Abstract
The 3′ end of the VWF gene was screened in the affected members of 3 different families with type 2A (phenotype IID) von Willebrand disease (vWD). Exons 49 to 52 of the VWF gene were amplified and screened for mutations by chemical cleavage mismatch detection. Mismatched bands were detected in exon 52 of 2 patients and in exon 51 of a third patient. Using direct DNA sequencing, a heterozygous G8562A transition leading to a Cys2008Tyr substitution was found in all the patients in family 1, and a T8561A transversion leading to a Cys2008Ser substitution was found in both patients from family 2. In a patient from a third family, an 8-base deletion from nucleotide 8437 to 8444 was identified in exon 51. The 2 mutations in exon 52 were reproduced by in vitro site-directed mutagenesis of full-length von Willebrand factor (vWF) cDNA and transiently expressed in COS-7 cells. The corresponding recombinant VWFs for these 2 mutations exhibited the typical aberrant vWF:Ag multimer pattern seen in the plasma of the patients. These 3 mutations demonstrate the importance of other carboxy-terminal cysteines in addition to the reported Cys2010 residue, in the normal dimerization of vWF, and their essential role in the assembly of normal multimeric vWF.
Introduction
von Willebrand factor (vWF) is a multimeric plasma glycoprotein with a major role in primary hemostasis. It is synthesized by endothelial cells1 and megakaryocytes.2After translation in the endoplasmic reticulum as a prepropolypeptide, the signal peptide is cleaved to produce the pro-vWF monomer, which dimerizes through intermolecular disulfide bonds at or near the cysteine-rich carboxy terminal.3 The tail-to-tail dimers are transported to the Golgi apparatus or the post-Golgi compartments,4 where further disulfide bonds form near the amino-terminal of the subunits.5 Both these disulfide bridges are critical in the final multimerization to yield vWF multimers that measure more than 20 million daltons. These large multimers are the most efficient molecules in mediating platelet adhesion. The multimeric pattern of vWF, seen on high-resolution sodium dodecyl sulfate–agarose electrophoresis, shows that each multimer band actually consists of a triplet.6 Limited proteolysis at position Tyr842-Met843 of the mature vWF subunit, produces, in addition to the intact subunit, an N-terminal and a C-terminal fragment contained within vWF multimers.7
Mutations in the VWF gene can cause quantitative deficiencies of vWF (mild type 1 or severe type 3 von Willebrand disease [vWD]) and qualitative abnormalities of vWF (type 2 vWD). In the most recent classification, type 2 vWD is further subdivided into 4 groups, designated types 2A, 2B, 2M, and 2N.8 Functional studies and studies with proteolytic fragments using binding assays and specific monoclonal antibodies to the vWF protein have helped the understanding of the relation between the structure and the function of this highly complex adhesive protein.9 Further investigations have identified mutations causing the various type 2 vWD subtypes and localized them to specific regions of the VWDgene.10 The type 2A subgroups show different vWF antigen (vWF:Ag) multimer patterns, including a rare phenotype previously designated as type IID vWD, which is inherited in an autosomal dominant manner.11,12 This phenotype is unique in having an aberrant vWF:Ag multimer pattern as a result of defective posttranslational processing and vWF multimer assembly, and, in contrast to type IIA and IIB phenotypes, there is little evidence of in vivo proteolysis.13 These differences are manifested in major variations between the vWF:Ag multimer patterns of these 3 phenotypes. Schneppenheim et al14 have reported a new class of mutations in types 2 and 3 vWD, grouped in the C-terminal region of vWF, which are responsible for dimerization and subsequent multimerization of vWF. They also identified a Cys2010Arg mutation in 2 unrelated families with type IID vWD phenotype15 and concluded that the cysteine at position 2010 is essential for normal dimerization of vWF subunits through disulfide bonding of C-terminal domains.
We have previously reported a family with type IID vWD phenotype.12 Here we report the identification of a candidate mutation in 4 members in 4 generations of this family and one other candidate mutation in the same codon in 2 members in 2 generations of another unrelated family. In a third patient, we have also identified an 8-base deletion from nucleotide 8437 to 8444 (nucleotides are numbered from the A of the initiator methionine codon) affecting codons 1966 to 1969 (amino acid residues are numbered from the first amino acid of the mature vWF subunit). This results in a frameshift and in the creation of a cysteine residue at the new codon 1968 and a termination codon at position 1974. The translational products of this heterozygous mutation, expressed in a dominant form, result in a truncated vWF subunit 73 amino acids smaller than the wild type.
Patients, materials, and methods
Coamatic FVIII chromogenic assay kit was purchased from Chromogenix AB (Mölndal, Sweden). Two types of agarose were used for multimer analysis: for the stacking gel, Seakem agarose HGT(P), purchased from FMC Bioproducts (Rockland, ME), and for the separating gel, agarose type VII, purchased from Sigma (Poole, Dorset, United Kingdom). Polyclonal rabbit anti–human vWF antibody was purchased from Dako (High Wycombe, United Kingdom).
Genomic DNA was prepared from leucocytes using the Applied Biosystems 340A nucleic acid extractor (PE Applied Biosystems, Warrington, United Kingdom), and the primers were synthesized on a 391 DNA synthesizer (PE Applied Biosystems). Bioline UK (London, United Kingdom) supplied Bioline BioTaq polymerase. Long-range Taq polymerase was from Roche Diagnostics, (Lewes, United Kingdom). PerkinElmer DNA thermal cyclers were used in all polymerase chain reaction (PCR) reactions (PE Applied Biosystems). PCR products were purified using the Geneclean II kit (Bio 101 Inc, Vista, CA). DNA sequencing was carried out using an ABI Prism 310 Genetic Analyzer with big dye terminator cycle sequencing chemistry (PE Applied Biosystems). Restriction enzymes were purchased from Promega (Southampton, United Kingdom).
vWF expression vector pSVHvWF1 was kindly provided by Dr A. Inbal (Sheba Medical Centre, Israel). Shuttle vector pSP70 was from Promega. Site-directed mutagenesis was performed using the unique site elimination (USE) mutagenesis kit from Amersham Pharmacia Biotech (St Albans, United Kingdom). All tissue culture reagents were supplied by Life Technologies (Paisley, United Kingdom). The stirred ultrafiltration device was from Flowgen (Ashby de la Zouch, United Kingdom). Plasmid DNAs for transfection were purified using the Wizard Purefection plasmid DNA purification system (Promega). All other consumables were purchased from Sigma unless otherwise stated.
Blood samples
Four patients from a large family12 with type IID vWD phenotype (family 1) were available and consented to genetic investigation. These were the propositus (now deceased), his niece, her daughter, and her newly born daughter. Two patients in family 2, a mother and a daughter, were the only available members for investigation. In family 3, only one affected member was available for study. Blood was obtained by venipuncture; it was mixed 9 parts blood to 1 part trisodium citrate and was centrifuged for platelet-poor plasma and platelet-rich preparation.
Phenotypic analysis
Bleeding time analysis was carried out by the Ivy method in families 1 and 2 and by the Simplate method in family 3. FVIII coagulant activity (VIII:C) was measured on freshly separated plasma using a chromogenic assay. The von Willebrand factor antigen (vWF:Ag) was measured by enzyme-linked immunosorbent assay (ELISA)16 and ristocetin cofactor activity (vWF:Ricof) by fixed platelet aggregometry17 on frozen and thawed platelet-poor plasma. Plasma vWF:Ag multimeric analysis was carried out on platelet-poor plasma, and platelet vWF:Ag from washed platelets was obtained from platelet-rich preparation as previously described.18 A type IID phenotype plasma sample, obtained as part of an international multicenter comparison of von Willebrand factor multimer sizing techniques,19 was used for comparison with vWF:Ag multimers of patients examined in this study.
Mutation screening
High-molecular-weight genomic DNA was prepared from leucocytes and used for the PCR amplification of VWF gene sequences. Specific primer sequences for exons 49, 50, and 52 were in accordance with Schneppenheim et al,15 and those for exon 51 were in accordance with Zhang et al.20 Optimized annealing temperatures for the PCR reactions were 64°C, 63°C, 68°C, and 62°C for exons 49 to 52, respectively. Amplified products were purified before mutation screening by the chemical cleavage mismatch detection (CCMD) technique.21 Identified mismatched bands were investigated by direct DNA sequencing, using the dideoxy chain termination method. The identified candidate mutations were authenticated by ScaI (family 1) and SacI (family 2) restriction enzyme digestion according to the manufacturer's instructions, followed by electrophoresis on a 2% agarose gel.
Expression of von Willebrand factor in COS-7 cells
vWF expression vector pSVHvWF1 consists of the full-length vWF cDNA inserted into the expression vector pSV7d,22 as described elsewhere.23 Full-length EcoRI VWF cDNA fragment was ligated into the shuttle vector pSP70, producing the vector pSP70VWFEcoRI. An EcoRV fragment (nucleotide 8486 in exon 51 to nucleotide 67 in pSP70), was then ligated into another pSP70 vector, generating the shuttle vector pSP70VWFEcoRV. To introduce the 2 nucleotide substitutions (G8562A and T8561A) for the candidate mutations C2008Y and C2008S, respectively, site-directed mutagenesis24 was performed on the resultant shuttle vector using the USE mutagenesis kit. The USE selection primer (SDMpSP70HpaI; Table 1) contains a mutation in a unique, nonessential restriction site (HpaI), providing the basis for selection. Mutagenic primers (SDMC2008Y and SDMC2008s) (Table 1) introduced the desired target candidate mutations.
Oligonucleotide primers used for Unique Site Elimination site-directed mutagenesis
| Name . | Sequence . | Position . |
|---|---|---|
| SDM C2008Y | 5′-GTG CAG GAC CAG TAC TCC TGC TGC TC-3′ | 8549-8574 |
| (in vWF cDNA) | ||
| SDM C2008S | 5′-CAG GAC CAGAGC TCC TGC TGC TCT-3′ | 8552-8575 |
| (in vWF cDNA) | ||
| SDMpSP70HpaI | 5′-CGA TAA GCC AGG GTACCC TGC ATT AAT GAA TCG-3′ | 78-110 |
| (in pSP70) |
| Name . | Sequence . | Position . |
|---|---|---|
| SDM C2008Y | 5′-GTG CAG GAC CAG TAC TCC TGC TGC TC-3′ | 8549-8574 |
| (in vWF cDNA) | ||
| SDM C2008S | 5′-CAG GAC CAGAGC TCC TGC TGC TCT-3′ | 8552-8575 |
| (in vWF cDNA) | ||
| SDMpSP70HpaI | 5′-CGA TAA GCC AGG GTACCC TGC ATT AAT GAA TCG-3′ | 78-110 |
| (in pSP70) |
Underlined nucleotides indicate base substitutions.
Clones containing the desired base substitutions were directly sequenced as described above to verify the presence of the candidate vWD mutation and to exclude any errors from in vitro amplification.EcoRV fragments containing the candidate vWD mutations were then cloned into the vector pSP70EcoRI. Finally, the full-lengthEcoRI fragments of vWF cDNA containing the desired candidate mutations were cloned back into the full-length vector, pSVHVWF1, generating the expression vectors pSVHC2008Y and pSVHC2008S. The correct size and orientation of the inserts were verified by restriction analysis and direct sequencing as described above. Because the third mutation was in fact an 8–base pair (bp) deletion, the expression vector pSVHdel51 was produced by subcloning a segment of patient cDNA produced by long-range PCR into the vector pSVHVWF1 to produce the expression vector pSVHdel51. Plasmid DNA for transfection was purified using the Wizard Purefection Plasmid DNA purification system (Promega).
The monkey kidney cell line COS-7 was transfected with 50 μg pSVHvWF1, pSVHC2008Y, pSVHC2008S, or pSVHdel51 using a diethyl aminoethyl (DEAE)–dextran–based technique.25 26Cotransfection experiments were performed using a total of 50 μg plasmid DNA comprising a 1:1 or a 1:9 ratio of wild-type (WT) and mutant constructs. COS-7 cells were grown in Dulbecco modified essential medium (DMEM) supplemented with 2 mM L-glutamine, 50 IU/mL penicillin, 50 μg/mL streptomycin, and 10% (vol/vol) fetal calf serum. Cells in the log phase of growth were used to seed tissue culture flasks so that the cells were 60% to 70% confluent the next day. These cells were transfected with 50 μg plasmid DNA in Tris-buffered saline, supplemented with (500 μg/mL) DEAE-dextran. After incubation for 1 hour at 37°C, the transfection media were removed and the cells were incubated for another 4 hours in DMEM containing 100 mM chloroquine, before recovery overnight in DMEM. Cells were cultured for 72 hours in AIM-V serum-free medium supplemented with L-glutamine, penicillin, and streptomycin as above. Conditioned medium from cells mock transfected with the shuttle vector pSP70 was used as a control. Benzamidine, N-ethylmaleimide, EDTA, and phenylmethylsulfonyl fluoride were added to the conditioned medium before centrifugation to remove cellular debris. The medium was concentrated using a stirred ultrafiltration device before storage at −20°C. vWF:Ag levels were measured by ELISA, and the multimeric structure was determined as described above.
Results
Patient diagnosis and phenotypic results
Clinical symptoms of the patients are summarized in Table2, and the results of the hemostatic tests that resulted in the diagnosis of vWD are shown in Table3. Autosomal dominant transmission of the disease was observed only in families 1 and 2. The plasma vWF:Ag multimer pattern that classified the patients as having type IID vWD is shown in Figure 1.
Clinical symptoms in affected members of the families investigated
| Family . | Patient . | Generation . | Sex . | Degree of bruising . | Prolonged bleeding from cuts . | Epistaxes . | Bleeding after dental extraction . | Gastrointestinal tract bleeding . | Menorrhagia . |
|---|---|---|---|---|---|---|---|---|---|
| F1 | P1 | II:1 | M | + | Unknown | None | ++ | ++ | N/A |
| F1 | P2 | III:1 | F | ++ | + | None | + | None | ++ |
| F1 | P3 | IV:1 | F | ++ | + | + | + | None | ++ |
| F1 | P4 | V:1 | F | No clinical symptoms to date (baby) | |||||
| F2 | P1 | I:1 | F | No clinical symptoms to date | |||||
| F2 | P2 | II:1 | F | + | Unknown | + | Unknown | ++ | ++ |
| F3 | P1 | — | F | — | Unknown | Unknown | Unknown | None | +++ |
| Family . | Patient . | Generation . | Sex . | Degree of bruising . | Prolonged bleeding from cuts . | Epistaxes . | Bleeding after dental extraction . | Gastrointestinal tract bleeding . | Menorrhagia . |
|---|---|---|---|---|---|---|---|---|---|
| F1 | P1 | II:1 | M | + | Unknown | None | ++ | ++ | N/A |
| F1 | P2 | III:1 | F | ++ | + | None | + | None | ++ |
| F1 | P3 | IV:1 | F | ++ | + | + | + | None | ++ |
| F1 | P4 | V:1 | F | No clinical symptoms to date (baby) | |||||
| F2 | P1 | I:1 | F | No clinical symptoms to date | |||||
| F2 | P2 | II:1 | F | + | Unknown | + | Unknown | ++ | ++ |
| F3 | P1 | — | F | — | Unknown | Unknown | Unknown | None | +++ |
Phenotypic data for affected members of the families investigated
| Family . | Patient . | Generation . | Bleeding time (min) . | FVIII:C (U/mL) . | vWF:Ag (U/mL) . | vWF:Ricof (U/mL) . | vWF:Ag Multimer . | |
|---|---|---|---|---|---|---|---|---|
| Plasma . | Platelet . | |||||||
| F1 | P1 | II:1 | 17 | 1.42 | 2.02 | 0.25 | Abnormal | Abnormal |
| F1 | P2 | III:1 | 17 | 0.64 | 1.16 | 0.33 | Abnormal | Abnormal |
| F1 | P3 | IV:1 | 19 | 0.58 | 0.89 | 0.30 | Abnormal | Abnormal |
| F1 | P4 | V:1 | Unavailable | 1.14 | 1.05 | 0.59 | Abnormal | Not done |
| F2 | P1 | I:1 | Unavailable | Unavailable | Unavailable | Unavailable | Abnormal | Not done |
| F2 | P2 | II:1 | >20 | 0.66 | 0.49 | 0.27 | Abnormal | Abnormal |
| F3 | P1 | — | >20 | 0.78 | 0.54 | 0.27 | Abnormal | Abnormal |
| Family . | Patient . | Generation . | Bleeding time (min) . | FVIII:C (U/mL) . | vWF:Ag (U/mL) . | vWF:Ricof (U/mL) . | vWF:Ag Multimer . | |
|---|---|---|---|---|---|---|---|---|
| Plasma . | Platelet . | |||||||
| F1 | P1 | II:1 | 17 | 1.42 | 2.02 | 0.25 | Abnormal | Abnormal |
| F1 | P2 | III:1 | 17 | 0.64 | 1.16 | 0.33 | Abnormal | Abnormal |
| F1 | P3 | IV:1 | 19 | 0.58 | 0.89 | 0.30 | Abnormal | Abnormal |
| F1 | P4 | V:1 | Unavailable | 1.14 | 1.05 | 0.59 | Abnormal | Not done |
| F2 | P1 | I:1 | Unavailable | Unavailable | Unavailable | Unavailable | Abnormal | Not done |
| F2 | P2 | II:1 | >20 | 0.66 | 0.49 | 0.27 | Abnormal | Abnormal |
| F3 | P1 | — | >20 | 0.78 | 0.54 | 0.27 | Abnormal | Abnormal |
C indicates coagulation; vWF, von Willebrand factor; Ag, antigen; Ricof, ristocetin cofactor.
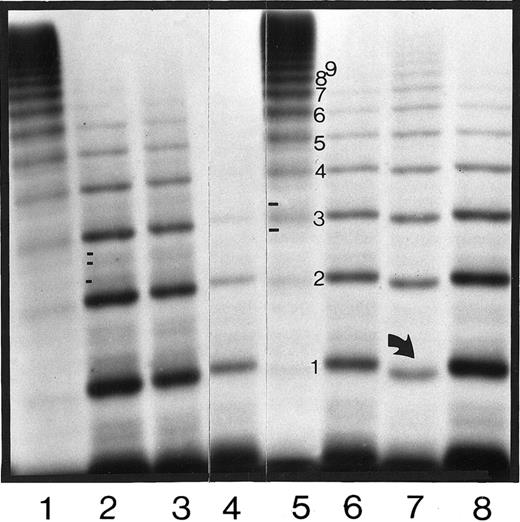
Autoradiograph of plasma vWF:Ag multimers of the affected members of families 1, 2, and 3.
Plasma samples electrophoresed in a 1.8% SDS-agarose gel and visualized by sodium iodide 125–labeled anti-vWF:Ag monoclonal antibody showing the abnormal multimer band patterns in F1-P1, F1-P2, F1-P3, F2-P1, and F3-P1 in lanes 2, 3, 4, 6, and 7, respectively. A type IID phenotype international reference plasma11 19 is shown in lane 8, and 2 normal plasmas are shown in lanes 1 and 5. The direction of electrophoresis is from top to bottom. The arrow in lane 7 indicates the faster migrating central band 1 in F3-P1. Differences in the minor satellite bands between normal and IID plasma (2 bands in normal and 3 bands in IID plasma) are shown in lanes 2 and 5, respectively.
Autoradiograph of plasma vWF:Ag multimers of the affected members of families 1, 2, and 3.
Plasma samples electrophoresed in a 1.8% SDS-agarose gel and visualized by sodium iodide 125–labeled anti-vWF:Ag monoclonal antibody showing the abnormal multimer band patterns in F1-P1, F1-P2, F1-P3, F2-P1, and F3-P1 in lanes 2, 3, 4, 6, and 7, respectively. A type IID phenotype international reference plasma11 19 is shown in lane 8, and 2 normal plasmas are shown in lanes 1 and 5. The direction of electrophoresis is from top to bottom. The arrow in lane 7 indicates the faster migrating central band 1 in F3-P1. Differences in the minor satellite bands between normal and IID plasma (2 bands in normal and 3 bands in IID plasma) are shown in lanes 2 and 5, respectively.
The diagnosis of type IID phenotype was confirmed in the patient from family 3 by an abnormal multimer pattern similar to what the other type IID patients had, but with some significant differences. More multimer bands were present in the patient from family 3, and were of uniform intensity, but, more important, they migrated faster than did multimers from the other type IID families (Figure 1). The faster rate of migration in the patient from family 3 is best seen in the first 2 multimer bands, especially when using a higher resolving 1.8% agarose gel (Figure 1).
Candidate mutations
Mutation screening was carried out by CCMD, followed by direct DNA sequencing of exons 49 to 52 of the VWF gene. CCMD of exon 52 PCR products led to the detection of a mismatched hydroxylamine band in the affected members of families 1 and 2. In the patient from family 3, multiple mismatched bands were identified in exon 51 with both hydroxylamine and osmium tetroxide.
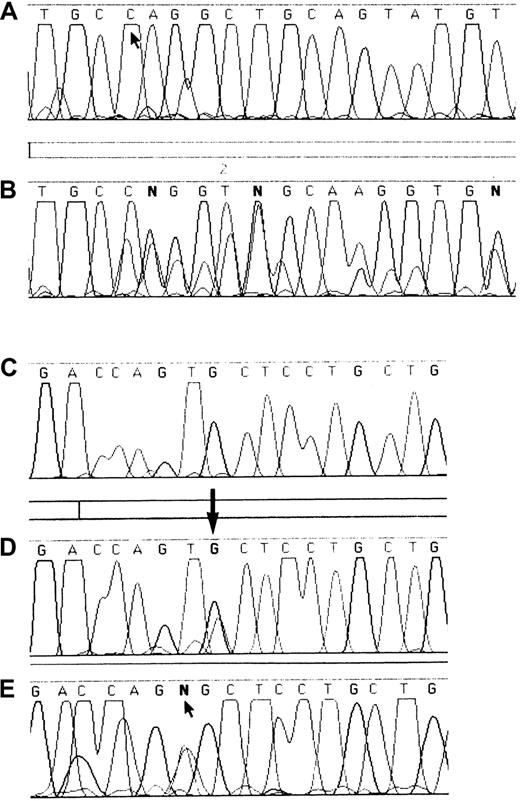
Mutations causing the mismatched bands were identified by direct DNA sequencing and, when possible, were authenticated by restriction enzyme analysis. In family 1, DNA sequencing identified a G8562A transition in a heterozygous form, leading to a Cys2008Tyr substitution. In family 2, DNA sequencing identified a T8561A transversion predicting a Cys2008Ser substitution (Figure 2). In patient 3, exon 51 was sequenced and a heterozygous 8-base deletion from nucleotide 8437 to 8444 was identified. The deletion affecting codons 1966 to 1969 resulted in a frameshift leading to the creation of a cysteine residue at the new 1968 position and a termination codon at 1974 (Figure 2).
DNA sequence analysis of exons 51 and 52 of the
VWF gene in WT and affected members of families 1, 2, and 3. Panel A shows the WT sequence of exon 51 from T8434 to T8457, with an arrow indicating nt 8437 where the deletion begins in patient F3P1. Panel B shows the DNA sequence of the patient from family 3, with a series of superimposed bases as a result of the heterozygous deletion beginning at position 8437. Panels C to E show the sequence of exon 52 from nucleotide 8555 to 8571. Panel C shows the WT sequence. Panel D shows the sequence from one affected member from family 1, with a heterozygous G8562A transition. Panel E shows the sequence from patient F2P2, showing a T8561A transversion.
DNA sequence analysis of exons 51 and 52 of the
VWF gene in WT and affected members of families 1, 2, and 3. Panel A shows the WT sequence of exon 51 from T8434 to T8457, with an arrow indicating nt 8437 where the deletion begins in patient F3P1. Panel B shows the DNA sequence of the patient from family 3, with a series of superimposed bases as a result of the heterozygous deletion beginning at position 8437. Panels C to E show the sequence of exon 52 from nucleotide 8555 to 8571. Panel C shows the WT sequence. Panel D shows the sequence from one affected member from family 1, with a heterozygous G8562A transition. Panel E shows the sequence from patient F2P2, showing a T8561A transversion.
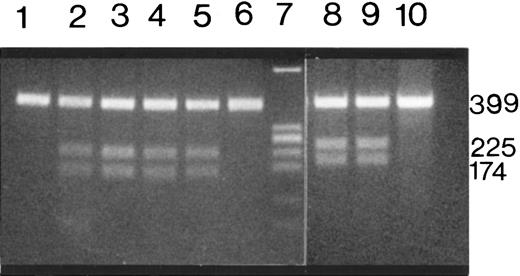
Mutations in exon 52 were further investigated using restriction enzyme digestion followed by electrophoresis. Mutations in families 1 and 2 created a new ScaI and a new SacI restriction site, respectively. When the PCR products from the 4 patients in family 1 were digested with ScaI, an uncut 399-bp fragment (normal allele) and cut 225- and 174-bp fragments (abnormal allele) were present, demonstrating the heterozygous nature of the mutation in these patients. PCR products from the mother and daughter in family 2 were digested with SacI; both uncut and cut fragments were present, indicating heterozygosity for this mutation in this family (Figure 3). Exon 52 of the VWFgene in 50 unrelated persons was also amplified and digested withScaI and SacI, and the new restriction sites identified in patients from family 1 and 2 were not found in any of the 100 normal alleles.
Restriction enzyme analysis of exon 52 PCR products from affected members of families 1 and 2.
ScaI-restricted PCR products of patients F1-P1, F1-P2, F1-P3, F1-P4 (lanes 2 to 5) and a healthy person (lane 6). Lanes 8 and 9 contain the SacI-digested PCR products from 2 affected members of family 2 (F2-P1-and F2-P2). Lanes 1 and 10 contain unrestricted PCR products from F1-P1 and F2-P2. Amino acid substitutions of the 2 mutations in exon 52 create a newScaI and SacI restriction site, respectively, and the 399-bp PCR product is restricted to 225-bp and 174-bp fragments. All the patients show the 225-bp and 174-bp fragments in addition to a 399-bp PCR product, whereas the 2 normal samples show only the unrestricted 399-bp PCR product. Lane 7 contains the plasmid pBR322 digested with EcoRI and AvaII as a molecular-weight marker.
Restriction enzyme analysis of exon 52 PCR products from affected members of families 1 and 2.
ScaI-restricted PCR products of patients F1-P1, F1-P2, F1-P3, F1-P4 (lanes 2 to 5) and a healthy person (lane 6). Lanes 8 and 9 contain the SacI-digested PCR products from 2 affected members of family 2 (F2-P1-and F2-P2). Lanes 1 and 10 contain unrestricted PCR products from F1-P1 and F2-P2. Amino acid substitutions of the 2 mutations in exon 52 create a newScaI and SacI restriction site, respectively, and the 399-bp PCR product is restricted to 225-bp and 174-bp fragments. All the patients show the 225-bp and 174-bp fragments in addition to a 399-bp PCR product, whereas the 2 normal samples show only the unrestricted 399-bp PCR product. Lane 7 contains the plasmid pBR322 digested with EcoRI and AvaII as a molecular-weight marker.
Expression of recombinant VWF in COS-7 cells
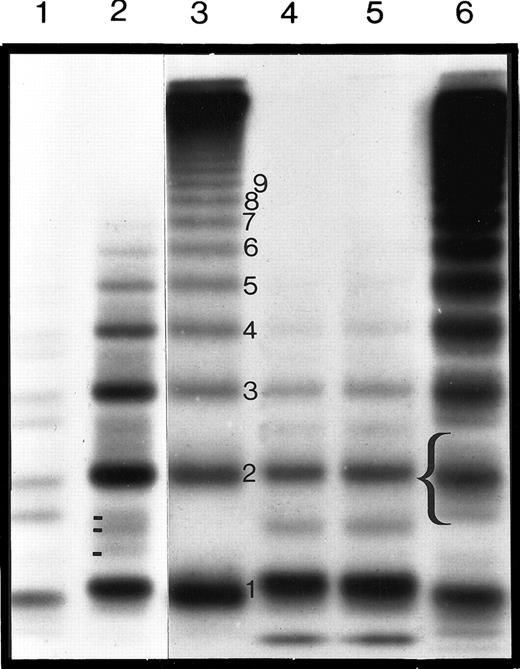
Wild-type (pSVHvWF1) and mutant (pSVHC2008Y, pSVHC2008S, and pSVHdel51) full-length cDNAs were transiently expressed in COS-7 cells under the control of the SV40 late promoter in 6 separate transfection experiments. Concentrations of vWF:Ag in the conditioned medium of COS-7 cells transfected with pSVHC2008Y and pSVHC2008S were not significantly reduced compared to that of cells transfected with pSVHvWF1, and they reached maximal levels of 0.1 U/mL after concentration. However, the level of vWF:Ag in the conditioned medium of cells transfected with pSVHdel51 was below the level of reliable quantitation by ELISA (more than 0.01 U/mL). Conditioned medium from COS-7 cells mock-transfected with pSP70 did not contain any detectable vWF:Ag. Analysis of the recombinant vWF:Ag (rvWF:Ag) by sodium dodecyl sulfate (SDS)–agarose gel electrophoresis showed that whereas the wild-type rVWF contained the full range of molecular-weight multimers, pSVHC2008Y and pSVHC2008S rVWF showed a reduction in the high- and intermediate-molecular-weight multimers with aberrant triplet structure. This is consistent with the candidate mutation as the causative mutation of the observed type IID phenotype in these patients (Figure 4). Whereas in type IID plasma 3 minor bands could be seen between the major bands, only one minor band was visible in the type IID rVWF. The minor triplet bands seen in plasma vWF are not present in the WT rVWF (Figure 4). Multimeric analysis of rVWF from cells transfected with pSVHdel51 failed to show any vWF:Ag multimers, which may have been a result of the low levels of vWF:Ag or the instability of any vWF expressed.
Autoradiograph of the multimeric analysis of plasma and rVWF from COS-7 cells after transfection with WT or type IID vWF cDNA.
The autoradiograph shows plasma and rVWF samples electrophoresed on a high-resolution 1.8% SDS-agarose gel and visualized by autoradiography. Electrophoresis is from top to bottom and shows the high-, intermediate-, and low-molecular-weight multimers, together with the triplet structure of each of the multimer bands. Normal plasma is shown in lane 6, typical type IID vWD plasma in lane 2, and type IIA vWD plasma in lane 1. Wild-type rVWF, C2008S rVWF, and C2008Y rVWF are in lanes 3, 4, and 5, respectively. The bracket indicates the triplet structure of the second major multimer band, and different minor satellite bands (2 in normal and 3 in IID plasma) are also indicated. Note that no minor satellite bands can be seen in the WT rVWF.
Autoradiograph of the multimeric analysis of plasma and rVWF from COS-7 cells after transfection with WT or type IID vWF cDNA.
The autoradiograph shows plasma and rVWF samples electrophoresed on a high-resolution 1.8% SDS-agarose gel and visualized by autoradiography. Electrophoresis is from top to bottom and shows the high-, intermediate-, and low-molecular-weight multimers, together with the triplet structure of each of the multimer bands. Normal plasma is shown in lane 6, typical type IID vWD plasma in lane 2, and type IIA vWD plasma in lane 1. Wild-type rVWF, C2008S rVWF, and C2008Y rVWF are in lanes 3, 4, and 5, respectively. The bracket indicates the triplet structure of the second major multimer band, and different minor satellite bands (2 in normal and 3 in IID plasma) are also indicated. Note that no minor satellite bands can be seen in the WT rVWF.
The vWF:Ag levels of rVWF by COS-7 cells after cotransfection with WT and mutant cDNA sequences did not vary significantly compared with cells transfected with single WT or mutant cDNA. When WT or mutant cDNA was expressed alone in COS-7 cells, the multimeric pattern reflected the expected expressed cDNA (Figure5, lanes 1 and 2). In the cotransfection experiments, a pattern emerged similar to that observed when WT or mutant supernatants were mixed (Figure 5, lane 3). If the proportion of WT:mutant cDNA was varied, this was reflected in the relative proportions of WT or mutant pattern seen on the multimer gel.
Autoradiograph of the multimeric analysis of rVWF from COS-7 cells after cotransfection of WT and type IID vWF cDNA.
The autoradiograph shows rVWF samples electrophoresed in 1.4% SDS-agarose gel and visualized by autoradiography. Electrophoresis is from top to bottom and shows the high-, intermediate-, and low-molecular-weight multimers. rVWF from cells transfected with WT and IID cDNA are shown in lanes 1 and 2, respectively. Lane 3 shows rVWF from COS-7 cells transfected with both WT and IID cDNA.
Autoradiograph of the multimeric analysis of rVWF from COS-7 cells after cotransfection of WT and type IID vWF cDNA.
The autoradiograph shows rVWF samples electrophoresed in 1.4% SDS-agarose gel and visualized by autoradiography. Electrophoresis is from top to bottom and shows the high-, intermediate-, and low-molecular-weight multimers. rVWF from cells transfected with WT and IID cDNA are shown in lanes 1 and 2, respectively. Lane 3 shows rVWF from COS-7 cells transfected with both WT and IID cDNA.
Discussion
Because it has been shown that the underlying cause of the autosomal dominant type IID vWD phenotype is defective dimerization at the C-terminal of vWF,15 we targeted the 3′ end of theVWF gene and identified 3 different mutations in our type IID patients. The first mutation, identified in 4 patients in 4 generations of family 1, was a heterozygous cysteine-to-tyrosine substitution at position 2008. The second mutation was a cysteine-to-serine substitution at the same codon, in the mother and daughter of family 2. These 2 mutations were confirmed by restriction enzyme analysis and were found to be absent in 100 normal unrelated alleles screened. These mutations were only 2 amino acids upstream in exon 52 of the Cys2010Arg substitution previously described in 2 unrelated patients with a similar type IID vWD phenotype.15 We also identified a third mutation in a single patient with type IID vWD phenotype, but with slight differences in the vWF multimer pattern. The multimeric characteristics included loss of high-molecular-weight multimers, aberrant type IID vWD multimer pattern, faster migrating central bands of each of the multimers, dissimilar central multimer band intensity compared to typical type IID vWD multimers, and lack of the very intense first multimer band (Figure1, lane 7). The faster migration of the multimer bands suggests that they are of lower molecular weight than those in the other 2 families. This mutation therefore leads to the synthesis of a smaller subunit. Parents and other relatives of this patient were unavailable for study but were said to be asymptomatic, suggesting the possibility of a de novo origin of the mutation in this patient. This mutation is an 8-base deletion causing a frameshift in exon 51, affecting codons 1966 to 1969, and resulting in early termination of the polypeptide chain at position 1974, the loss of 10 cysteine residues, and the creation of a new cysteine residue in the new 1968 codon. Sequencing data (Figure 2) show that this mutation appears to be heterozygous at the DNA level. The abnormal gene product suggests that only the mutant allele is expressed, perhaps indicating that it is dominant in its expression, or that the apparently WT allele is not expressed.
These 3 mutations are within 90 amino acids of the cysteine knot, or the CK domain, in the C-terminal of vWF that has been shown to be essential for dimerization of vWF multimers.15,27 The sulfhydryl groups—16 of 17 cysteine residues encoded for by exons 49 to 52—are bonded in normal vWF28; therefore, the loss of one sulfhydryl group through substitution results in the loss of one intersubunit bond. If this bond lies in a critical conformational position, then the folding of the C-terminal end of the protein would be incorrect, thereby affecting dimerization. Schneppenheim et. al15 suggested that the type IID vWD phenotype arises from a single mutation involving Cys2010 within the C-terminal sequence of Cys2008-Ser-Cys-Cys2010 that abolishes dimerization and, hence, that it is the only cysteine responsible for the type IID phenotype. However, the families we report here have mutations involving another cysteine (Cys2008) residue or an 8-base deletion in exon 51, which produces a similar phenotype.
Recently, Katsumi et al,29 using a recombinant C-terminal cysteine knotlike domain of human vWF, have suggested that Cys2008, Cys2010, and Cys2048 are the only cysteine residues that can form the intersubunit bonds between vWF monomers in the cysteine knotlike domain at the vWF C-terminal, a common motif that is found in the dimerization region of several proteins. They reported that the substitution of Cys2008 or Cys2010 completely abrogated dimerization, whereas the mutation of Cys2048 did not completely prevent dimerization. In an earlier report, Voorberg et al30demonstrated that intermolecular disulfide bonding of the amino terminal domains of vWF proceeded independently of disulfide bridge formation between C-terminal domains—that is, dimerization and multimerization of vWF molecules were 2 independent processes. This hypothesis supports the theory that the aberrant multimer pattern seen in the plasma of the 3 patients with type IID vWD is the result of vWF multimerization in the absence of normal vWF dimerization. However, other single-base substitutions affecting cysteine residues, such as Cys2043Arg31 and Cys1991Trp,15 which were shown to prevent C-terminal dimerization, did not produce a type IID multimer pattern. Another reported frameshift mutation caused by a heterozygous single-base deletion in a CC dinucleotide (8565/6delC) in exon 5232 gave rise to a rare type IIE vWD phenotype rather than a type IID pattern. Therefore, it appears that even though all these mutations involve cysteine residues at the C-terminal and are potentially important for dimerization, only the substitution of some residues may result in a type IID phenotype.
The continuous in vivo proteolysis of vWF in healthy persons and the enhanced proteolysis that occurs between Tyr842 and Met843 in type IIA and IIB vWD are now well documented. Zimmerman et al,13using vWF subunit analysis, showed the presence of a 176-kd fragment associated with proteolysis in plasma from patients with these 2 phenotypes and in plasma from healthy persons. However, this fragment is almost absent in the types IIC, IID, and IIE phenotypes, all of which have an aberrant structure of individual oligomers. In their model for a vWF subunit molecule, Fowler et al33 proposed that proteolysis causes vWF fragmentation and triplet band formation. They suggested that the site for proteolysis is situated at the C-terminal end of the vWF molecule, which is also responsible for dimerization. The deletion seen in exon 51 of our unusual patient, P1F3, with the resultant truncated subunit and the additional amino acid loss in the C-terminal domain, may explain the dissimilar central multimer band intensity and the faster migration of the central band compared with the aberrant IID vWF multimer pattern seen in the other patients.
The 2 mutations involving Cys2008 were reproduced by in vitro site-directed mutagenesis of full-length vWF cDNA and were transiently expressed in COS-7 cells. Both corresponding rVWFs exhibited aberrant vWF:Ag multimers similar to those seen in the plasma of these patients, except that they lacked the intermediate minor bands. The difference between the number of minor multimer bands in rVWF and the corresponding patient plasma could be the result of the lack of proteolysis in the in vitro culture system, as evidenced by the lack of triplet structure seen in WT rVWF compared to normal plasma VWF. Variations in the intensity of the multimer bands in rVWF may be explained by the variability of expression levels between transient transfections. All 3 mutations affect cysteine residues and may account for the observed impaired dimerization. In spite of the expected dimerization defect, the expression of mutant full-length vWF resulted in the synthesis of a dimeric protein, suggesting intact disulfide bonding at the amino-terminus of the mutant vWF. Small amounts of tetramers were also observed in the mutants studied, suggesting a degree of dimerization, possibly based on the additional C-terminal cysteine residues involved in intersubunit bonding. Therefore, the mutations resulted in partial, rather than total, inability for multimerization between the subunits. However, the ability of the mutant subunits to multimerize was limited, as shown by the fact that only 6 to 8 multimers were seen. The other consistent feature was that between each multimer, a minor band was seen (Figure 1). We postulate that these are intermediary stages of assembly, with uneven numbers of monomers, whereas the main multimer bands have even numbers of monomers. In the coexpression experiments, this was explored further by including the option for normal and mutant monomers. The multimer pattern resembles the superimposition of both patterns. It is not possible to comment on the degree to which mutant and WT monomers may interact; additional experiments are needed to clarify this.
Study of rVWF from COS-7 cells after transfection with the mutation from patient P1F3 proved more difficult because of the low levels of rVWF transiently expressed by the transfected cells. This may be owing to the lower level of expression of this rVWF or to the fact that any mutant rVWF expressed without WT is unstable. Coexpressed material showed a multimer pattern similar to that seen in plasma but, again, with a larger number of high-molecular-weight multimers. It is clear that this truncated vWF molecule, resulting from the early termination of the polypeptide chain, results in the secretion of a smaller vWF subunit, as evidenced by the faster migration in the multimeric analysis of plasma. The frameshift completely alters subsequent codons and leads to the loss of 10 cysteine residues to be replaced by a single cysteine residue before the premature termination codon. This could give rise to alternative intermolecular disulfide bonding, producing the type IID phenotype seen in the patient.
We have shown that 2 separate mutations with different amino acid substitutions of Cys2008 in exon 52 in 2 unrelated families (families 1 and 2) result in aberrant vWF:Ag dimerization (as in the reported Cys2010 mutation15) and thereby in a variant vWD phenotype. This emphasizes the importance of both these cysteine residues for normal assembly of vWF. The third patient (P1F3), whose variant phenotype differs from that of these other patients, has a different mechanism for producing defective dimerization (a frameshift deletion resulting in early termination of translation in exon 51 and truncation of the protein). The truncation of the protein results in the loss of 10 cysteine residues and the creation of a new cysteine site so that the truncated protein is still able to dimerize with the remaining cysteines. Given the difference in multimers and the molecular basis for the patient phenotype, perhaps this should be regarded as a separate subtype rather than as subtype IID.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
M. Said Enayat, Department of Haematology, The Birmingham Children's Hospital NHS Trust, Steelhouse Lane, Birmingham, B4 6NH, United Kingdom; e-mail:said.enayat@bhamchildrens.wmids.nhs.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal