Myelodysplastic syndrome (MDS) is a slowly progressing hematologic malignancy associated with a poor outcome. Despite the relatively high incidence of MDS in the elderly, differentiation of MDS from de novo acute myeloid leukemia (AML) still remains problematic. Identification of genes expressed in an MDS-specific manner would allow the molecular diagnosis of MDS. Toward this goal, AC133 surface marker–positive hematopoietic stem cell (HSC)-like fractions have been collected from a variety of leukemias in a large-scale and long-term genomics project, referred to as “Blast Bank,” and transcriptome of these purified blasts from the patients with MDS were then compared with those from AML through the use of oligonucleotide microarrays. A number of genes were shown to be expressed in a disease-specific manner either to MDS or AML. Among the former found was the gene encoding the protein Delta-like (Dlk) that is distantly related to the Delta-Notch family of signaling proteins. Because overexpression of Dlk may play a role in the pathogenesis of MDS, the disease specificity of Dlk expression was tested by a quantitative “real-time” polymerase chain reaction analysis. Examination of the Blast Bank samples from 22 patients with MDS, 31 with AML, and 8 with chronic myeloid leukemia confirmed the highly selective expression of the Dlk gene in the individuals with MDS. Dlk could be the first candidate molecule to differentiate MDS from AML. The proposal is made that microarray analysis with the Blast Bank samples is an efficient approach to extract transcriptome data of clinical relevance for a wide range of hematologic disorders.
Introduction
Myelodysplastic syndrome (MDS) is a slowly progressing leukemic disorder that predominantly affects elderly people.1,2 Individuals with MDS exhibit cytopenia in at least one lineage of peripheral blood (PB) cells. However, the bone marrow (BM) of many affected individuals is hyper- to normocellular, suggesting the presence of ineffective hematopoiesis. Another characteristic feature of MDS is the dysplasia of at least one lineage (myeloid, erythroid, or megakaryocyte-platelet lineage) of BM cells, such as neutrophils with pseudo-Pelger anomaly, hypersegmented neutrophils, ringed sideroblasts, and micromegakaryocytes.3
The clinical course of MDS can be divided into several phases. Patients in the indolent, chronic phase exhibit a decrease in the number of PB cells and are diagnosed with either refractory anemia (RA) or RA with ringed sideroblasts (RARS). Individuals with dysplastic change in BM and monocytosis in PB are diagnosed with chronic myelomonocytic leukemia (CMMoL). After a variable interval, however, some individuals with MDS undergo a progressive transformation to overt leukemia. As the number of blast cells increases, the patients are diagnosed with RA with an excess of blasts (RAEB; 5%-19% blasts in BM), RAEB in transformation (RAEB-t; 20%-29% blasts in BM), and, finally, MDS-associated acute leukemia (> 20% blasts in BM).
MDS-associated leukemia is rarely cured by conventional chemotherapy. Intensive treatment with anticancer drugs often results in prolonged myelosuppression, which is one of the main causes of disease-related death.4 Individuals with acute leukemia that results from MDS therefore have a poor prognosis, which contrasts with the somewhat better outcome of de novo acute myeloid leukemia (AML) in this older group of patients. It is thus essential to differentiate MDS-associated leukemia from de novo AML to select the optimal therapeutic strategy.
This task is complicated, however, by the fact that some patients with de novo AML may exhibit dysplastic changes in BM during the course of their disease.5-7 It is thus extremely difficult to diagnose elderly individuals with overt leukemia if their BM exhibits dysplasia. The occurrence of a period of cytopenia before the development of leukemia indicates that the patient should be treated for MDS-associated leukemia. Without the clinical history, however, there are currently few other criteria with which to differentiate MDS-associated leukemia from de novo AML with dysplasia. The identification of genes that are expressed specifically in MDS blasts should facilitate the diagnosis and treatment of elderly people with overt leukemia, as well as clarify whether AML with dysplastic changes is truly a clinical entity distinct from MDS-associated leukemia.
The development of DNA microarrays or DNA chips has revolutionized the analysis of gene expression profiles. Such DNA microarrays can contain tens of thousands of test complementary DNAs (cDNAs) or oligonucleotides and, with a single hybridization step, allow a systematic comparison of the expression of the corresponding genes between 2 given samples.8 The completion of the human genome sequencing project will increase further the analytical power of this technology. The use of DNA microarrays has already allowed the identification of candidate mammalian genes related to carcinogenesis or cell differentiation.9-11
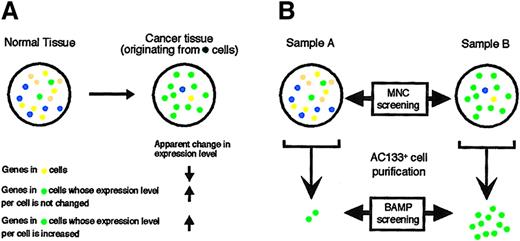
The DNA microarray technique is so powerful, however, that it yields “pseudopositive” results in many instances. For example, for the identification of genes whose expression is specifically induced or inhibited in cancer cells, a simple comparison of gene expression profiles between cancerous and normal tissues with the use of a DNA microarray is not a good approach. Normal tissue is composed of many cell types, which include both tissue-specific cells as well as cells that contribute to nonspecific components such as the circulatory system. In contrast, although cancerous tissue also comprises a variety of cell types, a particular cell type (the cancer cell) becomes predominant. The ratio of the various cell types thus significantly differs between normal and cancerous tissues. If, in the example shown in Figure 1A, the cancer has arisen from the green cell type, then the proportion of the other cell types becomes reduced in the cancerous tissue compared with that in the normal tissue. Furthermore, although the messenger RNA (mRNA) copy number per cell of genes expressed in these other cell types may not differ between cancerous and normal tissues, a comparison of the 2 tissues en bloc would incorrectly suggest that the expression of these genes is decreased in the cancer. Conversely, because of the expansion of the green cell population, such a comparison would suggest incorrectly that genes expressed in the cancer cells whose mRNA copy number is not affected by transformation are activated in the cancer tissue. It would not be an easy task to differentiate these pseudopositive genes from genes whose expression level is truly affected by carcinogenesis.
Changes in gene expression profile due to a shift in cell population composition.
(A) The normal tissue is composed of various cell types, whereas the cancer tissue is composed predominantly of the cancerous cell type (green). Expansion of the cancer cells thus results in a decrease in the proportion of the other cell types. The overall level of expression in the cancer tissue of genes that are selectively expressed in one type of noncancer cell (yellow) would thus appear decreased (arrow), compared with that in the normal tissue, only because of the corresponding decrease in the yellow cell population. Conversely, expansion of the green cells gives rise to an apparent increase in the tissue expression level of genes specifically expressed in this cell type, regardless of whether the number of the corresponding transcripts per cell is actually increased or not. (B) Previous comparisons of transcriptomes in the studies of blood cells have been performed with MNCs of sample A and sample B, irrespective of their cellular compositions (MNC screening). In the present study, transcriptomes were compared between leukemic blasts (green) to reduce the occurrence of pseudopositive and pseudonegative results generated by a shift in cell population composition. For this approach (BAMP screening), the target leukemic blasts were purified by affinity chromatography on a column containing anti-AC133.
Changes in gene expression profile due to a shift in cell population composition.
(A) The normal tissue is composed of various cell types, whereas the cancer tissue is composed predominantly of the cancerous cell type (green). Expansion of the cancer cells thus results in a decrease in the proportion of the other cell types. The overall level of expression in the cancer tissue of genes that are selectively expressed in one type of noncancer cell (yellow) would thus appear decreased (arrow), compared with that in the normal tissue, only because of the corresponding decrease in the yellow cell population. Conversely, expansion of the green cells gives rise to an apparent increase in the tissue expression level of genes specifically expressed in this cell type, regardless of whether the number of the corresponding transcripts per cell is actually increased or not. (B) Previous comparisons of transcriptomes in the studies of blood cells have been performed with MNCs of sample A and sample B, irrespective of their cellular compositions (MNC screening). In the present study, transcriptomes were compared between leukemic blasts (green) to reduce the occurrence of pseudopositive and pseudonegative results generated by a shift in cell population composition. For this approach (BAMP screening), the target leukemic blasts were purified by affinity chromatography on a column containing anti-AC133.
It is therefore important to develop new means of sample preparation or data normalization for DNA microarray experiments that will facilitate the identification of changes in gene expression that are of biologic relevance. Ideally, populations of cells with the same phenotype and background (cell origin, differentiation state, expression of surface markers), differing only in that one population is transformed and the other is not, should be purified from the cancerous and normal tissues before microarray analysis (Figure 1B). Any differences in the gene expression profiles between the 2 cell populations would thus likely be related to carcinogenesis. We propose that such an approach be termed BAMP (background-matched population) screening or comparison, and we have now applied this technique to identify genes useful for the diagnosis of MDS.
AC133,12 also known as PROML1,13 was recently identified as a cell surface protein whose expression is restricted to a blood cell population highly enriched in hematopoietic stem cells (HSCs) and to the retina. AC133 is expressed in a population of CD34high, CD38low, c-Kit+ blood cells, which is known to contain the HSCs,14 and this protein is thus one of the most specific markers for HSCs currently available.
We set out to purify and store AC133+ blastic cells from a variety of leukemia patients as a part of our large-scale and long-term genomics project, referred to as “Blast Bank,” to characterize leukemia-specific gene expression. The samples in the Blast Bank should all be at virtually identical stages of differentiation and show similar profiles of surface-marker expression, irrespective of their specific leukemic origin. In the present study, we have compared the gene expression profiles of cells in the Blast Bank derived from the individuals with MDS or de novo AML. Our results indicate that several genes are preferentially expressed in MDS, and they should facilitate the development of tools for the molecular diagnosis of this condition.
Materials and methods
Preparation of Blast Bank samples
The PB or BM aspirates were obtained from subjects after written informed consent was obtained. Mononuclear cells (MNCs) were purified from the specimens by the Ficoll-Hypaque density gradient centrifugation, labeled with AC133 MicroBeads (Miltenyi Biotec, Auburn, CA), and subjected to chromatography on miniMACS magnetic cell separation columns (Miltenyi Biotec) according to the manufacturer's instructions. Portions of the MNC and AC133+ cell preparations were stained with Wright-Giemsa solution, or analyzed with a FACScan flow cytometer (Becton Dickinson, Mountain View, CA) for the expression of CD34, CD38, and AC133. The number of blasts purified from the BM of leukemia patients was more than 100 times that purified from the BM of healthy volunteers (not shown). The contribution of normal HSCs to the Blast Bank samples of leukemic patients should therefore be negligible.
RNA preparation and DNA microarray analysis
Total RNA was extracted from the Blast Bank samples and amplified by the method of Van Gelder and colleagues15modified by Y. Nakamura (Institute of Medical Sciences, University of Tokyo, Japan). For the comparison of MNC screening and BAMP screening, RNA (2 μg each) was used to synthesize cDNA labeled with Cy3 or Cy5 (Amersham Pharmacia Biotech, Uppsala, Sweden), and hybridized to a cDNA microarray (IntelliGene Human Cancer CHIP; Takara Biomedicals, Shiga, Japan). To obtain expression profiles of Blast Bank samples, biotin-labeled complementary RNA (cRNA) was synthesized from the sample RNA (2 μg) by using the ExpressChip labeling system (Mergen, San Leandro, CA), and allowed to hybridize with an oligonucleotide microarray (HO-3) that contains oligonucleotides based on genes that mostly encode transcription factors as well as with our custom-made array containing oligonucleotides for membrane proteins, growth factors, and proteins involved in redox regulation (both obtained from Mergen). A total of 2304 genes were studied for expression profiling. Hybridized slides were then incubated with streptavidin, antistreptavidin first antibody, and finally Cy3-conjugated second antibody (all from Mergen) according to the manufacturer's instruction. Detection of the signals and statistical analysis of the digitized data were carried out with a GMS 418 array scanner (Affymetrix, Santa Clara, CA) and GeneSpring 3.2.2 software (Silicon Genetics, Redwood, CA), respectively. The gene list with GenBank accession numbers for our custom-made array can be obtained on request.
Real-time polymerase chain reaction
Portions of the unamplified cDNAs were subjected to polymerase chain reaction (PCR) with SYBR Green PCR Core Reagents (PE Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. The incorporation of the SYBR Green dye into the PCR products was monitored in real time with an ABI PRISM 7700 sequence detection system (PE Applied Biosystems), resulting in the calculation of threshold cycle, or CT value, that defines the PCR cycle number at which an exponential growth of PCR product begins. TheCT values for β-actin and Dlk were used to calculate the abundance of Dlk transcripts relative to that of β-actin mRNA. The oligonucleotide primers for PCR were 5′-CCATCATGAAGTGTGACGTGG-3′ and 5′-GTCCGCCTAGAAGCATTTGCG-3′ for β-actin cDNA, and 5′-CTGAAGGTGTCCATGAAAGAG-3′ and 5′-GCTGAAGGTGGTCATGTCGAT-3′ for Dlk cDNA.
Results
MNC screening and BAMP screening
Isolation of AC133+ cells from MNCs of leukemia patients by labeling with microbead-conjugated antibodies to (anti-) AC133 and chromatography on a magnetic cell separation column yielded a highly pure cell population (> 95% AC133+ cells in most instances). These cells were constantly c-Kithigh, CD45high, and CD16− (data not shown), being compatible with the previous characterization of AC133+cells. However, expression level of CD38 varied from sample to sample (from very low to moderately high) among different patients. It is, therefore, possible that the AC133+ fraction has still heterogeneity to some extent, which may have a relationship with the nature of diseases.
To date, we have collected 166 Blast Bank samples that contain 8 cases of healthy volunteers, 32 cases of AML, 10 cases of acute lymphoid leukemia (ALL), 42 cases of MDS (including MDS-associated leukemia), 25 cases of chronic myeloid leukemia (CML), 13 cases of aplastic anemia, and 36 cases of other diseases.
Most previous studies of gene expression analysis in leukemia cells were performed with MNCs prepared from PB or BM. It was therefore important to determine the extent to which the results obtained with BAMP screening differ from those obtained with MNC screening. To address this issue, we applied both screening approaches to the samples from the same pair of individuals and compared the resulting gene expression profiles (Figure 1B).
We first purified AC133+ cells from the MNCs of PB from healthy volunteers. Flow cytometry revealed that the MNCs and AC133+ cells comprised 7.6% and 80.5% CD34+cells, respectively (Figure 2A). We next purified AC133+ cells from the MNCs of a patient with MDS-associated leukemia (ID no. 7); flow cytometry revealed that the MNCs and AC133+ cells comprised 66.2% and 99.1% CD34+ cells, respectively (Figure 2B). In both instances, the AC133-based purification step yielded homogeneous mid-sized cells with a high ratio of nucleus to cytoplasm, although the AC133+ cells from the MDS patient exhibited unusual lobulated nuclei (Figure 2A,B).
Comparison of MNC and BAMP screening for healthy volunteers and a patient with MDS.
(A) MNCs isolated from the PB of healthy volunteers were subjected to anti-AC133 affinity chromatography, followed by fluorescence-activated cell sorting with anti-AC133. The eluted (AC133+) cells and the original MNCs were analyzed for surface expression of CD34 and CD38 by flow cytometry (upper panels). The percentage of CD34+cells in each sample is indicated. Wright-Giemsa staining of each cell preparation is also shown (lower panels, original magnification ×100). (B) Purification of AC133+ cells from a patient with MDS-associated leukemia. MNCs isolated from the PB of the patient (ID no. 7) were subjected to anti-AC133 affinity chromatography. The MNCs and AC133+ cells were then analyzed for surface expression of CD34 and CD38 and subjected to Wright-Giemsa staining as described for panel A, original magnification ×100. (C) Total RNA isolated from the MNCs of healthy volunteers and the patient with MDS was used to synthesize cDNA labeled with Cy5 and Cy3 dyes, respectively. A mixture of the labeled cDNA preparations was then subjected to hybridization with a cDNA microarray containing fragments of 382 cancer-related genes (upper panel). Red and green spots thus indicate genes specifically expressed in the healthy volunteers or in the patient, respectively; yellow spots correspond to genes expressed to similar extents in both samples. RNA prepared from purified AC133+ cells of the healthy volunteers and the patient was similarly analyzed (lower panel). A part of the scanned image is demonstrated. Some spots, including those numbered, exhibited opposite patterns of expression by MNC screening and BAMP screening.
Comparison of MNC and BAMP screening for healthy volunteers and a patient with MDS.
(A) MNCs isolated from the PB of healthy volunteers were subjected to anti-AC133 affinity chromatography, followed by fluorescence-activated cell sorting with anti-AC133. The eluted (AC133+) cells and the original MNCs were analyzed for surface expression of CD34 and CD38 by flow cytometry (upper panels). The percentage of CD34+cells in each sample is indicated. Wright-Giemsa staining of each cell preparation is also shown (lower panels, original magnification ×100). (B) Purification of AC133+ cells from a patient with MDS-associated leukemia. MNCs isolated from the PB of the patient (ID no. 7) were subjected to anti-AC133 affinity chromatography. The MNCs and AC133+ cells were then analyzed for surface expression of CD34 and CD38 and subjected to Wright-Giemsa staining as described for panel A, original magnification ×100. (C) Total RNA isolated from the MNCs of healthy volunteers and the patient with MDS was used to synthesize cDNA labeled with Cy5 and Cy3 dyes, respectively. A mixture of the labeled cDNA preparations was then subjected to hybridization with a cDNA microarray containing fragments of 382 cancer-related genes (upper panel). Red and green spots thus indicate genes specifically expressed in the healthy volunteers or in the patient, respectively; yellow spots correspond to genes expressed to similar extents in both samples. RNA prepared from purified AC133+ cells of the healthy volunteers and the patient was similarly analyzed (lower panel). A part of the scanned image is demonstrated. Some spots, including those numbered, exhibited opposite patterns of expression by MNC screening and BAMP screening.
RNA was purified from the MNCs and AC133+ cells and was used to generate Cy5 dye-labeled cDNA (red fluorescence) or Cy3-labeled cDNA (green fluorescence) for samples from the healthy volunteers or the MDS patient, respectively. The corresponding cDNA preparations from the normal controls and the MDS patient were mixed and exposed to a cDNA microarray containing fragments of 382 cancer-related genes (Figure 2C). The MNC comparison and the BAMP comparison revealed distinct differences in the transcriptome profiles between the healthy volunteers and the patient with MDS. Indeed, the mRNA abundance ratio (reflecting a difference in gene expression between healthy volunteers and the patient with MDS) for many spots displayed opposite patterns in the MNC and BAMP comparisons (shown numbered in Figure 2C).
Screening of MNC indicated that expression of the CD34 gene, a marker for immature blood cells, was increased in the patient with MDS compared with that in healthy volunteers (Cy5 fluorescence intensity, 519 arbitrary units [U]; Cy3 fluorescence intensity, 42 323 U). However, BAMP screening showed that the CD34gene is expressed at similar levels in the 2 AC133+ cell populations (Cy5 fluorescence intensity, 6 749 546 U; Cy3 fluorescence intensity, 5 543 512 U). The “pseudo” increase inCD34 gene expression apparent in the MNC screening therefore likely reflected the expansion of the CD34+ cell population, comprising mostly leukemic blasts, in the BM of the leukemic patient, not an increase in the CD34 mRNA copy number per cell. These results indicate that comparison of samples by BAMP screening provides distinct information from that obtained with MNC screening, and that the former approach is more suitable for extracting data of biologic relevance.
BAMP comparison between MDS and AML blasts
For the identification of MDS-specific genes, we prepared a custom-made microarray that contains oligonucleotides corresponding to 1152 human genes encoding membrane proteins, growth factors, and proteins involved in redox regulation. For our analysis, membrane proteins should be a target to be focused on because diagnosis by flow cytometry with antibodies to MDS-specific cell surface markers, if found, would be of great clinical value. We also chose, for the array, genes encoding proteins involved in redox regulation, because such molecules in addition to membrane proteins should play important roles in the acquisition of tolerance to chemotherapeutic reagents.
Any oncogenic events within cells should exert their effect, at least, partially through the regulation of gene transcription. Therefore, to gain insights into the pathogenesis of leukemias, we also used a commercially available microarray (HO-3) containing oligonucleotides based on 1152 genes encoding mainly transcriptional factors. Expression profile for a total of 2304 genes was thus obtained for every Blast Bank sample.
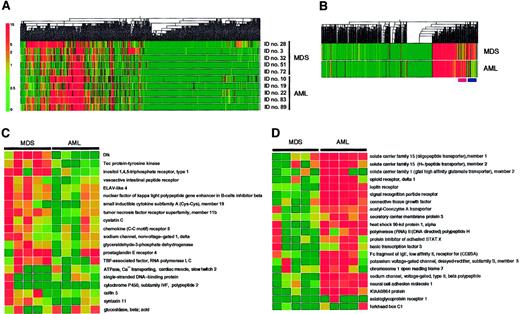
With these arrays, we have compared the transcriptome of leukemic blasts between the individuals with MDS (3 patients with MDS-associated leukemia and 2 patients with RAEB) and those with de novo AML (3 patients with subtype M1 and 2 patients with M4). The expression profile of the genes spotted on the arrays was visualized by construction of a “gene tree,” or dendrogram, that clusters genes with similar expression patterns (Figure3A). Almost half the genes were transcriptionally silent. Substantial diversity was apparent among the patients, however, in the extent of expression for the transcriptionally active genes. To identify disease-specific genes, we calculated the mean value for the expression intensity of each gene for the MDS group and the AML group, and then generated a new dendrogram based on these calculated values (Figure 3B). This “average tree” revealed the presence of clusters of MDS-specific genes and of AML-specific genes.
Expression profiles of 2304 genes in the MDS and AML blasts.
(A) Hierarchical clustering of 2304 genes based on their expression profiles in Blast Bank samples of individuals with AML or MDS. Each column represents a single gene on the microarray, and each row a separate patient sample. The fluorescence intensity of each gene was normalized by the mean fluorescence value of spots in each hybridization, and is shown color-coded as indicated on the left. Gray indicates the data of blank spots. (B) Mean expression values for each gene were calculated for MDS and AML samples and used to generate a dendrogram. The positions of the clusters of MDS-specific genes and AML-specific genes are indicated at the bottom by pink and purple bars, respectively. (C) Expression profiles of MDS-specific genes. Each row corresponds to a single gene, with the columns indicating the corresponding expression level in different samples. (D) Expression profiles of AML-specific genes. The gene names and accession numbers as well as expression intensity data for the genes shown in panels C and D are available on request. ELAV indicates embryonic lethal, abnormal vision; TBP, TATA box-binding protein.
Expression profiles of 2304 genes in the MDS and AML blasts.
(A) Hierarchical clustering of 2304 genes based on their expression profiles in Blast Bank samples of individuals with AML or MDS. Each column represents a single gene on the microarray, and each row a separate patient sample. The fluorescence intensity of each gene was normalized by the mean fluorescence value of spots in each hybridization, and is shown color-coded as indicated on the left. Gray indicates the data of blank spots. (B) Mean expression values for each gene were calculated for MDS and AML samples and used to generate a dendrogram. The positions of the clusters of MDS-specific genes and AML-specific genes are indicated at the bottom by pink and purple bars, respectively. (C) Expression profiles of MDS-specific genes. Each row corresponds to a single gene, with the columns indicating the corresponding expression level in different samples. (D) Expression profiles of AML-specific genes. The gene names and accession numbers as well as expression intensity data for the genes shown in panels C and D are available on request. ELAV indicates embryonic lethal, abnormal vision; TBP, TATA box-binding protein.
The expression profiles of the MDS-specific genes are shown in greater detail in Figure 3C. Genes shown to be highly MDS-specific include those encoding Dlk,16 Tec,17 and inositol 1,4,5-trisphosphate receptor type 1.18 Dlk, also known as Pref-1 (preadipocyte factor-1),19 FA1 (fetal antigen 1),20 and SCP-1 (stromal cell protein-1; GenBank accession number, D16847), is a transmembrane protein belonging to the superfamily of epidermal growth factor–like proteins; it is also distantly related to the Delta-Serrate-Notch family of signaling molecules. The differentiation of 3T3-L1 cells into adipocytes is accompanied by a decrease in the abundance of Dlk mRNA, whereas forced expression of Dlk in these cells inhibits adipocyte differentiation. Dlk thus appears to contribute to the determination of cell fate or differentiation by mediating cell-to-cell contact, an ability shared by the Delta-Serrate-Notch family of membrane proteins.21 Tec is a nonreceptor protein tyrosine kinase that is activated in response to various growth stimuli.22 Although Tec is expressed in a wide spectrum of blood cells, its mRNA was especially abundant in MNCs from the BM of MDS patients.17 Our present data further support the disease-specific expression of this kinase.
The AML-specific genes identified by the BAMP screening include those for member 123 and member 224 of solute carrier family 15, member 2 of solute carrier family 1,25 opioid receptor delta 1,26 and the leptin receptor27(Figure 3D). The members of the solute carrier families of proteins transport small peptides across cell membranes in a proton-dependent manner, and, thus, they may play a role in determining drug sensitivity of blasts. The leptin receptor was previously shown to be expressed in MNCs from the BM of AML patients.28
The expression of Dlk may be important not only for the diagnosis of MDS, but also for the molecular pathogenesis of the disease. Dlk transcripts have been detected in stromal cell lines capable of supporting the proliferation of HSCs, but not in those that fail to maintain HSC growth.29 Moreover, forced expression of Dlk enabled the latter cells to support HSC proliferation, indicating that Dlk contributes directly to this ability. These observations thus suggest that Dlk might support hematopoiesis by mediating cell-to-cell contact between HSCs and stromal cells in the microenvironment of BM. Given the supposed role of Dlk in differentiation block, overexpression of this protein in HSCs might directly contribute to the increased proliferation of these cells, the unusual phenotypes of differentiated cells, and the ineffective hematopoiesis that is characteristic of individuals with MDS.
Quantitation of Dlk gene expression by real-time PCR
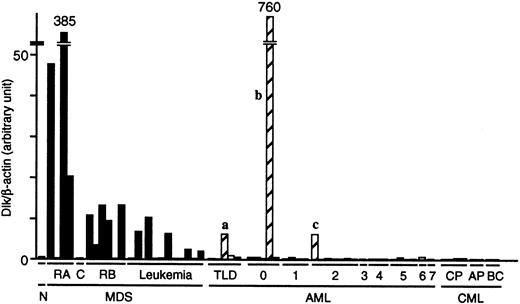
To confirm the preferential expression of the Dlk gene in MDS blasts, we prepared cDNAs from the Blast Bank samples of 22 patients with MDS, 31 with AML (all samples in the Blast Bank for both diseases at the timing of examination), and 8 with CML, and then subjected these cDNAs to “real-time” PCR analysis with primers specific for Dlk and β-actin. The abundance of Dlk mRNA relative to that of β-actin mRNA in the blasts from most MDS patients was markedly greater than that in the blasts from most AML patients (Figure4). The number of individuals for which the relative abundance of Dlk mRNA was more than twice that in the healthy control was 12 of 22 (55%) for MDS patients and 3 (designated a, b, and c) of 31 (10%) for AML patients (P < .0001). Interestingly, prominent dysplasia was apparent for all 3 lineages of BM cells from AML patient a and for 2 lineages of BM cells from patient c. Moreover, the mature neutrophils of patient c exhibited pseudo-Pelger anomaly, a hallmark of MDS. Therefore, despite the lack of clinical history before diagnosis, it is likely that patients a and c did not have de novo AML, but rather had undergone leukemic transformation from an early stage of MDS. The presence of few mature cells in the BM and PB of patient b (leukemic blasts constituted > 98% of cells in both specimens) rendered it impossible to assess blood cell dysplasia. Cytogenetic analysis failed to identify any chromosomal anomaly in the BM cells of patient b. Inclusion of patients a and c in the MDS group further enhances the potential of Dlk as a molecular marker of MDS.
Quantitation of Dlk mRNA in the Blast Bank samples from patients with MDS, AML, or CML.
The cDNA prepared from the blasts of 22 patients with MDS, 31 with AML, and 8 with CML was subjected to real-time PCR with primers specific for Dlk or β-actin. The ratio of the abundance of Dlk transcripts to that of β-actin transcripts (Dlk/β-actin) was calculated as 2n, where n is theCT value of β-actin minus theCT value of Dlk, and was normalized by the value of the sample from healthy volunteers (N). C indicates CMMoL; RB, RAEB; TLD, AML with trilineage dysplasia; CP, chronic phase; AP, accelerated phase; and BC, blast crisis. The numbers 0 to 7 refer to the AML subtypes M0 to M7. The letters a, b, and c refer to patients discussed in the text.
Quantitation of Dlk mRNA in the Blast Bank samples from patients with MDS, AML, or CML.
The cDNA prepared from the blasts of 22 patients with MDS, 31 with AML, and 8 with CML was subjected to real-time PCR with primers specific for Dlk or β-actin. The ratio of the abundance of Dlk transcripts to that of β-actin transcripts (Dlk/β-actin) was calculated as 2n, where n is theCT value of β-actin minus theCT value of Dlk, and was normalized by the value of the sample from healthy volunteers (N). C indicates CMMoL; RB, RAEB; TLD, AML with trilineage dysplasia; CP, chronic phase; AP, accelerated phase; and BC, blast crisis. The numbers 0 to 7 refer to the AML subtypes M0 to M7. The letters a, b, and c refer to patients discussed in the text.
We did not detect Dlk mRNA in the individuals with CML, either in its indolent chronic phase or terminal blast crisis. These negative data were not attributable to a low quantity of or degradation of the sample mRNAs, because the PCR product of β-actin mRNA was detected in amounts similar to those apparent with samples from patients with the other diseases. The expression of the Dlk gene is thus highly selective for the blasts from individuals with MDS.
Discussion
Our data indicate that purified cell subsets are more informative than are unfractionated cell populations for comparison of gene expression profiles by DNA microarray analysis. The cell fractionation protocol should be designed so that the characteristics of the samples for comparison, with the exception of the characteristic of interest (such as malignant transformation or drug resistance), are matched as closely as possible.
The hematopoietic system is a good target for this BAMP screening approach. Hundreds of cell-surface antigens have been identified on blood cells, and flow cytometric analysis based on these markers is routinely and extensively performed in clinical hematology laboratories. It is thus possible to isolate almost any specific population of blood cells by flow cytometry with a combination of antibodies specific for such cell-surface markers. A similar approach can be applied to solid tumors with the use of laser-dissection microscopy. Combination of this technique with amplification methods for RNA should allow microarray analysis of the gene expression profile of any given cell type.30 However, whereas purification of specific cell types from fresh blood in numbers of 105 or 106 is relatively straightforward, collection of the same number of cells by laser-dissection microscopy is a much more demanding task. Furthermore, it is usually necessary to fix and stain specimens, procedures that may damage cellular mRNA, before laser-dissection microscopy.
Populations of leukemic blasts usually contain immature, HSC-like cells as well as cells with committed phenotypes. The nature of the latter type of cells depends on the specific disease, and the difference in lineage commitment of these populations should thus contribute substantially to any differences in the transcriptomes detected in comparisons among leukemic blasts. In contrast, the immature, HSC-like subset of leukemic blasts that exhibits a CD34high, CD38low profile of surface antigen expression is common to many patients with different types of leukemia. We chose AC133 as the antigen on which to base our purification of such a disease-independent cell fraction. An AC133+ cell population can also be purified from healthy volunteers, thus allowing a direct comparison of AC133+ cells from such individuals with AC133+cells from patients with leukemia.
In addition to our comparison of MDS versus AML, our Blast Bank collection should prove to have a wide range of other applications in hematology. First, BAMP comparison of specimens between healthy individuals and leukemia patients is likely to shed light on the molecular events that underlie leukemogenesis. Second, in some leukemias including CML and MDS, overt leukemia is preceded by an indolent, chronic phase. Comparison of specimens obtained at different stages of such leukemias should facilitate characterization of the mechanism of disease progression. Third, collection of Blast Bank specimens over the long term would allow the sampling of leukemia patients at both drug-sensitive and drug-resistant stages; comparison of such samples should help to identify genes that contribute to the development of resistance to chemotherapeutic agents.
In conclusion, with the use of BAMP screening, we have identified a candidate MDS-specific gene. Further analysis of a larger number of patients is required to clarify whether the expression of theDlk gene is indeed a useful marker for the diagnosis of MDS.
We thank F. Takaku, H. Mizoguchi, and S. Sugano for critical reading of the manuscript; Y. Nakamura and T. Tanaka for advice on RNA amplification; and S. Kajigaya for helpful suggestions.
Supported in part by a Grant-in-Aid for Research on the Human Genome, Tissue Engineering, and Food Biotechnology and a Grant-in-Aid for Research on the Second-Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health and Welfare of Japan; by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan; and by the Science Research Promotion Fund of the Promotion and Mutual Aid Corporation for Private Schools of Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hiroyuki Mano, Division of Functional Genomics, Jichi Medical School, 3311-1 Yakushiji, Kawachi-gun, Tochigi 329-0498, Japan; e-mail: hmano@jichi.ac.jp.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal