platelet activation, washed platelets and purified neutrophils were mixed together in a ratio of 50:1, approximating the platelet-neutrophil ratio found in human blood. In the presence or absence of proteinase inhibitors, neutrophil-platelet suspensions were treated with cytochalasin B and fMLP, TRAP, or both, and were incubated at 22°C for 15 minutes. Platelets and neutrophils were then pelleted and lysed, and the nature and amounts of PSGL-1 and P-selectin associated with the cell pellets were assessed by Western blot analysis using monoclonal antibodies. Figure 5A illustrates that levels of PSGL-1, but not of P-selectin, were reduced in platelet-neutrophil extracts when cytochalasin B and fMLP were present at levels known to cause neutrophil degranulation. Incubation with fMLP alone was not sufficient to cause the loss of PSGL-1 from platelet-neutrophil extracts (data not shown). The loss of PSGL-1 was detected with both monoclonal antibody PL-1 and the anti–N-terminal peptide polyclonal antibody (data not shown), and the loss could be prevented by the inclusion of proteinase inhibitors. Western blots probed with AK4 failed to detect any loss of P-selectin from platelet-neutrophil lysates under any conditions of incubation, including the inclusion of TRAP to activate platelets and to cause surface expression of P-selectin. Furthermore, there was no evidence of generation of a 97-kd fragment of P-selectin in the platelet-neutrophil cell lysates (Figure 5A), characteristic of purified P-selectin proteolysis mediated by cathepsin G (Figure 5B). Consistent with this, surface-expressed P-selectin on TRAP-activated platelets was resistant to proteolysis by purified cathepsin G or elastase (data not shown), suggesting a platelet-specific mechanism for the protection of P-selectin by these proteinases. Similar results were observed in platelet-neutrophil experiments using PAF or LPS to induce degranulation after 1 hour (data not shown).
Identification of the cathepsin G cleavage site in the N-terminus of PSGL-1
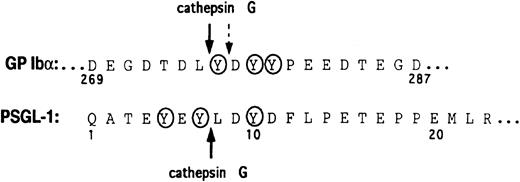
To determine potential N-terminal cleavage site(s) on PSGL-1 for cathepsin G, a synthetic peptide corresponding to residues Thr-3–Glu-17 of mature PSGL-1 was digested, and the fragments were analyzed by reverse-phase HPLC and amino acid sequence analysis. The cathepsin G cleavage site was identified as between Tyr-7 and Leu-8 (Figure 6).
Amino acid sequences of GP Ibα and the N-terminus of PSGL-1.
The cathepsin G cleavage sites are shown with arrows. A secondary cathepsin G cleavage site in GP Ibα is indicated by the broken arrow. Tyrosine sulfation sites are circled.
Amino acid sequences of GP Ibα and the N-terminus of PSGL-1.
The cathepsin G cleavage sites are shown with arrows. A secondary cathepsin G cleavage site in GP Ibα is indicated by the broken arrow. Tyrosine sulfation sites are circled.
Discussion
Arrest of circulating neutrophils on endothelium activated by inflammatory stimuli is crucial for targeting of these cells to sites of injury or infection. The initial contact of neutrophils with activated endothelial cells involves rolling and is mediated predominantly by the interaction of endothelial P-selectin and its counter-receptor on neutrophils, PSGL-1.1-4,8 The importance of selectin-mediated neutrophil rolling in inflammation has been demonstrated in selectin-deficient mice, in which neutrophil rolling is impaired, limiting subsequent adhesion and transmigration.5,6 One mechanism for regulating the interaction between PSGL-1 and P-selectin is down-regulation of PSGL-1 surface expression. For example, Davenpeck et al15 showed that a 50% reduction in PSGL-1 surface expression decreased the capacity of neutrophils to roll on P-selectin by 90%. Down-regulation of PSGL-1 can occur by its redistribution on the membrane surface after neutrophil activation, potentially affecting P-selectin binding.13,14 In resting neutrophils, PSGL-1 is primarily expressed on the tips of the microvilli.14 On neutrophil activation with fMLP, PSGL-1 is redistributed to the uropod of the activated neutrophil.14 Another potential mechanism for down-regulation is shedding or proteolysis of PSGL-1. PSGL-1 is particularly sensitive to regulation by proteolysis because key residues involved in the recognition of P-selectin are found within the N-terminal 10 amino acids of the mature protein.9,37Evidence for this mechanism comes from the recent studies of Davenpeck et al,15 who demonstrated an EDTA-sensitive shedding of PSGL-1 after the treatment of neutrophils with PAF or PMA, possibly because of a metalloproteinase or some other divalent cation-dependent mechanism.
The aim of the present study was to determine whether physiologically relevant proteinases released when neutrophils become activated might also be involved in regulating the PSGL-1–P-selectin interaction. Released elastase and cathepsin G contribute significantly to the inflammatory disease process in ischemia–reperfusion injury38 and in sepsis.39 Treatment of neutrophils with purified cathepsin G or elastase resulted in a rapid loss of P-selectin binding. The susceptibility of PSGL-1 to proteolysis was demonstrated indirectly using inhibitors of neutrophil elastase and cathepsin G to prevent the loss of PSGL-1 from the surfaces of degranulated neutrophils and directly using purified cathepsin G and elastase and isolated PSGL-1 or peptides with sequence-matching PSGL-1. We were also able to confirm, at least in part, the findings of Davenpeck et al15 that there was an EDTA-sensitive component of PSGL-1 loss from neutrophils treated with PMA, high concentrations of PAF, or the combination of PAF and LPS. We were unable, however, and in contrast to their findings, to confirm that lower concentrations of PAF resulted in the loss of PSGL-1 by a proteolytic or shedding mechanism. The reason for this discrepancy is unclear, but our findings are consistent with those of Rainger et al,18 who found that PAF did not cause elastase release from neutrophils even when adherent to P-selectin. The results presented here are also in agreement with reports from others20 describing a loss of P-selectin binding to elastase-treated neutrophils.
Concomitant with these studies, we also detected a susceptibility of P-selectin to proteolysis by the same neutrophil proteinases, using purified platelet P-selectin. However, when neutrophils were mixed with platelets under conditions that induced degranulation, and in the presence of TRAP to activate platelets and cause P-selectin surface expression, we saw evidence of proteolysis of only PSGL-1. Several reports have demonstrated an ability of cathepsin G40-42and elastase42 43 to act on platelets in washed platelet systems in a rapid proteolytic fashion; however, platelet P-selectin has not previously been shown to be a substrate for cathepsin G or elastase. Results presented here suggest that in purified systems, cathepsin G and elastase can both use PSGL-1 and P-selectin as substrates. In mixtures of platelets and neutrophils, only PSGL-1 appears to be a primary target of proteolysis. This may represent a mechanism to modulate PSGL-1 adhesive function on neutrophils under conditions of cell activation.
Additional studies are clearly necessary to evaluate in different inflammatory situations whether the primary mechanism for down-regulation of PSGL-1 function is its redistribution or its shedding or a combination of both. Our findings suggest that shedding of PSGL-1 as a mechanism for down-regulation of function may be limited to severe inflammatory events such as sepsis, in which both LPS and PAF play a pivotal role in the disease process.35 Although not all neutrophil agonists have been examined, it would be anticipated that PSGL-1 redistribution may provide a common mechanism for regulating PSGL-1 function. For example, neutrophil activation in response to fMLP results in a profound redistribution of PSGL-1 surface expression,14 without any apparent PSGL-1 shedding (this study). A rate limiting role for elastase has been proposed for neutrophil extravasation from inflamed microvessels, but not for PAF-induced neutrophil adhesion to inflamed endothelium.17 Because PAF appears critical for regulating the change in adhesive phenotype from rolling through P-selectin–PSGL-1 to firm adhesion through CD18 integrins,13 it is probable that the down-regulation of PSGL-1 function during the inflammatory sequence of rolling, adhesion, and extravasation primarily involves its surface redistribution. These findings are consistent with our observation that physiological concentrations of PAF alone result in no apparent shedding of PSGL-1. This conclusion is supported by the data of Rainger et al,18 who found that neutrophils adherent to P-selectin released elastase in response to fMLP, C5a, and IL-8, but not to PAF. Thus, the mechanism for regulation of PSGL-1 function may predominantly depend on the combination of stimuli to which neutrophils are exposed within a particular inflammatory event.
We have previously shown that mocarhagin, a cobra venom metalloproteinase, not only cleaved PSGL-1 but also cleaved the platelet von Willebrand factor receptor, GP Ibα, between Glu-282 and Asp-283 within a highly negatively charged sequence containing 3 sulfated tyrosines.9,22 In both substrates, cleavage was immediately N-terminal to an aspartate residue and to the C-terminal side of sulfated tyrosine(s). Loss of P-selectin binding to mocarhagin-treated neutrophils or recombinant PSGL-1 was associated with loss of a 10–amino acid peptide from the N-terminus and provided evidence that this region was critical for ligand binding in addition to the sialylated, fucosylated, core-2 O-glycan at Thr-16.9-12,44 Cathepsin G is a component of neutrophil granules16 and, like mocarhagin, has been shown to cleave GP Ibα within the sulfated tyrosine sequence at a site close to but not identical with the mocarhagin site.22,23 Cleavage to the C-terminal side of Tyr-7 would result in loss of the potential tyrosine sulfation sites at Tyr-5 and Tyr-7, implying that one or both of these residues or the Gln-1–Tyr-7 sequence is critical for P-selectin binding. This suggests that Tyr-10, which would remain on PSGL-1 after cleavage at Tyr-7–Leu-8 (see the early time points in Figure 3), may be insufficient in isolation to support P-selectin binding. This conclusion is in accord with the recent crystal structure of a P-selectin–PSGL-1 binding site complex that indicates direct involvement of Tyr-7 in recognition of P-selectin.37 The Tyr-7/Leu-8 cleavage site on PSGL-1 is analogous to the cathepsin G cleavage sites on GP Ibα (Figure 6 and Andrews et al21). Cleavage occurs immediately adjacent to a (sulfated) tyrosine and within an exclusively negatively charged sequence.
In conclusion, the present study demonstrates that P-selectin binding to neutrophils is irreversibly down-regulated after stimulation of neutrophils to undergo secretion or when they are treated with purified cathepsin G or neutrophil elastase. These proteinases cleave partially purified PSGL-1 on a comparable time scale to loss of P-selectin binding, strongly suggesting that proteolysis of PSGL-1 is the likely mechanism for secretion-dependent inhibition of P-selectin binding. Our current findings suggest cathepsin G– and neutrophil elastase–mediated proteolysis of PSGL-1 may, therefore, constitute an autocrine mechanism for the down-regulation of PSGL-1 function on activated neutrophils in severe inflammatory events.
We thank Ms Carmen Llerena and Ms Liesel Fitzgerald for outstanding technical assistance.
Supported by the National Health and Medical Research Council of Australia.
E.E.G. and M.D. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Michael C. Berndt, Baker Medical Research Institute, PO Box 6492, St Kilda Rd, Central, Melbourne, Australia; e-mail: michael.berndt@baker.edu.au.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal