Abstract
Functional consequences of 12 mutations—10 missense, 1 splicing defect, and 1 frameshift mutation—were characterized in the uroporphyrinogen decarboxylase (URO-D) gene found in Utah pedigrees with familial porphyria cutanea tarda (F-PCT). All but one mutation altered a restriction site in the URO-D gene, permitting identification of affected relatives using a combination of polymerase chain reaction and restriction enzyme digestion. In a bacterial expression system, 3 of the missense mutants were found in inclusion bodies, but 7 were expressed as soluble proteins. Enzymatic activity of soluble, recombinant mutant URO-D genes ranged from 29% to 94% of normal. URO-D mRNA levels in Epstein-Barr–virus transformed cells derived from patients were normal (with the exception of the frameshift mutation) even though protein levels were lower than normal, suggesting that missense mutations generally cause unstable URO-Ds in vivo. The crystal structures of 3 mutant URO-Ds were solved, and the structural consequences of the mutations were defined. All missense mutations reported here and by others were mapped to the crystal structure of URO-D, and structural effects were predicted. These studies define structural and functional consequences of URO-D mutations occurring in patients with F-PCT.
Introduction
Subnormal activity of uroporphyrinogen decarboxylase (URO-D) in hepatocytes is responsible for the most common form of porphyria in humans, porphyria cutanea tarda (PCT).1,2 PCT occurs with a prevalence of 1 to 5 per 25 0001 2 and is clinically characterized by skin fragility, bullous lesions, and hypertrichosis on sun-exposed areas. Photosensitivity is mediated by uroporphyrin and by partially decarboxylated porphyrins that accumulate in the liver, circulate in the plasma, and are excreted in the urine.
Mutations at the URO-D locus, transmitted as an autosomal dominant trait, are found in approximately one third of patients with PCT.1,2 PCT associated with URO-D mutations is designated familial PCT (F-PCT). Patients with F-PCT display approximately half-normal URO-D activity and half-normal concentration of URO-D protein in all tissues. More than 30 differentURO-D mutations have been identified in unrelated patients.3-14 Most carriers of mutant URO-Dalleles do not express clinical phenotypes unless additional factors are present that further reduce the activity of URO-D in the liver.15,16 Factors shown to be important include alcohol abuse, exposure to the hepatitis C virus, use of medicinal estrogens, and development of hepatic siderosis (often caused by mutation of the hemochromatosis gene).15,16 Depletion of hepatic iron stores through phlebotomy therapy corrects the clinical and biochemical manifestations of F-PCT.2
URO-D catalyzes the sequential decarboxylation of the 4 acetate side chains of uroporphyrinogen to form coproporphyrinogen. Isomer I or isomer III of uroporphyrinogen may serve as substrate, but only coproporphyrinogen III is a substrate for the next enzyme in the pathway, coproporphyrinogen oxidase. The mechanism of decarboxylation is unknown, but the enzyme does not require cofactors or prosthetic groups for activity. Human URO-D is a homodimer with a monomeric molecular weight of approximately 41 000 d.17 The protein is encoded by a single gene located on chromosome 1,1p34.18,19 The URO-D gene spans 3.5 kb and contains 10 exons.20 The crystal structure of human URO-D has been determined at 0.16 nm (1.6 Å) resolution.21 Structurally, URO-D is a member of the α8/β8 barrel family,21 a family first characterized by triosephosphate isomerase. There is one active-site cleft per monomer, located adjacent to its neighbor in the dimer to create a single extended cleft that likely is large enough to accommodate 2 substrate molecules in proximity or to allow reaction intermediates to shuttle between monomers. To define the functional consequences of mutations at the URO-D locus, we sequenced both alleles ofURO-D in 14 patients with F-PCT. Twelve mutations were identified, 10 missense mutations, 1 splice site mutation, and 1 nucleotide insertion mutation, leading to a frameshift. The 10 missense mutations were recombinantly expressed, and the physical and functional characteristics of the mutant proteins were defined.
Patients and methods
Patients
Patients were evaluated at the University of Utah General Clinical Research Center under a protocol approved by the Institutional Review Board. All participants gave written informed consent. They had skin lesions characteristic of PCT and markedly increased secretion of porphyrins in the urine (range, 1025 to 4520 μg/24 h). Uroporphyrin and heptacarboxyl porphyrin were identified as the dominant urinary porphyrins by high-performance liquid chromatography.22 A diagnosis of F-PCT was based on the demonstration of approximately half-normal URO-D activity in erythrocyte lysates.23Erythrocyte URO-D activity ranged from 40% to 60% of normal.
Sequencing of URO-D loci
Genomic DNA was used as a template for polymerase chain amplification (PCR) of the URO-D locus. Conditions and primers used are listed in Table 1. An automated ABI 377 sequencer (Applied Biosystems, Foster City, CA) was used for sequencing of the PCR products.
PCR primers and conditions for sequencing of theURO-D locus
| EXON . | Forward primer/reverse primer . | Mg 2+ (mM) . | AT (no. cycles) . |
|---|---|---|---|
| 1 | TTCCCCTGCCACTGACAGG | 1.50 | 62 (35) |
| aggaaacagctatgaccCGGAGTTCAGGTTGGGAGGT | |||
| 2-4 | TGGATTGAGCTCGCAGTTACAGACA | 1.25 | 62 (35) |
| caggaaacagctatgaccGACTGGACTGATAGAAGG | |||
| 5, 6 | caggaaacagctatgaccGACTGGACTGATAGAAGG | 2.00 | 60 (5) A |
| TGGTGTCTGAATACTTAGAG | 62 (30) B | ||
| 7-9 | caggaaacagctatgaccGTATGTGGGGGAAACTTC | 1.50 | 62 (35) |
| CTGTGTTAGTGGTACTTGAGGGTAAAG | |||
| 8, 9 | caggaaacagctatgaccGGAGGGCAGCAGAAGTACAG | 1.50 | 62 (5) A |
| CTAGTATGGACCCAGGCATG | 64 (30) B | ||
| 10 | GCCTTACATCCGTGATGTGGC | 1.50 | 60 (5) A |
| caggaaacagctatgaccTTGTCCTGGAAACGATC | 63 (30) B |
| EXON . | Forward primer/reverse primer . | Mg 2+ (mM) . | AT (no. cycles) . |
|---|---|---|---|
| 1 | TTCCCCTGCCACTGACAGG | 1.50 | 62 (35) |
| aggaaacagctatgaccCGGAGTTCAGGTTGGGAGGT | |||
| 2-4 | TGGATTGAGCTCGCAGTTACAGACA | 1.25 | 62 (35) |
| caggaaacagctatgaccGACTGGACTGATAGAAGG | |||
| 5, 6 | caggaaacagctatgaccGACTGGACTGATAGAAGG | 2.00 | 60 (5) A |
| TGGTGTCTGAATACTTAGAG | 62 (30) B | ||
| 7-9 | caggaaacagctatgaccGTATGTGGGGGAAACTTC | 1.50 | 62 (35) |
| CTGTGTTAGTGGTACTTGAGGGTAAAG | |||
| 8, 9 | caggaaacagctatgaccGGAGGGCAGCAGAAGTACAG | 1.50 | 62 (5) A |
| CTAGTATGGACCCAGGCATG | 64 (30) B | ||
| 10 | GCCTTACATCCGTGATGTGGC | 1.50 | 60 (5) A |
| caggaaacagctatgaccTTGTCCTGGAAACGATC | 63 (30) B |
All reactions were performed in a total volume of 50 μL with 0.1 mM dNTP, 2.5 U Taq DNA polymerase (Life Technologies, Gaithersburg, MD), and 5 to 10 pmol each primer. PCR was performed in a HYBAID Thermal Reactor (Labnet, Woodbridge, NJ) with an elongation temperature of 72° and a denaturation temperature of 95°. When 2 annealing temperatures (AT) are given, A is the primary AT used for the specified number of cycles and B is the AT used for the completion of the PCR reaction. Oligonucleotide sequences are given 5′ to 3′, with the uppercase letters representing the portion that is complementary to regions of the URO-D locus. Lowercase portions of the primers correspond to the M13 reverse primer that were incorporated into the PCR product to facilitate sequencing.
Expression of URO-D proteins
Missense mutations were reconstructed in an Escherichia coli expression system. Mutations were introduced into a wild-typeURO-D cDNA using the pAlter (Promega, Madison, WI) site-directed mutagenesis system.17 MutatedURO-D genes were subcloned into an expression plasmid, allowing incorporation of a histidine tag at the amino terminus. Recombinant proteins were then purified using metal-chelate affinity chromatography (Qiagen, Valencia, CA),17 yielding 2 to 4 mg highly purified URO-D from a 1-L bacterial culture. Recombinant proteins were approximately 95% pure as judged by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. Wild-type URO-D protein from this expression system had the same specific activity as URO-D purified from human red blood cells.17
Crystallization and structural analysis
Three mutant URO-D proteins (G156D, F232L, I260T) were purified and concentrated to 6 mg/mL in 50 mM Tris, pH 7, 10% glycerol, 1 mM β-mercaptoethanol. These proteins were crystallized by slow equilibration in a buffer containing methylpentanediol as described.17 Crystals were isomorphous with those used previously to determine the crystal structure of URO-D.21Radiographic diffraction data were collected from crystals maintained at 100°K. Diffraction data from home and synchrotron radiographic sources were processed and scaled with Denzo and Scalepack (Hill Research, Charlottesville, VA).24 The structure of native URO-D (PDB code, 1URO)21 was used as the starting point to phase diffraction data obtained from crystals of mutant proteins. Models were refined with X-Plor25 and rebuilt with the program O (Table2).26
Radiographic refinement statistics for URO-D mutants
| Mutant . | Gly156Asp . | Phe232Leu . | Ile260Thr . |
|---|---|---|---|
| Resolution (nm) (highest resolution shell) | 0.2-0.22 (0.228-0.220) | 0.2-0.23 (0.238-0.230) | 0.2-0.19 (0.193-0.190) |
| No. observed reflections | 274 282 | 262 230 | 330 724 |
| No. unique reflections | 23 423 | 20 018 | 34 362 |
| Completeness (%) (highest resolution shell) | 97.1 (99.2) | 99.0 (99.7) | 96.0 (71.7) |
| Rmerge*(highest resolution shell) | 0.084 (0.39) | 0.078 (0.40) | 0.070 (0.34) |
| Mosaicity | 0.51 | 0.36 | 0.38 |
| R-factor, %† | 0.195 | 0.200 | 0.202 |
| R-free‡ (5% of data) | 0.238 | 0.243 | 0.235 |
| Mutant . | Gly156Asp . | Phe232Leu . | Ile260Thr . |
|---|---|---|---|
| Resolution (nm) (highest resolution shell) | 0.2-0.22 (0.228-0.220) | 0.2-0.23 (0.238-0.230) | 0.2-0.19 (0.193-0.190) |
| No. observed reflections | 274 282 | 262 230 | 330 724 |
| No. unique reflections | 23 423 | 20 018 | 34 362 |
| Completeness (%) (highest resolution shell) | 97.1 (99.2) | 99.0 (99.7) | 96.0 (71.7) |
| Rmerge*(highest resolution shell) | 0.084 (0.39) | 0.078 (0.40) | 0.070 (0.34) |
| Mosaicity | 0.51 | 0.36 | 0.38 |
| R-factor, %† | 0.195 | 0.200 | 0.202 |
| R-free‡ (5% of data) | 0.238 | 0.243 | 0.235 |
Rmerge = ΣI − 〈I〉/ΣI, where I is the intensity of an individual measurement and 〈I〉 is the average intensity from multiple observations.
R-factor = ΣFobs − Fcalc/Fobs, where Fobs and Fcalc are the observed and the calculated structure-factor amplitudes, respectively.
R-free is the R-factor for a 5% subset of reflections not used in refinement.
Enzymatic activity assays for URO-D
Growth of Epstein-Barr virus–transformed lymphocytes
Lymphocytes were isolated from venous blood and transfected with Epstein-Barr virus (EBV) as described by Pelloquin et al.27 Transformed cells were cultured in suspension at 37°C, 5% CO2, in RPMI 1640 with L-glutamine, 20% fetal calf serum and 50 μg/mL gentamicin (Life Technologies, Frederick, MD). Cells were passaged twice a week. Steady- state levels of URO-D protein in transformed cell lines were determined by Western blotting using a monospecific polyclonal rabbit anti–URO-D antibody.28 An antibody to the B56 subunit of β-phosphatase29 was used to normalize for protein load.
RNA levels by ribonuclease protection assays
Total RNA was extracted from 1 × 107EBV-transformed lymphocytes using Trizol reagent (Life Technologies).URO-D transcript levels were determined from 30 μg total cellular RNA using the Ambion ribonuclease protection assay system (RPA II; Austin, TX). An antisense RNA probe to URO-D mRNA protected a 376-base fragment at the 5′ end of the message and was used in conjunction with a control probe that protected a 125-base region of human β-actin mRNA. Probes were hybridized with the sample RNA for 6 hours, then digested for 1 hour with RNase reagents supplied with the Ambion RPA II system. Protected fragments were then separated on a 5% polyacrylamide gel. Gels were dried and exposed to film for 30 minutes to 4 days to obtain proper exposures. The relative level ofURO-D mRNA was quantified using densitometry measured with a BioRad Molecular Imager FX equipped with the Quantity One software package (BioRad, Hercules, CA).
Immunoblot analysis
Protein was extracted from 2 × 106EBV-transformed cells after centrifugation (1000g, 15 minutes) and 2 washes in 1 × phosphate-buffered saline (PBS) (10 × PBS = 1.37 M NaCl, 27 mM KCl, 43 mM Na2HPO4, 14 mM KH2PO4, pH 7.5). Cells were sonicated for 2 minutes on ice and then spun at 10 000g for 10 minutes at 4°C. Supernatants were resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, NH). Filters were blocked with 5% nonfat dry milk, 1 × PBS, and 0.1% Tween 20 for 30 minutes, followed by 2-hour incubation with a rabbit anti–human URO-D antibody28 diluted 1:8000 and a rabbit anti-B56 phosphatase 2A subunit antibody29 diluted 1:2500 at room temperature. Blots were washed 3 × 15 minutes with 1 × PBS and 0.1% Tween 20 at room temperature. The secondary antibody was horseradish peroxidase–conjugated goat anti–rabbit immunoglobulin G (IgG)–diluted 1:5000 in the blocking buffer. Blots were developed using the Renaissance Western Chemiluminescence reagent (New England Nuclear, Boston, MA) and exposure to film. The relative level of URO-D protein was quantified by densitometry as described above.
Results
Identification of mutations
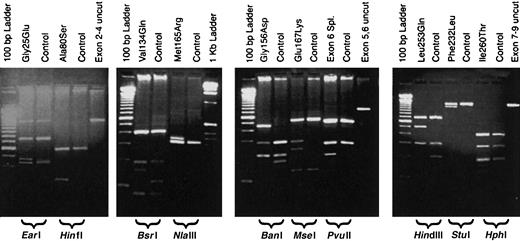
URO-D gene sequences from patient DNA samples were compared to the published sequence of URO-D (GenBank accession number AF047383). All mutations were confirmed by repeating both the PCR and the sequencing procedures. We identified 12 different mutations in 14 unrelated patients with F-PCT (Table3). Six of the 12 mutations have been reported elsewhere, and 2 were detected in more than one proband (Table3). The mutations resulted in altered restriction enzyme digestion patterns with one exception, a mutation characterized by insertion of a thymidine in codon 218 (Glu218insT). These mutations were easily detected by digesting one of the PCR products (Table 3, Figure1). Detection of the Glu218insT mutation required sequencing the exon 7-9 PCR product (Table 1). Representative results of restriction fragment length polymorphisms for 10 of the mutations are shown in Figure 1.
Characteristics of URO-D mutations
| Mutation . | Exon . | Location in cDNA . | RFLP . | Recombinant URO-D activity . |
|---|---|---|---|---|
| G25E | 2 | G → A 74 | EarI (+) | INS |
| A80S12 | 4 | G → T 238 | HinfI (+) | 33.4% |
| V134Q4 3-150 | 5 | G TG → C CA 399 | BsrI (+) | 93.7% |
| G156D | 5 | G → A 467 | BanI (−) | 28.8% |
| M165R13 | 6 | T → G 494 | NlaIII (−) | INS |
| E167K6 | 6 | G → A 499 | MseI (+) | 93.5% |
| Exon 6 splice30 | /6/ | G → C IVS6 | PvuII (+) | NT |
| R193P3-150 | 6 | G → C 578 | SfaNI(−) | INS |
| Glu218insT | 7 | Insert T 652 | None | NT |
| F232L | 7 | T → C 694 | StuI (−) | 43.0% |
| L253Q9 | 7 | T → A 758 | HindIII (+) | 75.0% |
| I260T | 8 | T → C 779 | HphI (+) | 60.5% |
| Mutation . | Exon . | Location in cDNA . | RFLP . | Recombinant URO-D activity . |
|---|---|---|---|---|
| G25E | 2 | G → A 74 | EarI (+) | INS |
| A80S12 | 4 | G → T 238 | HinfI (+) | 33.4% |
| V134Q4 3-150 | 5 | G TG → C CA 399 | BsrI (+) | 93.7% |
| G156D | 5 | G → A 467 | BanI (−) | 28.8% |
| M165R13 | 6 | T → G 494 | NlaIII (−) | INS |
| E167K6 | 6 | G → A 499 | MseI (+) | 93.5% |
| Exon 6 splice30 | /6/ | G → C IVS6 | PvuII (+) | NT |
| R193P3-150 | 6 | G → C 578 | SfaNI(−) | INS |
| Glu218insT | 7 | Insert T 652 | None | NT |
| F232L | 7 | T → C 694 | StuI (−) | 43.0% |
| L253Q9 | 7 | T → A 758 | HindIII (+) | 75.0% |
| I260T | 8 | T → C 779 | HphI (+) | 60.5% |
Superscript numbers indicate references for mutations previously reported.
Location in cDNA indicates where A of initial ATG is 1.
RFLP indicates restriction fragment length polymorphism; (+), adds the restriction site; INS, insoluble protein in inclusion bodies; (−), removes restriction site; and NT, not tested, no protein produced.
RFLP mutation was identified in 2 unrelated patients with PCT.
Restriction digestion patterns of PCR products detect mutations in the
URO-D gene. Genomic DNA from probands and controls was PCR amplified, digested with a restriction enzyme, and separated on a 2% agarose gel. Lanes are marked as either mutant or control (above), and the restriction enzyme used is indicated below the gel. A lane containing an undigested PCR product (control) is also included. Molecular markers are present on the outside lanes of each gel.
Restriction digestion patterns of PCR products detect mutations in the
URO-D gene. Genomic DNA from probands and controls was PCR amplified, digested with a restriction enzyme, and separated on a 2% agarose gel. Lanes are marked as either mutant or control (above), and the restriction enzyme used is indicated below the gel. A lane containing an undigested PCR product (control) is also included. Molecular markers are present on the outside lanes of each gel.
Catalytic activity of recombinant URO-D mutants
The 10 missense mutations were engineered into an E coli expression system. Seven of the recombinant proteins were stable and soluble. The other 3 proteins were expressed but were detected only in inclusion bodies and could not be solubilized. Enzymatic activity of the soluble purified proteins ranged from approximately 29% to 94% of wild-type URO-D (Table 3).
Steady-state levels of URO-D mRNA and protein in transformed lymphocytes
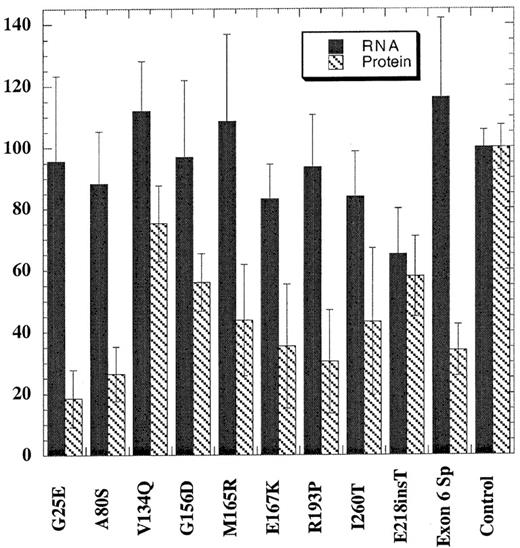
EBV-transformed cell lines were generated for only 10 of the mutations because lymphocytes from the probands with the Phe232Leu and Leu253Gln mutations failed to transform. Total RNA was prepared from cells and assayed for steady-state levels of URO-D mRNA using a ribonuclease protection assay (see “Patients and methods”). The quantity of URO-D mRNA in cells from patients with PCT was compared with cell lines derived from 6 nonporphyric patients with hemochromatosis. In the cell line with the insertion mutation (Glu218insT), the URO-D mRNA level was approximately half normal, indicating that transcription of the wild-type allele was not up-regulated (Figure 2). All other mutant cells lines contained normal levels of URO-D mRNA, indicating that transcripts were generated from both wild-type and mutant alleles. The splice mutant generated a stable, truncated transcript as previously reported.30 URO-D protein levels varied from 19% to 75% of control values (Figure 2). Subnormal amounts of URO-D protein in the presence of normal levels ofURO-D mRNA indicated that mutant proteins were probably unstable or insoluble.
URO-D protein and mRNA levels present in EBV-transformed lymphocytes.
Total URO-D mRNA and URO-D protein were quantified as described in “Patients and methods.” Densitometry values were normalized to the levels of β-actin for mRNA and B56 phosphatase 2A subunit for total protein. The average from 3 separate experiments is shown. Error bars represent the standard deviation. The control value represents the average of 6 nonporphyric EBV-transformed cell lines. Solid bars represent URO-D mRNA levels, and hatched bars represent URO-D protein level. In all mutants except one, the relative amount of URO-D mRNA exceeded the protein level. The insertion mutant (Glu218insT) generated approximately equivalent amounts of mRNA and protein.
URO-D protein and mRNA levels present in EBV-transformed lymphocytes.
Total URO-D mRNA and URO-D protein were quantified as described in “Patients and methods.” Densitometry values were normalized to the levels of β-actin for mRNA and B56 phosphatase 2A subunit for total protein. The average from 3 separate experiments is shown. Error bars represent the standard deviation. The control value represents the average of 6 nonporphyric EBV-transformed cell lines. Solid bars represent URO-D mRNA levels, and hatched bars represent URO-D protein level. In all mutants except one, the relative amount of URO-D mRNA exceeded the protein level. The insertion mutant (Glu218insT) generated approximately equivalent amounts of mRNA and protein.
Structure of recombinant mutants
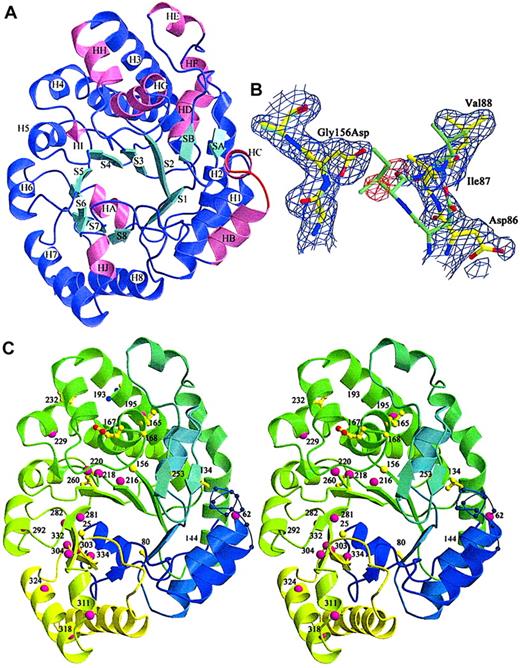
Three URO-D mutants (Gly156Asp, Phe232Leu, Ile260Thr) were produced in sufficient quantity for crystallization. Radiographic structures were compared with the wild-type structure to determine the structural effects of the mutations (Table 2). All mutant structures contained a common structural change, the disorder of a surface loop between HB and H1 (Figure 3A, residues 58-70).21 The structural explanation for this result was not apparent because this loop was remote from any of the mutations.
Location of mutations in the URO-D structure.
(A) View of a URO-D monomer approximately along the axis of the β-barrel, looking directly at the active site cleft. Secondary structural elements are labeled. β-Strands (cyan) forming the core of the barrel are labeled S1 to S8. Corresponding helices (purple) are labeled H1 to H8. Accessory helices are labeled HA to HJ (pink), and the accessory β-strands (cyan) are labeled SA and SB. The loop that became disordered in mutant structures (residues 56-62) between HB and H1 is shown in red. (B) Electron density (1ς 2Fo-Fc) covering the refined structure of Gly156Asp URO-D. The Gly156Asp structure is shown in yellow, and the wild-type URO-D structure is shown in green. The Asp side chain at position 156 causes the movement in nearby residues, 85 to 87, toward the active site cleft (right side of figure). Two water molecules (as depicted by the red difference density −3ς Fo-Fc) fill the gap left behind and form hydrogen bonds to the Asp. (C) Stereo diagram of the structure of a URO-D monomer, from blue to green to yellow, along the amino acid sequence. The disordered loop is shown as a thin blue line with spheres on the Cα positions. Numbers indicate the positions of mutant residues. Amino acids that were identified as mutant in this study are depicted as yellow ball-and-stick, and they are depicted as pink spheres for previously reported mutations.
Location of mutations in the URO-D structure.
(A) View of a URO-D monomer approximately along the axis of the β-barrel, looking directly at the active site cleft. Secondary structural elements are labeled. β-Strands (cyan) forming the core of the barrel are labeled S1 to S8. Corresponding helices (purple) are labeled H1 to H8. Accessory helices are labeled HA to HJ (pink), and the accessory β-strands (cyan) are labeled SA and SB. The loop that became disordered in mutant structures (residues 56-62) between HB and H1 is shown in red. (B) Electron density (1ς 2Fo-Fc) covering the refined structure of Gly156Asp URO-D. The Gly156Asp structure is shown in yellow, and the wild-type URO-D structure is shown in green. The Asp side chain at position 156 causes the movement in nearby residues, 85 to 87, toward the active site cleft (right side of figure). Two water molecules (as depicted by the red difference density −3ς Fo-Fc) fill the gap left behind and form hydrogen bonds to the Asp. (C) Stereo diagram of the structure of a URO-D monomer, from blue to green to yellow, along the amino acid sequence. The disordered loop is shown as a thin blue line with spheres on the Cα positions. Numbers indicate the positions of mutant residues. Amino acids that were identified as mutant in this study are depicted as yellow ball-and-stick, and they are depicted as pink spheres for previously reported mutations.
The most dramatic effect of a point mutation was seen with glycine 156, an absolutely conserved residue located on strand 3 of the β-barrel of the enzyme.21 In the Gly156Asp mutant protein, the carboxylate of the substituted side chain extended into a hydrophobic pocket surrounded by residues Ile87, Phe217, Leu161, Tyr164, and Val90. This apparently caused a shift in 2 additional conserved residues; Asp86 shifted approximately 0.29 nm (2.9 Å) and Ile87 shifted approximately 0.34 nm (3.4 Å). In addition, the side chain of Leu88 was displaced by approximately 0.18 nm (1.8 Å) (Figure 3B). The 3 side chains that moved in response to the Gly156Asp mutation lined the active site cleft. Movement of these side chains into the cleft explained the markedly impaired catalytic activity of this mutant protein (Table 3).
Crystal structures of the Phe232Leu and Ile260Thr mutants revealed no significant local perturbations surrounding the mutations, even though recombinant mutant proteins had reduced catalytic activity (Table 3). In the native structure, Phe232 is partially solvent accessible. In spite of only a minor structural change, the activity of recombinant Phe232Leu URO-D was reduced to 43% of wild type (Table 3). In the Ile260Thr structure, the extra carbon atom in isoleucine is replaced by a water molecule that forms a hydrogen bond to the threonine side chain. This relatively conservative mutation results in only a modest reduction of catalytic activity (Table 3).
Other URO-D mutations
Thirty URO-D mutations have been reported elsewhere.3-14 Ten of these mutant alleles are predicted to generate shorter proteins as a result of a premature stop codon or a variation in a splice site.3-13 The remaining 20 previously reported mutations are missense mutations listed in Table4 and mapped onto the URO-D monomer in Figure 3C. The structural location of these mutations and the predicted structural effects are described in Table 4.
Structural characterization of the reported missense mutations in URO-D
| Residue change . | Structural location . | Probable effect of mutation . |
|---|---|---|
| G25E | H1-S2 | Tight turn, restricted main chain |
| P62L | HC-H1 | Residue within disordered loop, unknown function or effect of mutation |
| A80S/G | S2 | No explanation |
| V134Q | H2 | Polar residue into a hydrophobic pocket |
| R144P | H2 | Salt bridge between R144 and Q63, helix-helix stability; P is also a helix breaker |
| G156D | S3-HG | Tight turn and restricted side-chain access |
| M165R | HG | Charged residue inserted into a hydrophobic pocket |
| E167K4-151 | HG | Charge reversal and the E167 links residues in a surface loop |
| G168R | HG-HH | Tight turn, restricted main chain |
| R193P | H3 | Pro insertion into middle of helix |
| L195F | H3 | No explanation |
| L216Q | S4 | Hydrophilic residue placed into hydrophobic core, near active site |
| E218K | S4 | Charge reversal and longer side chain, near active site |
| H220P4-151 | S4-HI | Histidine links residues in the intersection of several loops, proline is rigid and hydrophobic |
| F229L4-151 | H4 | No explanation |
| F232L4-151 | H4 | No explanation |
| L253Q | H4-S5 | No explanation |
| I260T | S5 | No explanation, water molecule replaces extra carbon atom |
| G281E/V | S6 | Mutant side chain protrudes into active site cleft |
| L282R4-151 | S6 | Charged residue inserted into a hydrophobic pocket, near active site |
| R292G4-151 | H6 | Arginine forms a salt bridge between H6 and S8 |
| G303S | S7 | Gly in conformationally strained conformation, side-chain conflicts |
| N304K4-151 | S2 | Tightly packed between H6 and H7, no room for longer Lys side chain |
| Y311C4-150 | HJ | Tyrosine packs against Arg179 side chain not supported by Cys, at dimer interface |
| G318R4-151 | H7 | Glycine lies on an internal turn of helix, large side chains disrupt packing |
| M324T4-151 | H7 | Side chain points toward protein disrupts packing of helix, dimer interface |
| R332H | S8 | No explanation |
| I334T | S8 | No explanation |
| Residue change . | Structural location . | Probable effect of mutation . |
|---|---|---|
| G25E | H1-S2 | Tight turn, restricted main chain |
| P62L | HC-H1 | Residue within disordered loop, unknown function or effect of mutation |
| A80S/G | S2 | No explanation |
| V134Q | H2 | Polar residue into a hydrophobic pocket |
| R144P | H2 | Salt bridge between R144 and Q63, helix-helix stability; P is also a helix breaker |
| G156D | S3-HG | Tight turn and restricted side-chain access |
| M165R | HG | Charged residue inserted into a hydrophobic pocket |
| E167K4-151 | HG | Charge reversal and the E167 links residues in a surface loop |
| G168R | HG-HH | Tight turn, restricted main chain |
| R193P | H3 | Pro insertion into middle of helix |
| L195F | H3 | No explanation |
| L216Q | S4 | Hydrophilic residue placed into hydrophobic core, near active site |
| E218K | S4 | Charge reversal and longer side chain, near active site |
| H220P4-151 | S4-HI | Histidine links residues in the intersection of several loops, proline is rigid and hydrophobic |
| F229L4-151 | H4 | No explanation |
| F232L4-151 | H4 | No explanation |
| L253Q | H4-S5 | No explanation |
| I260T | S5 | No explanation, water molecule replaces extra carbon atom |
| G281E/V | S6 | Mutant side chain protrudes into active site cleft |
| L282R4-151 | S6 | Charged residue inserted into a hydrophobic pocket, near active site |
| R292G4-151 | H6 | Arginine forms a salt bridge between H6 and S8 |
| G303S | S7 | Gly in conformationally strained conformation, side-chain conflicts |
| N304K4-151 | S2 | Tightly packed between H6 and H7, no room for longer Lys side chain |
| Y311C4-150 | HJ | Tyrosine packs against Arg179 side chain not supported by Cys, at dimer interface |
| G318R4-151 | H7 | Glycine lies on an internal turn of helix, large side chains disrupt packing |
| M324T4-151 | H7 | Side chain points toward protein disrupts packing of helix, dimer interface |
| R332H | S8 | No explanation |
| I334T | S8 | No explanation |
On the dimer interface.
Within 0.7 to 1.2 nm of the dimer interface.
Discussion
The structure of URO-D consists of a TIM (triosephosphate isomerase) barrel with 8 central β-strands surrounded by 8 α-helices (Figure 3A). The active site is at the C-terminal end of the β-barrel, and many of the invariant residues are concentrated in this region.21 Potential effects of amino acid substitutions at any particular position fall into 3 categories: substitutions affecting catalytic activity; structural restrictions to main chain conformation (usually substituted Gly or Pro residues), and side chain size or polarity changes. To date, no genetic mutation has been identified at a residue that lies directly in the active site. Most of the missense mutations previously described are postulated to result in loss of activity through perturbations of the structure surrounding the active site or perturbations of more distant structural features predicted to affect overall thermal stability, protease susceptibility, aggregation, or solubility of the enzyme. Predicted consequences of the missense mutations identified to date are shown in Table 4. Several of the mutations cause conservative changes in the protein sequence, and the structural consequences are predicted to be slight. Nonetheless, heterozygotes for these mutations have approximately half-normal activity for URO-D in erythrocyte lysates.
Two of the mutants we identified encode truncated proteins. In the Glu218insT mutation, the insertion of a thymidine in codon 218 introduces a stop codon (TGA) in place of the wild-type codon (GAG) for glutamic acid. EBV-transformed cells from the patient with this mutation were from the only cell line studied that had approximately half-normal levels of URO-D mRNA (Figure 2). This finding suggests that the mutant transcript is rapidly degraded through a nonsense-mediated decay mechanism.31,32 The exon 6 splice mutation, previously described,7,30 produces a truncated but stable mRNA. Deletion of exon 6 does not disrupt the reading frame, but the mutant protein is catalytically inactive and unstable.30 An EBV-transformed line from the proband with this mutation contained normal amounts of URO-D mRNA but half-normal amounts of protein (Figure 2).
Three mutations identified in this study—Gly25Glu, Met165Arg, and Arg193Pro—yielded insoluble recombinant proteins. In each case, structural considerations suggested that the mutation caused misfolding of the protein. The first of these mutations, Gly25Glu, was expected to destabilize the protein fold because this residue was located in a tight turn, between helix 1 and strand 2, with φ and ψ angles that favored only glycine with no obvious space to accommodate a larger side chain. The Met165Arg mutation was previously described by Mendez et al,13 who could detect no activity attributable to the mutant protein in an E coli expression system. Although Met165 was remote from the active site, it was packed in a hydrophobic pocket that appeared unable to accommodate a charged arginine side chain (Figure 3C). Thus, the Met165Arg substitution was expected to disrupt the protein's tertiary structure. The third insoluble recombinant protein was generated by the Arg193Pro mutation, which was expected to disrupt helix 3 (Figure 3A).
Three missense mutations (Ala80Ser, Gly156Asp, Phe232Leu) yielded recombinant URO-Ds with catalytic activities of approximately 40% or less. The Ala80Ser mutation was previously detected by Brady et al,12 but properties of the mutant enzyme were not determined. A second mutation at the same residue, Ala80Gly, has been described by McManus et al,9 who reported the catalytic activity of recombinant Ala80Gly URO-D to range from 19% to 39% of normal, values similar to the 33% activity we found for recombinant Ala80Ser URO-D. Ala80 is located at the N-terminal end of the S2 β-strand buried with other hydrophobic side chains below the active site. The predicted effect of the Ala80Ser mutation would be disruption of the hydrophobic core, causing conformational changes that could propagate to alter the structure at the base of the active site. Reduced catalytic activity of the Ala80Gly mutation might result through disruption of the structure by introduction of a void or as a result of increased mobility in the main chain observed to bond through hydrogen with the glutamine 38 side-chain. Hydrogen bonding with glutamine 38 appears to help order the invariant arginine 37 residue in the active site. In spite of these considerations, glycine is found in this position in the URO-D of barley, tobacco, and Synochoccusspp.21
The crystal structure of the Gly156Asp mutant revealed that this mutation perturbed nearby residues on the surface of the active site cleft (Figure 3B), thereby providing a plausible explanation for the reduced activity. In contrast, the crystal structure of the Phe232Leu mutant shows that this mutation, distant from the active site, did not alter the structure. It is unclear why the substitution of a leucine at this position led to reduced activity; leucine occupies this position in the URO-D of Caulobacter cresentus.21
Two mutations caused only a modest reduction of catalytic activity in the recombinant proteins Leu253Gln and Ile260Thr. Residue 253 is located at the transition between helix 4 and strand 5. The sequence of URO-D in this region varies considerably between species but is conserved in mammals.21 In contrast to our finding of 75% normal activity in purified recombinant Leu253Gln URO-D, McManus et al9 reported the identical mutant and found a marked reduction in activity of the mutant enzyme in a crude bacterial extract. The reason for this discrepancy is unclear. Although residue 253 is remote from the active site, the mutation results in the substitution of a hydrophilic glutamine residue for the hydrophobic, aliphatic leucine, a change that might have significant structural effects. The crystal structure of the Ile260Thr mutant revealed that the void generated by removal of the terminal isoleucine methyl is filled by incorporation of a buried water molecule, a change that has little effect on the overall structure but reduces catalytic activity by 40%.
Two missense mutations yielded recombinant URO-D with near normal catalytic activity, Val134Gln and Glu167Lys. Both have been previously described. Meguro et al4 also found near normal activity for the Val134Gln recombinant protein. The substitution of a glutamine for valine at position 134 resulted in the insertion of a polar residue in a hydrophobic pocket on helix 2 at a location remote from the active site. Catalytic activity of the Glu167Lys has not been previously measured, but the recombinant mutant URO-D has been shown to be rapidly degraded.6 Glutamic acid normally at position 167 links residues in a surface loop. The charge reversal resulting from substitution of a lysine would alter the structure of this surface loop and might target the protein for degradation.
URO-D is a dimer in the crystal21 and in solution.17 Formation of the dimer interface is critically important—directed mutations designed to yield monomeric proteins by disruption of the interface resulted in insoluble or unstable proteins when expressed in E coli (data not shown). Many of the clinically identified human mutations lie near the dimer interface, and it seems likely that the properties of these mutants (Table 4, footnote) resulted from disruption of the dimer interface. It is attractive to speculate that the active sites of adjacent monomers, which form a contiguous cleft in the dimer, cooperate in catalysis, perhaps by allowing reaction intermediates to shuttle between active sites for successive decarboxylations.
Patients with F-PCT are heterozygous for mutant URO-Dalleles and theoretically could produce 3 dimeric proteins: the wild-type homodimer, the mutant homodimer, and a mutant–wild-type heterodimer. The observation that URO-D activity in erythrocyte lysates from patients with F-PCT usually approximates 50% is consistent with the conclusion that most clinically relevant mutations yield unstable or unfolded proteins that are unable to form dimers. It is possible that the occasional detection of URO-D activity of 40% or less13 reflects formation of mutant–wild-type heterodimers with reduced catalytic activity or altered stability. Normal levels ofURO-D mRNA were detected in EBV-transformed lines (with the exception of the mutation causing the premature stop codon), yet URO-D protein levels varied widely. This is surprising in view of the half-normal protein levels found in red blood cells, suggesting that the capacity of the proteolytic machinery capable of recognizing mutantURO-D genes differs between these 2 cell types.
URO-D is not a rate-limiting step in the porphyrin biosynthetic pathway,1,2 and most carriers of mutant URO-Dalleles do not express a clinical phenotype. Homozygosity forURO-D null alleles is lethal,33 but homozygosity or compound heterozygosity for mutant alleles that encodeURO-D genes with some residual activity can be tolerated.1,2 In homozygotes or compound heterozygotes, the porphyric phenotype is pronounced and is usually evident in early childhood.2
The incidence of mutant URO-D alleles in the population is unknown, but it is unlikely to approach the approximately 30% incidence we and others observe in patients with PCT.2,15 The conclusion that heterozygosity for mutantURO-D alleles is a risk factor for the development of PCT15 is supported by the observation that heterozygous mice are more sensitive to porphyrinogenic stimuli than wild-type animals.33 In heterozygous mice and humans displaying porphyric phenotypes, hepatic URO-D protein level is half-normal, but catalytic activity is reduced to approximately 20%.33-35This finding strongly suggests that an inhibitor of hepatic URO-D is generated when the porphyric phenotype becomes manifest. The mechanism by which mutant URO-D alleles cause predisposition to the development of a URO-D inhibitor remains unresolved.
Supported by National Institutes of Health grants R-37 DK20503, RO-1 GM56775, MO-1 RR00064, and P-30 CA42014.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
John D. Phillips, Division of Hematology, University of Utah School of Medicine, 50 North Medical Dr, Salt Lake City, UT 84132; e-mail: john.phillips@hsc.utah.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal