Abstract
Human erythrocyte pyruvate kinase plays an important role in erythrocyte metabolism. Mutation on the gene results in pyruvate kinase deficiency and is an important cause of hereditary nonspherocytic hemolytic anemia. Because of difficulties in isolating the mutant enzymes from patients, these mutations have not been fully studied. In this study, a complementary DNA (cDNA) encoding the human erythrocyte pyruvate kinase was generated. The cDNA was cloned into several expression vectors, and the protein was expressed and purified. The tetrameric protein exhibited properties characteristic of authentic human erythrocyte pyruvate kinase, including response to substrate, phosphoenolpyruvate, activation by fructose 1,6-bisphosphate, and inhibition by adenosine triphosphate (ATP). The N-terminal segment of the protein was highly susceptible to proteolysis, but only 2 of the 4 subunits were cleaved and lacked 47 N-terminal amino acid residues. A mutant protein, R510Q, which is the most frequently occurring mutation among Northern European population, was also generated and purified. The mutant protein retained its binding capacity to and could be activated by fructose 1,6-bisphosphate and showed similar kinetics toward phosphoenolpyruvate and adenosine diphosphate as for the wild-type enzyme. Conversely, the mutant protein has a dramatically decreased stability toward heat and is more susceptible to ATP inhibition. The enzyme instability decreases the enzyme level in the cell, accounting for the clinically observed “pyruvate kinase deficiency” of patients who are homozygous for this mutation. This study provides the first detailed functional characterization of human erythrocyte pyruvate kinase. These findings will allow the establishment of a fine correlation between molecular abnormalities and the clinical expression of the disease.
Introduction
Pyruvate kinase (ATP:pyruvate 2-O-phosphotransferase [EC 2.7.1.40]; PK) is a key enzyme in glycolysis, a metabolic pathway present in nearly all organisms and in all kinds of cells. The enzyme catalyzes the conversion of phosphoenolpyruvate (PEP) to pyruvate with the production of adenosine triphosphate (ATP) from adenosine diphosphate (ADP). The reaction product, pyruvate, is located in a position connecting the metabolic pathways of carbohydrates, amino acids, and lipids. Therefore, a tight regulation of PK activity is of great importance not only for glycolysis itself but also for cell metabolism in general. PK has been largely conserved through evolution. The enzyme is usually a tetramer comprising 4 identical subunits, each consisting of approximately 500 to 600 amino acids depending on the source of the enzyme.1A high degree of structural homology has been reported among PKs from different species. Crystal structures have been known for PKs from cat muscle,2,3 rabbit muscle 4, Escherichia coli,5 yeast,6 and Leishmania mexicana.7 These structures resemble each other in that each subunit has 3 domains, an (α/β)8 barrel A domain, a β-stranded B domain, and an (α/β) open-sheet C domain. With the exception of prokaryotes,8 a fourth domain, corresponding to the N-terminus, is also present. The 4 identical subunits are arranged in a tetramer with D2 symmetry (ie, 3 twofold rotation axes intersecting each other at right angles).
Biophysical, biochemical, and kinetic properties have been extensively studied for PKs from different sources.1 One of the main features of this enzyme is its allosteric response to a large number of effectors. The enzyme can adopt at least 2 different conformations, an active R-state and an inactive T-state. Transition between these 2 states can be triggered on binding one or more effectors. For example, PEP homotropically activates the enzyme; likewise, binding of fructose 1,6-bisphosphate (FBP), an upstream glycolytic intermediate generated by phosphofructose kinase, and low pH favor the R-state.1,9 Conversely, the enzyme is inhibited by ATP, alanine, and phenylalanine.10 In addition, partial activation through proteolysis,11 N-terminal phosphorylation favoring the T-state12,13 as well as hormone-triggered dimerization for some forms of enzyme14have also been reported. Elucidation of the enzyme structure has greatly enhanced the understanding of the allosteric transition mechanism and identified possible residues and interactions involved in the transition. Mattevi et al5 compared the E coli T-state with the rabbit muscle R-state structures and proposed a model in which the domains approximate rigid bodies connected by flexible hinges. The R ↔ T transition involves concerted rotations of domains mediated by interdomain and intersubunit interactions within the tetramer. This model was further refined by the comparative study of the yeast and L mexicanacrystal structures.6 7
The human genome contains 2 structural genes encoding 4 PK isozymes, the muscle (M1), kidney and fetal (M2), liver (L), and erythrocyte (R) forms.1 The erythrocyte PK (RPK) is encoded by thePKLR gene and is exclusively expressed in erythrocytes.15 This isozyme plays a central role in erythrocyte metabolism, because it catalyzes one of the 2 major steps of ATP production in the cell. In fact, mature red blood cells lack mitochondria, and, therefore, the energy needed for maintaining the cell membrane charge and for cycling NAD+ is generated nearly exclusively through glycolysis. Mutations in the PKLRgene result in PK deficiency, which is among the causes of nonspherocytic hemolytic anemia. More than 100 mutations have been identified at the DNA level to date.16 Most of them are missense mutations involving a single nucleotide substitution. Nonsense mutations, deletions, and alterations of the splicing sites have also been reported.17-19 Patients carrying these mutations have been primarily found in Japan, Europe, and the United States, but increasing evidence indicates that the disease is spread worldwide. Clinical diagnosis of PK deficiency relies on the demonstration of quantitative and/or qualitative enzyme abnormalities in the cells. Protein properties like thermal stability, susceptibility to proteolysis, substrate affinity, and allosteric response to effectors have been used to characterize the mutant enzymes.20However, these methods often give controversial results because of the instability of most mutant enzymes and because of the difficulty in obtaining a sufficient amount of the protein. In addition, contamination with the isozyme from leukocytes and presence of hybrid enzyme from heterozygous or compound heterozygous patients also hamper the enzymatic characterization. Although detection of mutations at the gene level has been advancing rapidly,17,19,21,22 the lack of protein characterization makes it difficult to predict the phenotypic consequence of specific mutations. To analyze the mutations at the protein level and eventually to elucidate the protein crystal structure, we cloned the full-length human R-type PK complementary DNA (cDNA) and expressed, purified, and characterized the recombinant enzyme. We also produced a mutant protein, R510Q, that is the most frequently occurring mutation among Northern European populations and Caucasian Americans.23 R510Q represents the first characterized recombinant mutant protein of human RPK.
Materials and methods
Materials
M-MTL reverse transcriptase was purchased from GIBCO, BRL (Gaithersburg, MD). Restriction enzymes and Taqpolymerase were purchased from New England Biolabs (Beverly, MA). Oligonucleotides were synthesized by Life Technologies (Paisley, United Kingdom). Plasmid pCR2.1 was from Invitrogen (Carlsbad, CA), pBluescript II KS from Stratagene (Austin, TX), and pET-21a from Novagen (Abingdon, United Kingdom). FBP, PEP, lactate dehydrogenase, and nicotinamide adenine dinucleotide reduced form (NADH) were from Roche Molecular Biochemicals (Basel, Switzerland). Other chemicals were reagent grade and obtained from Sigma-Aldrich (St Louis, MO).
RNA extraction, cDNA synthesis, and subcloning of polymerase chain reaction product
Peripheral blood (10 mL) was drawn from a patient with hereditary spherocytosis and normal PK activity. Total reticulocyte RNA was obtained by ammonium chloride lysis24,25 and reverse transcribed to cDNA by using random hexamer primers and M-MTLV reverse transcriptase. The entire cDNA obtained was amplified by polymerase chain reaction (PCR) (94°C for 30 seconds, 57°C for 30 seconds, and 72°C for 110 seconds; 30 cycles) by using primers designed according to the cDNA sequence reported by Kanno et al.26 The forward primer was 5′ATTGGAGCTCAAGGAGGCTGAAAGCATGTCGATCCAGGAGA and included a SacI restriction site and a ribosome-binding site. The reverse primer was 5′CCGGAATTCAGGGGGGAGGGGCGTCTCAGGA and included an EcoRI site. The PCR product was purified, restricted, and cloned into SacI- andEcoRI-digested pCR2.1 vector. The recombinant plasmid obtained was designated pMI1. The insert was sequenced in an ABI PRISM 310 genetic analyzer (PE Applied Biosystems, Foster City, CA) by the Big Dye termination method.
Correction of missense mutations by site-directed mutagenesis
To correct the RPK-coding sequence from undesired mutations (see “Results”), the insert was excised from pMI1 and subcloned into theSacI/EcoRI pBluescript II KS to make pPV8. The missense mutations 195G>A and 1237T>A found in the RPK sequence were reversed in a single mutagenesis experiment by following the protocol of Nickoloff et al27 and by using the commercially available Chamaleon double-stranded site-directed mutagenesis kit (Stratagene). The primers used for reversing the point mutations were 5′CTGCCAGCTGCTATGGCAGACACCTTCCTGG and 5′GACTGCCAAGGGCA-ACTTCCCTGTGGAAGCGG (the underlined sequences indicate the reverted bases) for nucleotide 195 and 1237, respectively. The selection primer for destroying the vector's uniqueXhoI site was 5′ATACCGTCGACCACGAGGGGGGGCCCGG. Plasmids isolated from cultures of individual transformants were screened by XhoI digestion. More than 80% of unlinearized plasmids contained the correct gene, as confirmed by DNA sequencing of the entire RPK insert. The pBluescript II KS harboring the correct cDNA, termed pGG1, was checked for expression.
Construction of pAZ1 coding for RPK
Starting from pGG1 (see above), a new expression vector was generated. This plasmid, designated pAZ1, was obtained by (1) creating in pGG1 an artificial NdeI site upstream of the translational initiation site (CATATG), (2) excising the insert by NdeI/EcoRI digestion, and (3) inserting the restricted insert into a pET-21a. The mutagenic primer used to introduce into pGG1 the new restriction site was 5′CAAGGAGGCTGAACATATGTC-GATCCAGGA. The selection primer used to abolish the vector unique restriction siteAflIII was 5′CGCAGGAAAGACCTTGGGAGCAAAAGGCC. The insert was completely sequenced.
Construction of pAZ2 coding for mutant R510Q
To obtain the mutant protein R510Q, the pAZ1 was subjected to site-directed mutagenesis. The mutation primer was 5′GGTCCACTTATGCCAAGGAGTCTTCCCCTT; the selection primer, 5′CTGAATTCGAGCTCCGACGTCAAGCTTGCGGCCGCA, converted the unique restriction site SalI intoAatII. The plasmid containing the desired point mutation was named pAZ2.
Expression trials
E coli BL21(DE3) and BL21(DE3)pLys transformed with the expression vectors were grown at 37°C in Luria-Bertani (LB) medium containing 100 μg/mL ampicillin. When the culture optical density at 600 nm reached a value of 0.4 to 0.6, the expression was induced by addition of isopropyl-β-D-thiogalactopyranoside to a final concentration of 0.2 to 1 mM. The induction time ranged between 5 and 24 hours; the induction temperature ranged between 20°C and 37°C.
Enzyme purification
The enzyme was purified by a simple procedure that included an ammonium-sulfate precipitation and 2 chromatographic steps. Typically, 2 liters E coli BL21(DE3) cells transformed with pAZ1 or pAZ2 were grown at 37°C to an optical density at 600 nm of 0.5 in an LB medium containing 100 μg/mL ampicillin. After 12 hours' induction at 24°C in the presence of 0.2 mM isopropyl-β-D-thiogalactopyranoside, the cells were collected by centrifugation and suspended in 200 mL buffer A (50 mM potassium phosphate pH 6.5, 100 mM KCl, 1 mM EDTA, and 1mM phenylmethylsulfonylfluoride). The cell suspension was sonicated at 800 W for 6 minutes and cleared by centrifugation, and the supernatant was subjected to ammonium-sulfate treatment (45% saturation). The pellet collected by centrifugation was suspended in buffer B (as buffer A but pH 7.5), dialyzed, and applied to a 45 × 5-cm DEAE-Sepharose (Amersham Pharmacia Biotech, Uppsala, Sweden) column equilibrated in buffer B. The recombinant protein was eluted by buffer B, concentrated, and applied to a 2 × 100-cm Sephacryl S-200 (Amersham Pharmacia Biotech) column equilibrated with buffer A. The protein was eluted with buffer A. To prevent proteolysis during purification, inhibitor cocktails from Sigma and Roche were used. Protein concentration was determined according to Lowry et al,28 using bovine serum albumin as standard. The purity of the protein was assessed by native gel electrophoresis and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The stock solution of the purified enzyme was stored at −20°C in buffer A containing 30% glycerol.
Definition of major kinetic parameters
kcat or turnover number is the number of catalytic events per second per active site. Kmis the substrate concentration at which the reaction velocity is half maximal. S0.5 is used for reactions displaying sigmoidal kinetics and is defined as Km.kcat/S0.5 is a measure of how efficiently an enzyme converts substrate to product at low substrate concentrations. Hill coefficient (nH) is an empirical parameter related to the degree of cooperativity; values larger than unity indicate positive cooperativity among ligand binding sites.
Enzyme activity assay
The enzyme activity of RPK was determined at 37°C by lactate dehydrogenase–coupled spectrophotometric assay according to the method recommended by the International Committee for Standardization in Hematology.20 The standard reaction mixture contained 100 mM Tris pH 8.0, 100 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 0.2 mM NADH, 10 μg LDH, 10 mM PEP, and 1.5 mM ADP in a final volume of 1 mL. The reaction was started by adding enzyme solution (0.5-1 μg). One unit is the amount of enzyme catalyzing the oxidation of 1 μmol NADH/min under the above conditions.
Kinetic analyses
Enzymatic activity was assayed at 37°C by using various concentrations of PEP and ADP under conditions identical to those described above except for substrates and effectors. Kinetic parameters were determined as follows: for PEP at fixed concentration of 1.5 mM ADP; for ADP at 5 mM PEP; for FBP at 0.1 mM PEP and 1.5 mM ADP. In all assays, when ATP was used, an equimolar concentration of Mg2+ was added. In all cases the reaction was started by adding PEP, and the enzyme activity was assayed at least at 10 different concentrations of substrate or effector. All measurements were performed in triplicate by using a Jasco V-550 UV/VIS spectrophotometer. The plot of Lineweaver-Burk was used to determineVmax and apparent Kmvalues. The Hill plot was used to determine the apparentS0.5 and nH values.
Thermal stability assay
Thermal stability was measured by incubating the enzyme (100-200 μg/mL) at a given temperature in a solution consisting of 50 mM potassium phosphate pH 6.5, 100 mM KCl, and 1 mM EDTA. Samples (10 μL) were removed at intervals and immediately assayed as described above.
Protein sequencing
The N-terminal sequence of purified RPK was determined by adsorptive biphasic column technology, using an HPG1000 A protein sequenator (Hewlett-Packard) with the routine 3.0 chemistry method and PHT__4M HPLC method.
Analytical gel chromatography
To determine the molecular mass of native enzyme, the purified RPK was subjected to gel filtration on a Superose 12 HR 10/30 prepacked column (Amersham Pharmacia Biotech) equilibrated in 20 mM potassium-phosphate buffer pH 6.5, 50 mM KCl, and 1 mM EDTA. A total of 100 μL purified RPK solution (0.2 mg/mL) was applied to the column on an ÄKTA purifier system (Amersham Pharmacia Biotech) and eluted with the equilibration buffer at 0.5 mL/min. For column calibration the following proteins were used: catalase (240 kDa), aldolase (158 kDa), albumin (68 kDa), ovalbumin (45 kDa), chymotrypsinogen (25 kDa), and cytochrom c (12 kDa).
SDS-gel electrophoresis
SDS 10% polyacrylamide gel electrophoresis was performed according to the method of Laemmli.29 The markers used for molecular mass determination were phosphorylase b (94 kDa), albumin (68 kDa), E coli PK (50.3 kDa), ovalbumin (45 kDa), and trypsin inhibitor (20.1 kDa).
Isoelectrofocusing
The isoelectrofocusing of purified RPK was performed according to standard protocol of Righetti and Gianazza,30 using an immobilized pH gradient ranging from pH 4 to pH 10.
Results
cDNA synthesis and its expression
Reticulocytes from a patient with hereditary spherocytosis and normal RPK activity were the source of total RNA. Reverse transcription with specific primers and PCR amplification resulted in a cDNA of 1750 base pair (bp) containing the whole reading frame of PKLR gene, cloned into SacI- and EcoRI-restricted pCR2.1. Sequencing of the cloned fragment showed that 3 nucleotides were different from the published sequence.26 31 One substitution (132G>A) corresponded to a silent mutation; whereas 2 substitutions (195G>A and 1237G>A) caused changes in coded amino acids (Met65Ile and Phe413Ile, respectively). We suggest that the substitutions were introduced during PCR amplification reaction, and, thus, we corrected the 2 missense mutations. To this end, the insert was transferred into the SacI, EcoRI restriction sites of pBluescript II KS. After site-directed mutagenesis, the resulting recombinant plasmid, designated pGG1, contained the correct coding region of RPK, cloned under the control of the T7 promoter. Starting from pGG1, we subcloned the cDNA into several expression vectors. The best expression was obtained using pAZ1, a pET-21a derivative with the RPK gene inserted between theNdeI and EcoRI sites. The highest level of soluble protein was obtained on induction for 12 hours at 24°C. Under these conditions the level of expression was 20 to 25 mg/L of culture.
Enzyme purification
The RPK expressed in BL21(DE3) transformed with pAZ1 (for wild-type RPK) or pAZ2 (for R510Q mutant) was purified by the method described under experimental procedures. The intrinsic E coli PK was eliminated by using a concentration of ammonium sulfate (45% saturation) at which only the RPK was precipitated. The specific activity of purified RPK was about 350 U/mg. Typically, the yield was 10 mg/L of culture.
Assessment of the oligomeric state of the enzyme
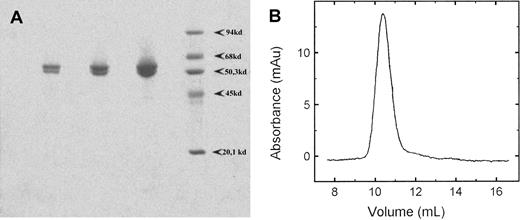
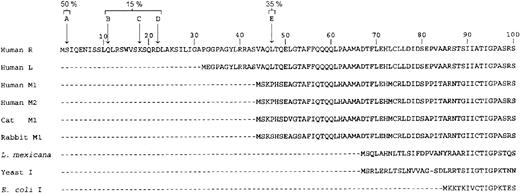
The purified protein was analyzed on a 10% SDS-PAGE. To our surprise we always observed 2 major bands on the gel (Figure1A). The slower-moving band had a molecular mass of 62 kDa, which corresponded to the full-length RPK subunit with a calculated molecular mass of 61874 Da (Swiss-prot No.O75758). The other band migrated as a 58 kDa. The observed heterogeneity can be explained either by the presence of contaminants or by partial proteolysis of the pure protein. To test the first possibility, we ran a sample on a native polyacrylamide gel. Only a single band was observed (data not shown). We also ran a sample on a Superose 12 HR column. The protein eluted from the column as a single peak, symmetric in shape. The peak was at a position of 240 kDa and corresponds to a tetramer of RPK (Figure 1B). This finding indicated that there was only one single species in the protein preparation. To assess if the heterogeneity was due to partial proteolysis, we determined the N-terminal sequence of the protein. Two major sequences were obtained, one corresponding to a full-length polypeptide (50%) and the second one starting at Thr48 (35%). The N-terminal residues and the percentage of each species, found by sequencing, are shown in Figure 2. We confirmed the homogeneity of the tetramer by isoelectrofocusing. An essentially single, narrow band focused at pH 6.8 on polyacrylamide slab gel in a 4 to 10 pH gradient (data not shown). From these results we concluded that the tetrameric protein comprises 2 full-length subunits and 2 partially proteolyzed ones. We tried different inhibitor cocktails to prevent proteolysis during purification, but without any success. Partial proteolysis has been reported for the RPK in human erythrocytes and has been attributed to aging of the cells.10 13
SDS-PAGE and analytical gel chromatography of recombinant human RPK.
(A) SDS-PAGE of purified recombinant human RPK. Three samples with increasing protein concentrations were run in parallel with molecular mass standards on a 10% gel and stained with Coomassie Blue R-250. (B) Elution profile of RPK from a Superose 12 HR 10/30 column. The position of the peak corresponds to a protein of 240 kDa, indicating a tetramer. Catalase (240 kDa), aldolase (158 kDa), albumin (68 kDa), ovalbumin (45 kDa), chymotrypsinogen (25 kDa), and cytochrome c (12 kDa) were used as molecular mass standards.
SDS-PAGE and analytical gel chromatography of recombinant human RPK.
(A) SDS-PAGE of purified recombinant human RPK. Three samples with increasing protein concentrations were run in parallel with molecular mass standards on a 10% gel and stained with Coomassie Blue R-250. (B) Elution profile of RPK from a Superose 12 HR 10/30 column. The position of the peak corresponds to a protein of 240 kDa, indicating a tetramer. Catalase (240 kDa), aldolase (158 kDa), albumin (68 kDa), ovalbumin (45 kDa), chymotrypsinogen (25 kDa), and cytochrome c (12 kDa) were used as molecular mass standards.
Sequence of the N-terminal portion of human RPK.
Arrows indicate the N-terminal residues found by sequencing of the purified RPK. A typical preparation of the protein comprised 50% A, 35% E, and 15% other species. For comparison, the human RPK sequence is aligned with sequences of other human PK isozymes and with PKs of known 3-dimensional structure.
Sequence of the N-terminal portion of human RPK.
Arrows indicate the N-terminal residues found by sequencing of the purified RPK. A typical preparation of the protein comprised 50% A, 35% E, and 15% other species. For comparison, the human RPK sequence is aligned with sequences of other human PK isozymes and with PKs of known 3-dimensional structure.
R510Q mutant form production
The mutant enzyme was expressed and purified with the same procedures described for the wild-type protein, but using the pAZ2 expression vector. The expression level and the oligomeric state of the protein were identical to those of the wild-type enzyme.
Steady-state kinetics of the recombinant wild-type RPK and mutant form R510Q
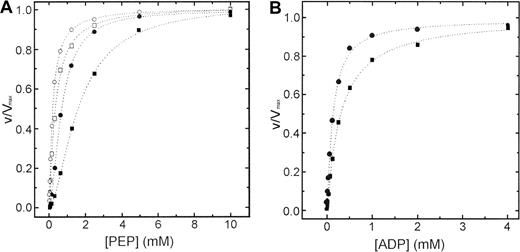
Steady state kinetics of wild-type and R510Q recombinant RPK as a function of PEP, in the absence and in the presence of FBP, are shown in Figure 3A. Parameters derived from these kinetics are summarized in Table 1. In the absence of the activator FBP, both proteins showed a typical homotropic response to PEP with a Hill coefficient of 1.6 (± 0.16) and 1.75 (± 0.19) for wild-type and R510Q, respectively. The presence of activator in the kinetics toward PEP shifted the sigmoidal curve of wild-type to a hyperbole, lowering the Hill coefficient to 1.05 (± 0.07). Similar effect was also exerted by FBP on R510Q (nH 1.08 ± 0.08). Inspection of the data in Table 1 shows that the S0.5 (PEP) values of mutant protein are slightly increased with respect to those of wild-type protein both in the absence or in the presence of 1 mM FBP (1.6 ± 0.11 versus 1.1 ± 0.04 in the absence of FBP, and 0.31 ± 0.02 versus 0.18 ± 0.02 in the presence of FBP).
Steady-state kinetics of wild-type and mutant R510Q RPK.
(A) Steady-state kinetics of recombinant wild-type (circles) and R510Q mutant (squares) RPK as a function of PEP. All experiments were performed at 37°C according to the published method.20Open symbols represent data points obtained in the presence of 1 mM FBP and filled symbols obtained without FBP. (B) Kinetics of wild-type (circles) and R510Q (squares) PKs against ADP. Concentration of PEP was fixed at 5 mM.
Steady-state kinetics of wild-type and mutant R510Q RPK.
(A) Steady-state kinetics of recombinant wild-type (circles) and R510Q mutant (squares) RPK as a function of PEP. All experiments were performed at 37°C according to the published method.20Open symbols represent data points obtained in the presence of 1 mM FBP and filled symbols obtained without FBP. (B) Kinetics of wild-type (circles) and R510Q (squares) PKs against ADP. Concentration of PEP was fixed at 5 mM.
Kinetic parameters of the wild-type and R510Q mutant erythrocyte pyruvate kinases
| . | PEP* . | ADP† . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FBP (−) . | 1 mM FBP . | ||||||||||
| kcat (s−1) . | S0.5 (mM) . | nH . | kcat/S0.5(s−1/mM) . | kcat (s−1) . | S0.5 (mM) . | nH . | kcat/S0.5(s−1/mM) . | kcat (s−1) . | Km (mM) . | kcat/Km(s−1/mM) . | |
| Wild-type | 355 ± 12 | 1.1 ± 0.04 | 1.60 ± 0.16 | 323 | 355 ± 11 | 0.18 ± 0.02 | 1.05 ± 0.07 | 1972 | 355 ± 13 | 0.17 ± 0.01 | 2088 |
| R510Q | 340 ± 10 | 1.6 ± 0.11 | 1.75 ± 0.19 | 212 | 304 ± 12 | 0.31 ± 0.02 | 1.08 ± 0.08 | 980 | 351 ± 10 | 0.24 ± 0.02 | 1463 |
| . | PEP* . | ADP† . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FBP (−) . | 1 mM FBP . | ||||||||||
| kcat (s−1) . | S0.5 (mM) . | nH . | kcat/S0.5(s−1/mM) . | kcat (s−1) . | S0.5 (mM) . | nH . | kcat/S0.5(s−1/mM) . | kcat (s−1) . | Km (mM) . | kcat/Km(s−1/mM) . | |
| Wild-type | 355 ± 12 | 1.1 ± 0.04 | 1.60 ± 0.16 | 323 | 355 ± 11 | 0.18 ± 0.02 | 1.05 ± 0.07 | 1972 | 355 ± 13 | 0.17 ± 0.01 | 2088 |
| R510Q | 340 ± 10 | 1.6 ± 0.11 | 1.75 ± 0.19 | 212 | 304 ± 12 | 0.31 ± 0.02 | 1.08 ± 0.08 | 980 | 351 ± 10 | 0.24 ± 0.02 | 1463 |
Results are means (SE) for 3 determinations from 4 different protein preparations. PEP, phosphoenolpyruvate; FBP, fructose 1,6-bisphosphate; ADP, adenosine diphosphate.
Kinetic parameters for PEP were obtained at fixed 1.5 mM ADP by fitting data to the Hill plot.
Kinetic parameters for ADP were obtained at fixed 5 mM PEP by fitting data to the Lineweaver-Burk plot.
Steady-state kinetics of the wild-type and R510Q mutant RPK as a function of ADP concentration are shown in Figure 3B. Parameters derived from these kinetics are summarized in Table 1. Both proteins exhibited hyperbolic response to ADP and similarKm values (0.17 ± 0.01 and 0.24 ± 0.02, for wild-type and R510Q, respectively). The addition of 1 mM FBP in the kinetics toward ADP did not alter the behavior of both enzymes (data not shown).
These data, on the whole, showed that there are no significant differences in the kinetic behavior between wild-type and the mutant form, suggesting that the mutation minimally perturbs the substrate binding sites. Moreover, recombinant wild-type RPK can be regarded as an authentic enzyme derived from red blood cells.10
Kinetics at physiologic concentrations of ATP and FBP
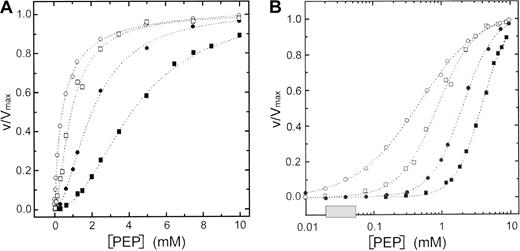
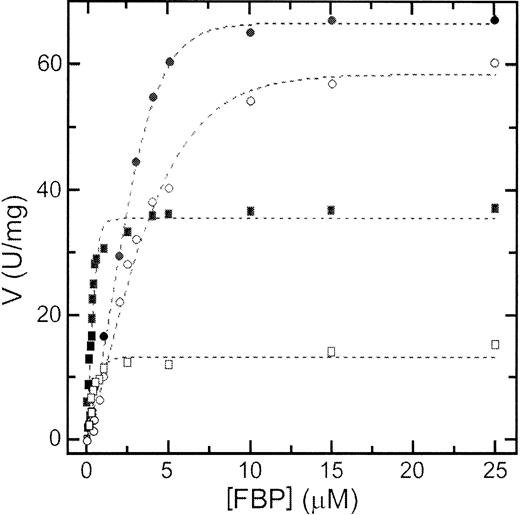
In human erythrocytes ATP is present at a concentration of about 1.5 mM, whereas FBP concentration is 30 μM.32 ATP inhibition combined with FBP activation may play a central role in regulating PK activity in erythrocytes. To evaluate the effect of ATP on the activity of RPK, we studied the kinetic behavior toward PEP in the presence of physiologic concentrations of ATP alone or ATP plus FBP (Figure 4A,B and Table2). In addition, we measured the activation by FBP at a subsaturating concentration of PEP in the absence and in the presence of ATP (Figure5 and Table 2).
Steady-state kinetics of wild-type and mutant R510Q RPK in the presence of ATP.
(A) Steady-state kinetics of recombinant wild-type (circles) and R510Q mutant (squares) RPK as a function of PEP in the presence of 1.5 mM ATP (filled symbols). Open symbols are data points obtained in the presence of 1.5 mM ATP plus 30 μM FBP. (B) Semilog plot of the data in panel A to show the difference between the wild-type and mutant protein at physiologic concentration of PEP (shaded box).
Steady-state kinetics of wild-type and mutant R510Q RPK in the presence of ATP.
(A) Steady-state kinetics of recombinant wild-type (circles) and R510Q mutant (squares) RPK as a function of PEP in the presence of 1.5 mM ATP (filled symbols). Open symbols are data points obtained in the presence of 1.5 mM ATP plus 30 μM FBP. (B) Semilog plot of the data in panel A to show the difference between the wild-type and mutant protein at physiologic concentration of PEP (shaded box).
Kinetic parameters of the wild-type and R510Q mutant erythrocyte pyruvate kinases in the presence of allosteric effectors
| . | PEP* . | FBP† . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.5 mM ATP FBP (−) . | 1.5 mM ATP 30 μM FBP . | ATP (−) . | 1.5 mM ATP . | |||||||||||
| kcat(s−1) . | S0.5 (mM) . | nH . | kcat/S0.5(s−1/mM) . | kcat (s−1) . | S0.5 (mM) . | nH . | kcat/S0.5(s−1/mM) . | kcat (s−1) . | S0.5 (μM) . | nH . | kcat (s−1) . | S0.5 (μM) . | nH . | |
| Wild-type | 349 ± 7 | 1.96 ± 0.15 | 2.09 ± 0.11 | 178 | 350 ± 10 | 0.42 ± 0.02 | 1.17 ± 0.11 | 833 | 64.9 ± 5 | 3.4 ± 0.02 | 1.56 ± 0.09 | 64.9 ± 5 | 6.7 ± 0.02 | 1.81 ± 0.09 |
| R510Q | 330 ± 8 | 3.92 ± 0.35 | 1.99 ± 0.14 | 84.2 | 305 ± 10 | 0.95 ± 0.05 | 2.02 ± 0.13 | 321 | 35.9 ± 6 | 0.5 ± 0.01 | 1.01 ± 0.11 | 14.4 ± 6 | 0.8 ± 0.01 | 1.10 ± 0.11 |
| . | PEP* . | FBP† . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.5 mM ATP FBP (−) . | 1.5 mM ATP 30 μM FBP . | ATP (−) . | 1.5 mM ATP . | |||||||||||
| kcat(s−1) . | S0.5 (mM) . | nH . | kcat/S0.5(s−1/mM) . | kcat (s−1) . | S0.5 (mM) . | nH . | kcat/S0.5(s−1/mM) . | kcat (s−1) . | S0.5 (μM) . | nH . | kcat (s−1) . | S0.5 (μM) . | nH . | |
| Wild-type | 349 ± 7 | 1.96 ± 0.15 | 2.09 ± 0.11 | 178 | 350 ± 10 | 0.42 ± 0.02 | 1.17 ± 0.11 | 833 | 64.9 ± 5 | 3.4 ± 0.02 | 1.56 ± 0.09 | 64.9 ± 5 | 6.7 ± 0.02 | 1.81 ± 0.09 |
| R510Q | 330 ± 8 | 3.92 ± 0.35 | 1.99 ± 0.14 | 84.2 | 305 ± 10 | 0.95 ± 0.05 | 2.02 ± 0.13 | 321 | 35.9 ± 6 | 0.5 ± 0.01 | 1.01 ± 0.11 | 14.4 ± 6 | 0.8 ± 0.01 | 1.10 ± 0.11 |
Results are means (SE) for 3 determinations from 4 different protein preparations. PEP, phosphoenolpyruvate; FBP, fructose 1,6-bisphosphate; ATP, adenosine triphosphate.
Kinetic parameters were obtained at fixed 1.5 mM ADP by fitting data to the Hill plot.
Kinetic parameters were obtained at fixed 0.1 mM PEP and 1.5 mM ADP by fitting data to the Hill plot.
Kinetics of wild-type and mutant R510Q RPK against FBP.
Kinetics of wild-type (circles) and R510Q (squares) RPK against FBP in the absence (filled symbols) and in the presence (open symbols) of 1.5 mM ATP. Concentration of PEP was fixed at 0.1 mM and the concentration of ADP at 1.5 mM.
Kinetics of wild-type and mutant R510Q RPK against FBP.
Kinetics of wild-type (circles) and R510Q (squares) RPK against FBP in the absence (filled symbols) and in the presence (open symbols) of 1.5 mM ATP. Concentration of PEP was fixed at 0.1 mM and the concentration of ADP at 1.5 mM.
In the presence of 1.5 mM ATP, the S0.5 and nH values of both wild-type and mutant enzymes are increased in respect to those obtained in the absence of ATP. These data suggest that ATP exerts an inhibitory effect on RPK activity and that the R510Q mutant is more severely affected than the wild-type enzyme. However, the ATP inhibition was almost completely counteracted by FBP in the case of the wild-type enzyme, but not in the case of the R510Q mutant. The combined effects of ATP and FBP on the wild-type and mutant enzyme can best be seen in Figure 4B. In the physiologic range of PEP, the mutant enzyme is about 20% active with respect to the wild-type enzyme (Figure 4B), suggesting an impairment of ATP production in a cell that relies on glycolysis to produce energy. Furthermore, as S0.5 values for FBP, either in the absence or in the presence of ATP, are lower than those for the wild-type enzyme, it is likely that the binding of FBP is not directly affected by the mutation (Table 2).
Thermal stability of wild-type and mutant proteins
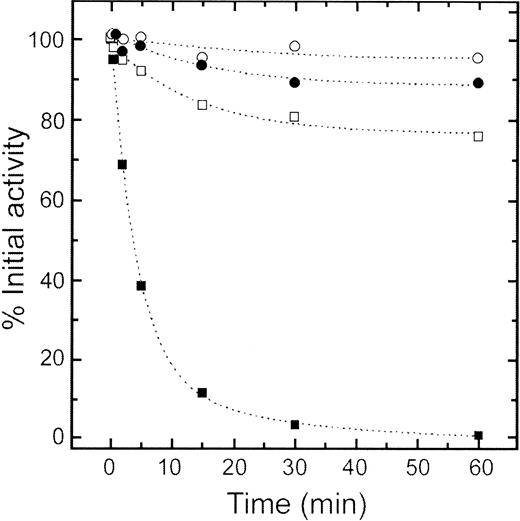
It has been reported that RPK from patients homozygous for the R510Q mutation has a thermal stability lower than that of the wild-type enzyme.23 Two patients carrying the R510Q mutation have been followed and characterized by us (see Zanella et al19 33). A summary of the relevant biochemical data concerning RPK of these patients is reported in Table3. In both instances the PK activity is markedly reduced and the enzyme is heat labile. The thermal stability of the purified recombinant wild-type and mutant proteins at 37°C, 53°C, and 63°C was tested in the presence and in the absence of 1 mM FBP. The mutant protein had a thermal stability dramatically lower than that of the wild-type protein. The difference was best seen at 53°C at which point the mutant lost nearly all its activity after 60 minutes of incubation, whereas the wild-type protein remained largely active (Figure 6). FBP (1 mM) stabilized both proteins but to a different extent, as shown by 95% ± 8% and 76.6% ± 6% residual activity for wild-type and R510Q mutant RPK, respectively. Thus, the observed in vivo instability of the enzyme obtained from the patients correlates with thermal stability properties revealed by the in vitro studies of the mutant recombinant protein.
Selected parameters of RPK of 2 patients homozygous for the R510Q mutation
| Case . | PK activity (IU/g Hb) . | Km PEP (mmol/L) . | Heat stability (%) . |
|---|---|---|---|
| 1 (40 years) | 1.3 | 0.78 | 5 |
| 2 (less than 1 year) | 2.1 | 1.40 | 19 |
| Reference values | 11.1-15.5 | 0.71-1.22 | 57-99 |
| Case . | PK activity (IU/g Hb) . | Km PEP (mmol/L) . | Heat stability (%) . |
|---|---|---|---|
| 1 (40 years) | 1.3 | 0.78 | 5 |
| 2 (less than 1 year) | 2.1 | 1.40 | 19 |
| Reference values | 11.1-15.5 | 0.71-1.22 | 57-99 |
Thermal inactivation of wild-type and mutant R510Q RPK.
Thermal stability of wild-type (circles) and R510Q mutant (squares) RPK. Protein samples (100 μg/mL) were incubated at 53°C in a 50 mM potassium phosphate buffer pH 6.5, 100 mM KCl, and 1 mM EDTA. Aliquots were drawn at intervals for measuring residual activity, which was expressed as percentage of initial activity. Filled symbols were data obtained in the absence of FBP, and open symbols were data obtained in the presence of 1 mM FBP.
Thermal inactivation of wild-type and mutant R510Q RPK.
Thermal stability of wild-type (circles) and R510Q mutant (squares) RPK. Protein samples (100 μg/mL) were incubated at 53°C in a 50 mM potassium phosphate buffer pH 6.5, 100 mM KCl, and 1 mM EDTA. Aliquots were drawn at intervals for measuring residual activity, which was expressed as percentage of initial activity. Filled symbols were data obtained in the absence of FBP, and open symbols were data obtained in the presence of 1 mM FBP.
Discussion
With the cloning, expression, and purification of RPK we have established a procedure to produce in a large amount the human enzyme that is difficult to isolate in homogeneous form from blood. Moreover, the full-length cDNA cloned in an expression vector allows easy preparation of the mutant forms by site-directed mutagenesis. The successful production of the R510Q mutant form supports such an expectation. R510Q represents the first human mutant of RPK expressed in E coli. A mutant isozyme of the liver has been previously obtained by Kanno et al.26
PK is known to be a tetramer comprising 4 identical subunits. However, the recombinant RPK was purified as a heterotetramer. Electrophoresis, gel filtration analysis, and isoelectrofocusing, as well as N-terminal sequencing, indicated that the heterotetramer comprises 2 full-length subunits and 2 proteolyzed chains. The presence of protease inhibitors during purification failed to prevent such proteolysis. However, absence of the inhibitors never led to a tetramer comprising fully proteolyzed subunits. All enzymatic preparations resulted in the same SDS electrophoretic pattern. Surprisingly, the size of the subunits and the proteolysis pattern of the enzyme expressed in E coliresembled those described for authentic RPK from human blood. Kahn and Marie10 as well as Marie and Kahn11 reported that PK purified from mature red blood cells was a heterotetramer comprising 2 L′ and 2 Lc subunits (L′2Lc2 form). The L′ subunits migrated as a 62-kDa protein that corresponded to the full-length subunits, whereas the Lc subunits represented the proteolyzed forms, as indicated by a molecular mass of 57 to 58 kDa. Furthermore, they demonstrated that the L′4 form, which exists and can be purified only from erythroblasts and young red blood cells, can be transformed into the L′2Lc2 form by in vitro mild trypsin digestion. Apparently, the subunits lacking the first 47 amino acids present in the recombinant enzyme and the Lc subunits of apparent 57 to 58 kDa, previously reported, represent the same species derived from the same precursor L′. The observation that the proteolytic process occurs specifically even in E coli favors an intrinsic susceptibility of the enzyme for cleavage. One aspect about the proteolysis should be noted: among the 4 subunits only 2 are proteolyzed and the other 2 appear to be resistant to digestion. This finding suggests that the N-terminal segments of the 4 subunits might be differently organized within the tetramer. The 2 resistant ones may be better structured and shielded from proteolysis than the other 2, which may be more flexible and exposed. An important question is whether the proteolysis of the N-terminal segments plays a role in enzyme regulation in the red blood cells. Evidence in favor of a functional role of the proteolysis comes from the study of Marie and Kahn,11 and Kahn et al,34 who purified the full-length homotetramer (L′4) from erythroblasts and the heterotetramer (L′2Lc2) from mature red blood cells. They found that the L′2Lc2 form has a lowerS0.5 and a higher Hill coefficient toward PEP than the L′4 enzyme. From a structural point of view, proteolysis of the N-terminal fragment may affect the local structure of the protein and, thus, affect the allosteric properties of the enzyme. The N-terminal fragment has been reported to form a domain (N-domain) that participates in intersubunit and interdomain interactions.3,7 Allosteric transitions involve movements of domains and subunits and depend on these interactions to relay allosteric signals. Proteolysis of the N-terminal region may provide the enzyme with a greater freedom for conformation transition. Moreover, cleavage of the N-terminal residues leads to the removal of the single phosphorylation site at Ser43.12 Both erythrocyte and liver human PK can be phosphorylated in vitro and in vivo.13,35,36 Phosphorylation causes the enzyme to shift from an active R-state to an inactive T-state.13 35Phosphorylation of liver enzyme is known to favor gluconeogenesis in the liver, but its physiologic significance in red cells is not clear. Further studies are needed to clarify the functional role, if any, of phosphorylation.
RPK deficiency is the most common cause of hereditary nonspherocytic hemolytic anemia.23 It is known that the deficiency is due to a defective enzyme encoded by a mutated PKLR gene. In most cases only a single nucleotide substitution is involved. 1529G>A is a frequently occurring mutation, found primarily in Northern European populations and Caucasian Americans.17,19 The mutation results in a change from arginine to glutamine (R510Q). Patients carrying the mutation can have clinical symptoms, ranging from a mild compensated hemolysis to severe anemia.23 We have characterized the mutant protein and compared it with the recombinant wild-type enzyme. The data revealed that the mutated RPK has very similar kcat values as the wild-type protein, and S0.5 toward PEP and ADP is slightly increased. How can a mutant with comparable kinetic properties cause severe RPK deficiency leading to hemolytic anemia in the patients? To answer this question we have to consider the kinetic data under physiologic conditions. In the presence of 1.5 mM ATP, the allosteric activator (30 μM) is not able to fully antagonize the inhibition by ATP (nH 2.02 ± 0.13) as does with the wild-type enzyme, and, at physiologic concentrations of PEP, the mutant has only about 20% of the activity of the wild-type enzyme. More importantly, the mutant enzyme presents a dramatically lowered thermal stability, compared with the wild-type protein in both ligand-free and FBP-bound states. As shown in Figure 6, after incubation at 53°C for 1 hour the mutant protein is totally inactivated, whereas the wild-type enzyme retains 85% of the initial activity. An unstable protein may not be able to persist for the average life span of the red blood cells. Direct evidence for this finding comes from clinical studies. Lenzner et al17 and Zanella et al19 33 studied several patients homozygous for the R510Q mutation. They found that, although the patients had different clinical manifestations, all showed lower than normal residual PK activity (between 10% and 25%). It should be noted that the mature red blood cells lack the machineries to synthesize proteins de novo. Once RPK becomes deficient, the ATP level will drop and the cell viability will be severely affected. Therefore, our data strongly support the notion that the decreased enzyme activity observed in patients homozygous for the R510Q mutation is primarily due to a lowered level of RPK as a consequence of instability rather than to the altered kinetic and regulatory properties of the enzyme.
The overexpression of the recombinant human RPK has allowed, for the first time, a thorough enzymologic investigation of the protein. The availability of a system to produce mutant forms of RPK will allow a deeper understanding of the molecular basis of nonspherocytic hemolytic anemia and to establish a better relationship between phenotype and genotype.
Supported by grants from the University of Pavia (Progetto di Ricerca d' Ateneo, “Nuove tecnologie molecolari e cellulari” and FAR2000), from Ministero dell' Università e della Ricerca Scientifica e Tecnologica (Progetto Genomica Funzionale), and from Allos Therapeutics Inc, Denver, CO.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Giovanna Valentini, Department of Biochemistry, University of Pavia, via Taramelli 3b, 27100 Pavia, Italy; e-mail:giovale@unipv.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal