Abstract
Gene transfer into hematopoietic stem cells (HSCs) is an ideal treatment strategy for many genetic and hematologic diseases. However, progress has been limited by the low HSC transduction rates obtained with retroviral vectors based on murine leukemia viruses. This study examined the potential of vectors derived from the nonpathogenic human foamy virus (HFV) to transduce human CD34+ cells and murine HSCs. More than 80% of human hematopoietic progenitors present in CD34+ cell preparations derived from cord blood were transduced by a single overnight exposure to HFV vector stocks. Mice that received transduced bone marrow cells expressed the vector-encoded transgene long term in all major hematopoietic cell lineages and in over 50% of cells in some animals. Secondary bone marrow transplants and integration site analysis confirmed that gene transfer occurred at the stem cell level. Transgene silencing was not observed. Thus vectors based on foamy viruses represent a promising approach for HSC gene therapy.
Introduction
Although vectors based on oncoviruses such as murine leukemia viruses (MLVs) can transduce murine hematopoietic stem cells (HSCs) efficiently when packaged with the ecotropic envelope protein,1-3 this pseudotype only infects rodent cells. In primate experiments, the poor performance of MLV vectors is thought to be due to low cellular receptor levels for the nonecotropic pseudotypes used4 or the quiescent state of HSCs, because mitosis is required for nuclear entry of MLV genomes.5 Several alternative retroviral vector systems are being evaluated in hematopoietic cells, including vectors pseudotyped with different envelope proteins or based on lentiviruses.6-8 Issues of receptor expression levels, cell cycle requirements, and possible safety risks still need to be resolved before these vectors are successfully used for HSC gene therapy. In an attempt to overcome the limitations of existing vector systems, we and others have developed vectors based on the spumavirus family of retroviruses (foamy viruses).9-12
Foamy viruses are nonpathogenic retroviruses with a wide tissue tropism that are commonly found in mammalian species.13,14Serologic and polymerase chain reaction (PCR)–based surveys have failed to identify naturally occurring foamy virus infections in human populations, and the original “human” foamy virus (HFV) isolate is now thought to be a chimpanzee virus variant.15-17 The genomic organization of HFV is more complex than that of oncoviruses; in addition to gag, pol, and envgenes, 3 open reading frames are located between env and the 3′LTR (bel 1-3) that are expressed from an internal viral promoter (Figure 1). The Bel-1 gene product (also known as Tas) is a transcriptional transactivator of the HFV LTR required for completion of the viral life cycle.18-20 The functions of the bel-2 andbel-3 encoded factors are unknown and dispensable for virus production in vitro.21,22 Among retroviruses, a unique property of foamy viruses is that reverse transcription occurs during virion formation rather than after cellular entry, so infectious particles contain DNA genomes.23,24 We recently developed an HFV vector production system that is independent of the Bel-1 transactivator and generates high titer stocks free of replication-competent HFV25 (Figure 1). Here we show that these improved HFV vector stocks efficiently transduce human CD34+ hematopoietic cells and murine HSCs.
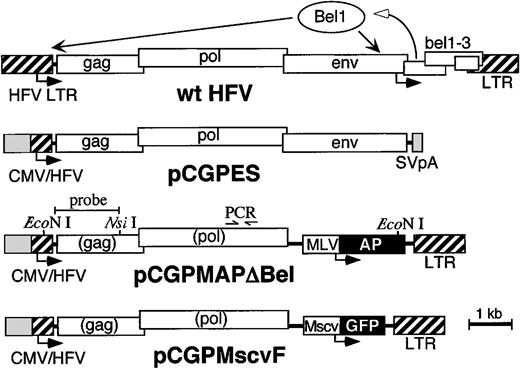
Bel-independent HFV vector system.
The structure of the wild-type (wt) HFV genome and relevant portions of packaging plasmid pCGPES and vector plasmids pCGPMAPΔBel and pCGPMscvF are shown. Arrows below each map represent transcription start sites. The bel1 gene in wt HFV is transcribed initially from the internal promoter, after which the Bel1 protein transactivates both the wt HFV LTR and internal promoters.18-20 In the Bel-independent plasmid constructs, cytomegalovirus (CMV) promoter sequences replace the 5′ HFV LTR U3 region, to generate a constitutive CMV/HFV fusion promoter.25 The packaging plasmid pCPGES expressesgag, pol, and env followed by an SV40 polyadenylation site (SV pA). The vector plasmids express AP or GFP reporter genes from internal MLV LTR or murine stem cell virus (MSCV) promoters, respectively. After reverse transcription, both LTRs in the vector provirus are derived from Bel-1–dependent HFV sequences so thegag and pol genes contained in vector genomes are not expressed in transduced cells. Recent studies show that only the most 5′ portion of gag and 3′ portion of pol are required in cis for vector production42 43 (our unpublished results, July 1998), so future vectors may contain significantly fewer viral sequences. The locations of relevant restriction sites, PCR primers, and the probe fragment used for Southern blot analysis are shown above the pCGPMAPΔBel map.
Bel-independent HFV vector system.
The structure of the wild-type (wt) HFV genome and relevant portions of packaging plasmid pCGPES and vector plasmids pCGPMAPΔBel and pCGPMscvF are shown. Arrows below each map represent transcription start sites. The bel1 gene in wt HFV is transcribed initially from the internal promoter, after which the Bel1 protein transactivates both the wt HFV LTR and internal promoters.18-20 In the Bel-independent plasmid constructs, cytomegalovirus (CMV) promoter sequences replace the 5′ HFV LTR U3 region, to generate a constitutive CMV/HFV fusion promoter.25 The packaging plasmid pCPGES expressesgag, pol, and env followed by an SV40 polyadenylation site (SV pA). The vector plasmids express AP or GFP reporter genes from internal MLV LTR or murine stem cell virus (MSCV) promoters, respectively. After reverse transcription, both LTRs in the vector provirus are derived from Bel-1–dependent HFV sequences so thegag and pol genes contained in vector genomes are not expressed in transduced cells. Recent studies show that only the most 5′ portion of gag and 3′ portion of pol are required in cis for vector production42 43 (our unpublished results, July 1998), so future vectors may contain significantly fewer viral sequences. The locations of relevant restriction sites, PCR primers, and the probe fragment used for Southern blot analysis are shown above the pCGPMAPΔBel map.
Materials and methods
Cell culture
BHK-2126 and 293T27 cells were grown in Dulbecco modified Eagle medium (DMEM; Gibco BRL, Bethesda, MD) supplemented with 10% fetal calf serum (FCS; Hyclone, Logan, UT) and antibiotics (penicillin and streptomycin). Human CD34+cells were isolated from umbilical cord blood by using a CD34+ cell isolation kit as per the manufacturer's instructions (Miltenyi Biotec, Auburn, CA). Transduction was by a single 10-hour exposure to HFV vector stocks in CH296-coated dishes (5 μg/cm2; Takara Biomedicals, Otsu, Japan) in DMEM supplemented with 20% FCS, 100 ng/mL Flt-3 ligand, and 100 ng/mL stem cell factor (SCF), after which the cells were either plated in soft agar colony assays for alkaline phosphate (AP) expression as described,12 plated in methylcellulose colony assays (Methocult H 4230; Stem Cell Technologies, Vancouver, BC, Canada) for expression of green fluorescent protein (GFP), or maintained in liquid culture (DMEM with 20% FCS, 100 ng/mL Flt-3 ligand, 100 ng/mL SCF, 20 ng/mL interleukin [IL]-3, and 20 ng/mL IL-6) for flow cytometry analysis. Colonies were cultured in DMEM with 20% FCS, 50 ng/mL SCF, 10 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF), 10 ng/mL IL-3, and 2 U/mL erythropoietin. All cytokines were human.
Murine bone marrow (BM) cells were isolated by flushing femurs with Hanks balanced salt solution and lysis of red blood cells (RBCs) by a 5-minute exposure to a hypotonic solution (155 mM NH4Cl, 7.3 mM NaHCO3, 126 μM EDTA). The remaining nucleated cells were then cultured in DMEM supplemented with 20% FCS, 20 ng/mL human IL-6, 50 ng/mL murine IL-3, and 50 ng/mL murine SCF for 48 hours before and after a single exposure to HFV vector stocks in CH296-coated dishes. Following transduction, the cells were harvested with a rubber policeman and washed with phosphate-buffered saline (PBS) before injection into irradiated recipients and plating in AP or GFP colony assays. Pretransplant colony-forming unit (CFU) transduction rates ranged from 20% to more than 90%. BM cells isolated from transplanted animals were plated directly in colony assays after lysis of RBCs. Colonies were cultured in the same medium as used during transduction.
Vector production
Plasmids pCGPES and pCGPMAPΔBel have been described.25 pCGPMscvF contains a murine stem cell virus (MSCV) promoter (SfcI-SmaI fragment of pMSCVneo28 and enhanced GFP gene from pEGFP (Clontech, Palo Alto, CA) in place of the MLV LTR and AP gene in pCGPMAPΔBel. CGPMAPΔBel and CGPMscvF HFV vector stocks were produced by calcium phosphate cotransfection of 293T cells with packaging plasmid pCGPES and vector plasmid pCGPMAPΔBel or pCGPMscvF, respectively, followed by collection of vector-containing media 72 hours later as described.25 The stocks were filtered through a 0.45-μm Durapore filter (Millipore, Bedford, MA), concentrated by centrifugation at 50 000g for 2 hours, resuspended in culture medium and used fresh for transduction. Stock titers (transducing units/mL) used for multiplicity of infection (MOI) calculations were determined on BHK-21 (CGPMAPΔBel vector) or 293T cells (CGPMscvF vector).
Mice
All transplants were performed with congenic mouse strains C57BL/6 (expressing the Ly5.2 antigen) and B6Ly5.2 (expressing the Ly5.1 antigen) purchased from the National Cancer Institute (Bethesda, MD). Twelve- to 16-week-old donor mice were treated with 120 mg/kg 5-fluorouracil (5-FU) by intraperitoneal injection 2 days before BM mononuclear cells were harvested. Eight- to 12-week-old recipient mice received 1100 rads from a dual cesium 137 gamma source (GammaCell 40, AEC, Kanata, ON, Canada) before infusion of 1 to 2 × 106 transduced BM cells by tail vein injection. Mice received acidified water with antibiotics (Baytril and amoxicillin) for 2 weeks after transplantation. Several mice receiving transplants had to be killed because of pathogen contamination in the animal care facility.
Peripheral blood analysis
Alkaline phosphate staining of peripheral blood smears obtained by retro-orbital puncture was performed by air-drying overnight, fixing with 0.5% gluteraldehyde in PBS for 10 minutes, washing 3 times for 10 minutes in PBS, heat inactivating for 1 hour at 68°C, and staining in X-Phos solution (100 μg/mL 5-bromo-4-chloro-3-indolyl phosphate, 1 mg/mL nitro blue tetrazolium) or Vector Red AP Substrate (Vector Laboratories, Burlingame, CA). Some smears were counterstained with Harris hematoxylin (Sigma Chemical, St Louis, MO) for 1 minute to highlight nuclear morphology. The percentage of AP+ white blood cells (WBCs) and RBCs was determined by Nomarski microscopy. Flow cytometric analysis of peripheral blood cells was performed on a FACScan (Becton Dickinson, San Jose, CA) set to identify platelets, WBCs, and RBCs according to their forward (FSC) and side scatter (SSC) characteristics as per the manufacturer's instructions, and either GFP fluorescence or AP expression with antiplacental AP antibody 8B6 from DAKO (Carpinteria, CA) conjugated with fluorescein isothiocyanate (FITC) by using the Fluoroscein-EX Protein Labeling Kit (Molecular Probes, Eugene, OR). For dual-label flow cytometry the following phycoerythrin (PE)-conjugated antibodies were used: B220 for B cells, anti-CD3 for T cells, Gr-1 for neutrophils, and Ter119 for RBCs (Pharmingen, San Diego, CA). Engraftment was monitored by flow cytometry for Ly5.1 and Ly5.2 antigens.
DNA studies
High molecular weight DNA was isolated from spleen and BM specimens with the DNA isolation kit D-5000 (Gentra, Minneapolis, MN), and analyzed by Southern blots.29 Locations of the probe fragment, relevant restriction enzyme sites, and PCR primers are shown in Figure 1. Vector copy numbers were quantitated by PhosphorImager (Molecular Dynamics, Sunnyvale, CA) after correction for sample loading by reprobing the same blot with a mouse genomic β-glucuronidase probe (from Mark Sands, Washington University, St Louis, MO). For PCR analysis, hematopoietic colonies were picked from soft agar cultures, immersed in 100 μL of a 50 μg/mL solution of proteinase K, incubated for 2 hours at 56°C, then for 30 minutes at 94°C. Twenty microliters of the sample was used for PCR analysis with forward primer AATGCTGGCATGGGAATAGT and reverse primer TAACTTCCTTGGGTGGCAAG. PCR conditions were: initial denaturation at 94°C for 3 minutes, followed by 35 cycles of 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1 minute. The samples were run in a 2% agarose gel and visualized after ethidium bromide staining under UV light. DNA integrity in each sample was confirmed by amplifying murine β-actin sequences30 under the same PCR conditions.
Results
Transduction of hematopoietic progenitor cells by HFV vectors
Helper-free HFV vectors expressing human placental AP or GFP were made by cotransfection of 293T cells with packaging and vector plasmids driven by a cytomegalovirus (CMV)-HFV fusion promoter (Figure 1). Using this vector system we produced viral stocks with titers ranging from 0.3 to 1.0 × 106 transducing units/mL that were further concentrated to approximately 107/mL by ultracentrifugation.
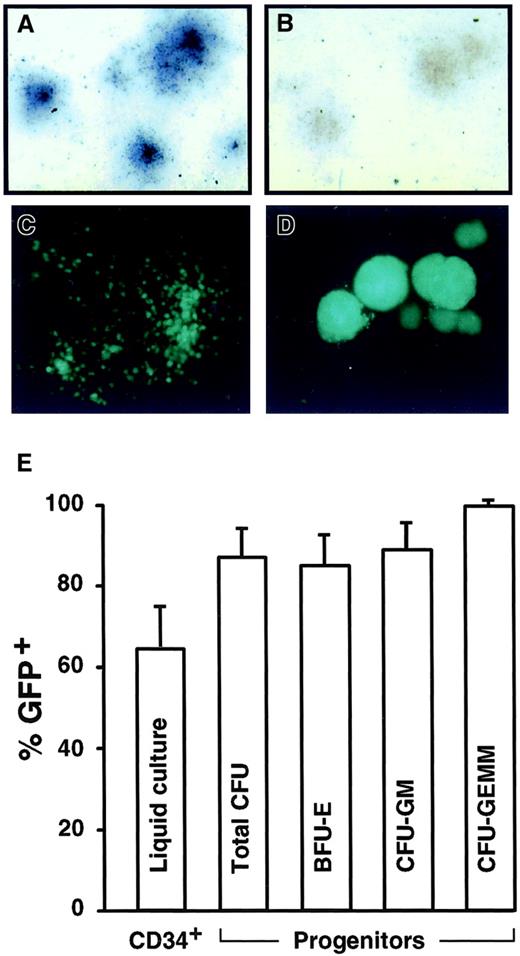
In prior experiments, transduction of primary hematopoietic cells by HFV vectors required cocultivation with vector-producing cells,12 so we first tested if different adhesion molecules would enable transduction with cell-free vector preparations. Poly-l-lysine, human fibronectin, and the recombinant fibronectin fragment CH296 were tested, but only fragment CH296 supported the transduction of murine hematopoietic progenitors (data not shown), as was previously observed with MLV vectors.31 32 To test if primitive human hematopoietic cells could be transduced, CD34+ cord blood cells were exposed for 10 hours to a single dose of cell-free HFV vector stocks on CH296-coated plates, then plated for progenitor colony assays or maintained for 5 days in liquid culture. As shown in Figure2, human CD34+ cells were efficiently transduced by both AP and GFP HFV vectors under these conditions, and transduction rates of over 80% were routinely obtained in colony assays.
Transduction of human CD34+ cells.
Examples of hematopoietic colonies derived from CD34+progenitors transduced by CGPMAPΔBel and stained with X-Phos to produce dark-colored AP+ cells (A) as compared to untransduced controls (B), or transduced by CGPMscvF and examined by fluorescence microscopy for GFP expression (C-D). Original magnification 25 × in panels A-D. (E) Transduction rates of CD34+ cells by GFP vector CGPMscvF as measured by flow cytometry of liquid cultures 5 days after infection, or by progenitor colony assays. The average results of 4 experiments at MOIs ranging from 5 to 23 are shown (with SDs). Total as well as burst-forming units-erythrocytes (BFU-E), colony-forming units-granulocyte/macrophage (CFU-GM), and colony-forming units-granulocyte, erythrocyte, macrophage, megakaryocyte (CFU-GEMM) colonies were enumerated.
Transduction of human CD34+ cells.
Examples of hematopoietic colonies derived from CD34+progenitors transduced by CGPMAPΔBel and stained with X-Phos to produce dark-colored AP+ cells (A) as compared to untransduced controls (B), or transduced by CGPMscvF and examined by fluorescence microscopy for GFP expression (C-D). Original magnification 25 × in panels A-D. (E) Transduction rates of CD34+ cells by GFP vector CGPMscvF as measured by flow cytometry of liquid cultures 5 days after infection, or by progenitor colony assays. The average results of 4 experiments at MOIs ranging from 5 to 23 are shown (with SDs). Total as well as burst-forming units-erythrocytes (BFU-E), colony-forming units-granulocyte/macrophage (CFU-GM), and colony-forming units-granulocyte, erythrocyte, macrophage, megakaryocyte (CFU-GEMM) colonies were enumerated.
Transgene expression in the peripheral blood cells of transplanted animals
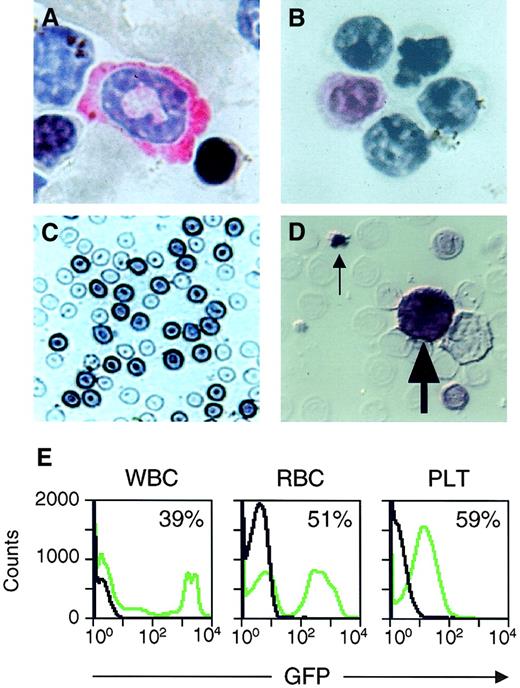
Murine BM transplantation assays were performed to determine if circulating peripheral blood cells would express vector-encoded transgenes and to document transduction at the HSC level. In these experiments, we used conditions originally established for MLV-based vectors,2 33 including pretreatment of BM donors with 5-FU, 2 days of ex vivo culture both before and after exposure to a single dose of HFV vector, followed by transplantation into lethally irradiated congenic recipients expressing distinct CD45 alleles. AP expression from vector CGPMAPΔBel was monitored in the peripheral blood of transplanted animals by histochemical staining of blood smears, which allowed us to identify AP+ neutrophils, lymphocytes, RBCs, and platelets (Figure3A-D). GFP expression from vector CGPMscvF was analyzed in WBCs, RBCs, and platelets by flow cytometry (Figure 3E). Transgene-expressing cells were observed in all blood cell lineages at 4 to 7 weeks after transplantation (Table1) and the combined mean transduction rates (± SD) from these experiments were 36% ± 22% for RBCs, 24% ± 17% for WBCs, and 26% ± 11% for platelets.
Analysis of transgene expression in peripheral blood cells.
(A-D) Photomicrographs of circulating peripheral blood cells from transplant recipients expressing CGPMAPΔBel-encoded AP. Staining with Vector Red and hematoxylin shows an AP+ neutrophil (A) and an AP+ lymphocyte (B) with pink-stained cytoplasm. Staining with X-Phos shows darkly stained AP+ RBCs (C) and an AP+ WBC (large arrow) and AP+ platelet (small arrow) present in a smear examined by Nomarski microscopy (D). Original magnification 400 × in panels A-D. (E) Examples of flow cytometry data from the peripheral blood of a transplant recipient expressing CGPMscvF-encoded GFP (green lines) and an untransduced control animal (black lines), with the percentage of GFP+ cells indicated.
Analysis of transgene expression in peripheral blood cells.
(A-D) Photomicrographs of circulating peripheral blood cells from transplant recipients expressing CGPMAPΔBel-encoded AP. Staining with Vector Red and hematoxylin shows an AP+ neutrophil (A) and an AP+ lymphocyte (B) with pink-stained cytoplasm. Staining with X-Phos shows darkly stained AP+ RBCs (C) and an AP+ WBC (large arrow) and AP+ platelet (small arrow) present in a smear examined by Nomarski microscopy (D). Original magnification 400 × in panels A-D. (E) Examples of flow cytometry data from the peripheral blood of a transplant recipient expressing CGPMscvF-encoded GFP (green lines) and an untransduced control animal (black lines), with the percentage of GFP+ cells indicated.
Transduced peripheral blood cells 4 to 7 weeks after transplantation
| Experiment . | Vector MOI . | Number of mice . | Reporter gene . | Weeks after transplant . | Average %(+) RBCs ±SD . | Average %(+) WBCs ±SD . | Average %(+) PLTs ±SD . |
|---|---|---|---|---|---|---|---|
| 1 | 0.6 | 4 | GFP | 4 | 38 ± 10 | 23 ± 8 | 14 ± 11 |
| 2 | 1.0 | 3 | AP | 6 | 11 ± 1 | 30 ± 22 | ND |
| 3 | 1.6 | 3 | AP | 6 | 9 ± 3 | 17 ± 20 | ND |
| 4 | 1.8 | 3 | AP | 4 | 61 ± 28 | 20 ± 18 | ND |
| 5 | 2.2 | 3 | GFP | 4 | 25 ± 7 | 4.5 ± 1.5 | 35 ± 22 |
| 6 | 2.6 | 4 | AP | 4 | 58 ± 22 | 55 ± 6 | ND |
| 7 | 5.0 | 1 | GFP | 4 | 62 | 5 | 28 |
| 8 | 5.2 | 1 | AP | 7 | 26 | 38 | ND |
| Experiment . | Vector MOI . | Number of mice . | Reporter gene . | Weeks after transplant . | Average %(+) RBCs ±SD . | Average %(+) WBCs ±SD . | Average %(+) PLTs ±SD . |
|---|---|---|---|---|---|---|---|
| 1 | 0.6 | 4 | GFP | 4 | 38 ± 10 | 23 ± 8 | 14 ± 11 |
| 2 | 1.0 | 3 | AP | 6 | 11 ± 1 | 30 ± 22 | ND |
| 3 | 1.6 | 3 | AP | 6 | 9 ± 3 | 17 ± 20 | ND |
| 4 | 1.8 | 3 | AP | 4 | 61 ± 28 | 20 ± 18 | ND |
| 5 | 2.2 | 3 | GFP | 4 | 25 ± 7 | 4.5 ± 1.5 | 35 ± 22 |
| 6 | 2.6 | 4 | AP | 4 | 58 ± 22 | 55 ± 6 | ND |
| 7 | 5.0 | 1 | GFP | 4 | 62 | 5 | 28 |
| 8 | 5.2 | 1 | AP | 7 | 26 | 38 | ND |
WBC values include lymphocytes, monocytes, and granulocytes. PLTs were not scored for AP because of difficulties identifying AP− PLTs.
MOI indicates multiplicity of infection; RBCs, red blood cells; WBCs, white blood cells; PLTs, platelets; GFP, green fluorescent protein; AP, alkaline phosphate; ND, not done.
Transduction of long-term repopulating cells
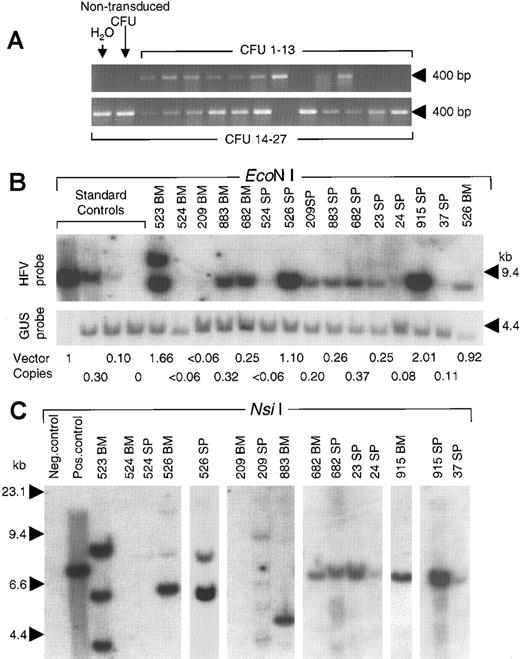
Transplant recipients were followed for up to 6 months to monitor transduction of long-term repopulating cells. Mice transplanted with CGPMAPΔBel-transduced BM cells that survived and maintained a minimum of 50% donor cell engraftment levels were analyzed. As shown in Table2, 4 of 7 of these animals had a significant number of AP+ peripheral blood RBCs and WBCs; the mean transduction rates were 24% for RBCs and 25% for WBCs. Similar AP expression levels were observed in myeloid colonies grown from BM progenitor cells harvested at this time and cultured in soft agar (mean 32%). We also tested colonies for the presence of the vector provirus by using PCR (Figure 4A). The percentages of PCR+ and AP+ colonies were similar in each animal, indicating that the lack of significant long-term expression in 3 of 7 animals was due to reconstitution from nontransduced HSCs as opposed to transgene silencing.
Long-term analysis of transplanted mice
| Mouse no. . | Months after transplant . | Peripheral blood . | Bone marrow . | |||
|---|---|---|---|---|---|---|
| % AP+ RBCs . | % AP+ WBCs . | %AP+ CFU (pos/total) . | %PCR+ CFU (pos/total) . | Provirus copy no. . | ||
| 523 | 6 | 42 | 66 | 69 (54/78) | 77 (20/26) | 1.66 |
| 524 | 6 | 0 | 2 | 0 (0/47) | 5 (1/22) | < 0.06 |
| 526 | 6 | 1 | 1 | 3 (1/37) | 9 (2/22) | 0.92 |
| 209 | 6 | 0 | 1 | 0 (0/60) | 0 (0/25) | < 0.06 |
| 883 | 6 | 15 | 6 | 36 (13/36) | 47 (9/19) | 0.32 |
| 682 | 6 | 15 | 31 | 29 (12/41) | 22 (6/27) | 0.25 |
| 915 | 4 | 95 | 70 | 91 (40/44) | 72 (10/14) | 2.01* |
| Mouse no. . | Months after transplant . | Peripheral blood . | Bone marrow . | |||
|---|---|---|---|---|---|---|
| % AP+ RBCs . | % AP+ WBCs . | %AP+ CFU (pos/total) . | %PCR+ CFU (pos/total) . | Provirus copy no. . | ||
| 523 | 6 | 42 | 66 | 69 (54/78) | 77 (20/26) | 1.66 |
| 524 | 6 | 0 | 2 | 0 (0/47) | 5 (1/22) | < 0.06 |
| 526 | 6 | 1 | 1 | 3 (1/37) | 9 (2/22) | 0.92 |
| 209 | 6 | 0 | 1 | 0 (0/60) | 0 (0/25) | < 0.06 |
| 883 | 6 | 15 | 6 | 36 (13/36) | 47 (9/19) | 0.32 |
| 682 | 6 | 15 | 31 | 29 (12/41) | 22 (6/27) | 0.25 |
| 915 | 4 | 95 | 70 | 91 (40/44) | 72 (10/14) | 2.01* |
CFU indicates colony-forming unit; pos/total, the number of AP+ or PCR+ colonies over the total number analyzed; PCR, polymerase chain reaction; for other abbreviations, see Table 1.
Performed on spleen DNA.
DNA analysis of vector proviruses.
(A) BM colonies grown in soft agar were individually picked and subjected to PCR with primers that amplify a 403-bp fragment of foamy virus vector DNA. Representative results from 27 colonies obtained at 6 months after transplantation are shown, 22 of which contained the vector genome. Negative controls included water and DNA from an untransduced BM colony. (B) Genomic DNA samples from BM and spleen (SP) of long-term and secondary transplant recipients (indicated by number) were digested with EcoN I and probed for vector sequences (Figure 1 has locations). Standards were untransduced mouse spleen DNA mixed with DNA from diploid fibroblasts containing a single integrated HFV vector provirus (9.8-kb EcoN I fragment) to generate the indicated copy numbers. The upper blot was probed with HFV sequences. To correct for sample loading, the same blot was reprobed with mouse genomic β-glucuronidase sequences to detect a 3.8-kb fragment (GUS probe). The calculated copy numbers based on this PhosphorImager analysis are shown below each lane (provirus copies per diploid genome). (C) Integration site analysis using Nsi I to assay for unique-sized junction fragments. Positive and negative control samples were 100% and 0% standards of panel B, respectively. The positions of size standards are indicated in each panel.
DNA analysis of vector proviruses.
(A) BM colonies grown in soft agar were individually picked and subjected to PCR with primers that amplify a 403-bp fragment of foamy virus vector DNA. Representative results from 27 colonies obtained at 6 months after transplantation are shown, 22 of which contained the vector genome. Negative controls included water and DNA from an untransduced BM colony. (B) Genomic DNA samples from BM and spleen (SP) of long-term and secondary transplant recipients (indicated by number) were digested with EcoN I and probed for vector sequences (Figure 1 has locations). Standards were untransduced mouse spleen DNA mixed with DNA from diploid fibroblasts containing a single integrated HFV vector provirus (9.8-kb EcoN I fragment) to generate the indicated copy numbers. The upper blot was probed with HFV sequences. To correct for sample loading, the same blot was reprobed with mouse genomic β-glucuronidase sequences to detect a 3.8-kb fragment (GUS probe). The calculated copy numbers based on this PhosphorImager analysis are shown below each lane (provirus copies per diploid genome). (C) Integration site analysis using Nsi I to assay for unique-sized junction fragments. Positive and negative control samples were 100% and 0% standards of panel B, respectively. The positions of size standards are indicated in each panel.
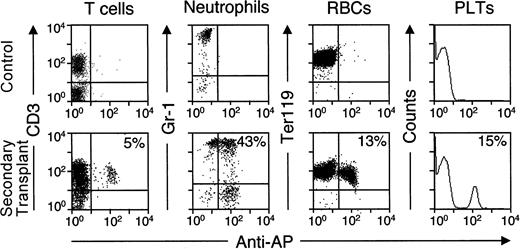
We performed secondary transplants with BM cells harvested from long-term primary recipients no. 915 and no. 682, both of which expressed AP. Although the reconstituting potential of the BM grafts was reduced following serial transplantation, AP+peripheral blood cells were detected in all 3 secondary recipients by histochemical staining at 4 weeks after transplantation (Table3). In addition, flow cytometry with anti-AP and lineage-specific antibodies demonstrated AP+neutrophils, lymphocytes, RBCs, and platelets in the peripheral blood cells of secondary transplant recipients from donor no. 682 at 8 weeks after transplantation (Figure 5). BM progenitors harvested from secondary recipient no. 23 at 8 weeks after transplantation were plated in colony assays and shown to express AP (20 of 50 colonies) and contain the vector genome as determined by PCR (5 of 18 colonies). The long-term transgene expression observed in primary transplant recipients, combined with the marking data from multiple cell types in secondary recipients, demonstrates that transduced HSCs persisted for several months in primary recipients and maintained their ability to reconstitute hematopoiesis in secondary recipients.
Analysis of secondary transplants at 4 weeks after bone marrow transplantation
| Secondary recipient . | Donor . | Months after primary transplant . | % AP+ WBCs in donor pretransplant . | % donor cells (Ly5.1+) . | % AP+ total RBCs . | % AP+ total WBCs . | % AP+ donor WBCs . |
|---|---|---|---|---|---|---|---|
| 23 | 682 | 6 | 31 | 61 | 10 | 17 | 28 |
| 24 | 682 | 6 | 31 | 55 | 7 | 13 | 24 |
| 37 | 915 | 4 | 70 | 15 | 8 | 5 | 33 |
| Secondary recipient . | Donor . | Months after primary transplant . | % AP+ WBCs in donor pretransplant . | % donor cells (Ly5.1+) . | % AP+ total RBCs . | % AP+ total WBCs . | % AP+ donor WBCs . |
|---|---|---|---|---|---|---|---|
| 23 | 682 | 6 | 31 | 61 | 10 | 17 | 28 |
| 24 | 682 | 6 | 31 | 55 | 7 | 13 | 24 |
| 37 | 915 | 4 | 70 | 15 | 8 | 5 | 33 |
The percent of AP+ WBCs in donor pretransplant peripheral blood samples was obtained from the primary recipient at the time of transplantation. The percent donor cells is the percentage of Ly5.1+ WBCs in the peripheral blood of the secondary recipient. The percent AP+ donor WBCs is (% AP+ total WBCs) / (% donor cells).
For abbreviations, see Table 1.
AP expression in peripheral blood cells of recipients of secondary transplants.
Eight weeks after transplantation of BM cells from primary recipient no. 682, peripheral blood cells from secondary recipients were stained with an FITC-conjugated anti-AP antibody and the indicated PE-conjugated lineage-specific antibodies. Examples of flow cytometry data are shown for different lineages, with the percentage of each cell type that expressed vector-encoded AP indicated. Platelets were identified according to their FSC and SSC characteristics instead of by using a specific antibody. Controls were blood samples from untransplanted mice.
AP expression in peripheral blood cells of recipients of secondary transplants.
Eight weeks after transplantation of BM cells from primary recipient no. 682, peripheral blood cells from secondary recipients were stained with an FITC-conjugated anti-AP antibody and the indicated PE-conjugated lineage-specific antibodies. Examples of flow cytometry data are shown for different lineages, with the percentage of each cell type that expressed vector-encoded AP indicated. Platelets were identified according to their FSC and SSC characteristics instead of by using a specific antibody. Controls were blood samples from untransplanted mice.
DNA analysis of transplant recipients
Southern blots were used to determine the copy number of vector proviruses in genomic DNA samples from long-term and secondary transplant recipients. BM and spleen DNA samples were digested withEcoNI, which cuts CGPMAPΔBel provirus genomes twice to generate an 8.2-kb restriction fragment (Figure 1) that was detected in each mouse found to express AP by histochemical analysis, including all 3 recipients of secondary transplants (Figure 4B). Except for one sample, the vector fragment was the predicted size of an intact provirus. The BM of mouse no. 523 had 2 provirus fragments, one of which was larger than expected, possibly due to transfer of an abnormal vector genome or a recombination event after transduction. Vector copy numbers ranged from 0.08 to 2.01 proviruses per diploid genome in transgene-expressing mice. In 2 of 3 mice with low AP expression levels (no. 524 and no. 209) no vector genomes could be detected in BM DNA samples (Table 2). Mouse no. 526 expressed AP at low levels despite a BM copy number of 0.92. In this mouse, it is not clear which BM cells contained vector proviruses and whether they expressed AP, because less than 10% of the myeloid progenitors that grew in BM colony assays contained vector sequences based on PCR (Table 2).
Vector integration sites were analyzed by digestion of DNA samples from long-term and secondary transplant recipients with NsiI, (Figure 4C), which cuts once in the vector genome and once in flanking chromosomal DNA to produce a distinct fragment for each provirus integration site (Figure 1). One to 5 vector-hybridizing bands were seen in the samples from AP-expressing mice, consistent with monoclonal or oligoclonal hematopoietic reconstitution by transduced cells. In most cases, common junction fragments were present in the spleen and BM samples from the same mouse suggesting that a common transduced precursor repopulated both organs (mice no. 526, no. 682, and no. 915). In the secondary transplant experiments, a single junction fragment present in the primary recipient's BM (mice no. 682 and no. 915) was the only fragment detected in secondary recipients (mice no. 23, no. 24, and no. 37, respectively). Thus a single transduced cell reconstituted hematopoiesis in both primary and secondary recipients, consistent with transduction of pluripotent HSCs.
Discussion
In summary, we have shown that vectors developed from human foamy virus can efficiently transduce human CD34+ hematopoietic progenitors and murine HSCs, and that recipients of transduced BM cells express the vector-encoded transgene in all hematopoietic cell lineages analyzed. Transduction at the HSC level was demonstrated by long-term transgene expression in transplant recipients, secondary transplantation experiments, and integration site analysis. Except for one cell clone with an aberrant provirus, all transduced cells contained an intact, integrated, foamy virus vector genome that appeared to be transcriptionally active.
Our results suggest that HSCs may also be susceptible to infection with wild-type foamy viruses. Although foamy viruses have been isolated from blood cells,34-36 it is not clear what role the hematopoietic system plays in natural infections. Infected HSCs could act as a reservoir of provirus-containing cells capable of massive expansion during the production of mature blood cells.
Murine BM transplantation has served as an important animal model for HSC transduction experiments. The results obtained with HFV vectors are comparable to the best results from prior studies with ecotropic MLV vectors, where over 50% of hematopoietic cells were transduced in some transplant recipients.1-3 Possible explanations for the high transduction rates we observed include improved transduction of nondividing cells or expression of cellular HFV receptor molecules on HSCs. HFV vectors may have transduced quiescent HSCs, because they transduce stationary phase cell cultures more efficiently than MLV vectors,10 HFV genomes enter the nuclei of G1/S phase-arrested cells,37 and transduction by cDNA-containing HFV particles is not dependent on adequate nucleotide pools.23,24 Future studies on whether pretreatment of BM donors with 5-FU and ex vivo cytokine stimulation are required may help resolve these issues. Although the cellular receptor(s) for HFV is not known, it must be ubiquitously expressed because HFV vectors can transduce cells from all vertebrate species tested.10 38
A common problem associated with MLV vectors has been the silencing of transgene expression over time.39,40 We did not observe significant silencing in our experiments, based on a comparison of vector marking and transgene expression levels in the hematopoietic progenitors of long-term and secondary transplant recipients, despite our use of an internal MLV LTR promoter previously found to silence in hematopoietic cells.3,40 41 Persistent expression of HFV vectors could be due to unidentified cis-acting functions present in the vector backbone that prevent silencing, or specific attributes of the MLV LTR promoter we used, such as the presence of a single internal copy, or the removal of the transfer RNA primer binding site. Further studies will be required to determine if specific attributes of the HFV vectors used can prevent transgene silencing.
High HSC transduction rates have not been achieved in primate transplantation studies with nonecotropic MLV pseudotypes, so it is not clear how well murine experiments with HFV vectors will predict results in humans. However, the amphotropic MLV vectors commonly used in primates transduce murine HSCs at significantly lower rates than their ecotropic counterparts,4 suggesting that the mouse model may be valid when the same vector system is used. Because HFV vectors transduce murine and human hematopoietic progenitors at similar rates, they may also transduce primate HSCs efficiently. Thus, vectors based on the nonpathogenic foamy viruses may be a safe and effective alternative to MLV or lentivirus vectors for use in human gene therapy.
We thank Roli Hirata for expert technical assistance.
Supported by U.S. National Institutes of Health grants (to D.W.R., G.V., and N.J.) and the Medical Research Council of Canada (to G.T.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David W. Russell, Division of Hematology, University of Washington, Box 357720, Seattle, WA 98195; e-mail:drussell@u.washington.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal