Abstract
The interleukin-13 receptor (IL-13R) complex is composed of 2 different chains, IL-13Rα1 (also known as IL-13Rα′) and IL-13Rα2 (also known as IL-13Rα). For a functional IL-13 receptor, the IL-13Rα1 chain forms a productive complex with the primary IL-4 binding protein (IL-4Rα also known as IL-4Rβ). However, the function of the IL-13Rα2 chain is not clear even though this chain binds IL-13 with high affinity. This study demonstrates that IL-13Rα2 can undergo internalization after binding to ligand without causing activation of its signaling pathways. These conclusions were drawn on the basis of (1) internalization of 125I–IL-13 in Chinese hamster ovarian (CHO-K1) and T98G glioblastoma cells transiently transfected with the IL-13Rα2 chain; (2) a recombinant chimeric fusion protein comprising IL-13 and a mutated form ofPseudomonas exotoxin (termed IL13-PE38QQR or IL-13 toxin) is specifically cytotoxic to IL-13Rα2–transfected CHO-K1 cells in a gene dose-dependent manner, whereas cells transfected with vector alone were not sensitive; and (3) IL-13 did not cause activation of signal transduction and activation of transcription 6 (STAT6) in IL-13Rα2–transfected cells. IL-13 efficiently caused activation of STAT6 protein in cells transfected with the IL-13Rα1 and IL-4Rα chains, and IL-13Rα2 inhibited this activation. Taken together, these observations indicate that internalization of IL-13Rα2 is signal independent and that this property of IL-13Rα2 can be exploited for receptor-directed cancer therapy.
Introduction
Interleukin-13 (IL-13) and IL-4 are related multifunctional cytokines with similar biological activities on human B cells and monocytes.1-7 The cognate receptors for these cytokines are complex and have been shown to share 2 chains with each other.8-12 The IL-4 receptor system is well characterized and has been shown to be composed of a primary IL-4 binding protein, IL-4Rα (also known as IL-4Rβ).13 This chain forms a heterodimer with either the IL-2Rγc chain (type I IL-4R) or IL-13Rα1 (also known as IL-13Rα′) (type II IL-4R) for signaling.11 14-17 In some situations, all 3 chains (IL-4Rα, IL-2Rγc, and IL-13Rα1) may constitute the IL-4R complex; however, whether all 3 chains are simultaneously required for IL-4 function is not known.
In contrast, the receptor for IL-13 is less well characterized. We have studied the structure of IL-13R in various cell types and reported that IL-13 binds to 2 isoforms of an approximate 65-kDa protein in human renal cell carcinoma cells.8-12 One of these proteins also binds IL-4.8,14 On the basis of the binding characteristics, cross-linking, and displacement of radiolabeled IL-4 and IL-13 in various cell types, we hypothesized that, as is the case for the IL-4R system, IL-13R may also be composed of 3 different types.9-12 More recently, 2 different chains of the IL-13R system have been cloned. The murine and human IL-13Rα1 chain was first cloned.14,18 This chain can bind IL-13 with low affinity, but, when coupled with the IL-4Rα chain, the heterodimer binds IL-13 with high affinity and mediates IL-13–induced signaling.14,18 The second chain of the IL-13R, termed IL-13Rα2 (also known as IL-13Rα), has also been cloned from a human renal cell carcinoma cell line (Caki-1). This chain shares approximately 50% homology with the IL-5R at the DNA level. It contains a very short intracellular domain and binds IL-13 with high affinity.19
On the basis of the above studies, we have proposed that the type I IL-13R complex is composed of both chains of IL-13R (IL-13Rα1 and IL-13Rα2) and IL-4Rα chain. Although IL-13 binds to all 3 chains, only the IL-13Rα1 and IL-4Rα chains form a productive complex. Because of this arrangement, IL-13 binds to these cells strongly. Only IL-13, not IL-4, is able to displace the binding of125I–IL-13. In the type II IL-13R system, IL-13Rα2 is not present, and the IL-13Rα1 chain forms a complex with the IL-4Rα chain. In these cells both interleukins compete for the binding of their radiolabeled cognaters.12 The structure of the type III IL-13R is similar to that of type II receptors except that these cells also express the IL-2Rγc chain. Although it appears that the IL-2Rγc chain does not bind IL-13, it does affect IL-13 binding and function in some cell types.20 21
After binding to their receptors, both IL-4 and IL-13 signal through phosphorylation-dependent activation of Jak kinases and signal transduction and activator of transcription (STAT) protein.11,17,22 In particular, STAT6 is activated in response to both IL-4 and IL-13.17,22,23 In the IL-4R system, activation of STAT6 protein was seen in cells transfected with IL-13Rα1 chain and IL-4Rα chain; however, no activation was observed in cells transfected with the IL-13Rα2 chain alone or when transfected with IL-4Rα chain.15 Whether IL-13 induces STAT6 activation by similar combination of receptor subunits is not known. It is also not known whether the IL-13Rα2 chain plays a major role in IL-13 functions. It has been reported that the extracellular domain of IL-13Rα2 is secreted in the plasma and urine of mice. However, in the context of human physiology, this protein has not been identified.24 25
To determine the function of IL-13Rα2, this chain was transfected either alone or in combination with other known chains of the IL-4R and IL-13R systems in Chinese hamster ovary (CHO-K1) cells and T98G glioblastoma cells. Transfectants were studied to determine whether (1) IL-13R is internalized after binding to125I–IL-13, (2) the intracellular domain of IL-13Rα2 plays a role in ligand internalization, (3) IL-13 can transmit signals in cells that are transfected with this chain, and (4) a chimeric protein composed of IL-13 and a mutated form ofPseudomonas exotoxin (termed IL13-PE38QQR)26 27is cytotoxic to cells that are transfected with the IL-13Rα2 chain. Our studies demonstrate, for the first time, that IL-13Rα2 can be internalized after binding to IL-13 without inducing intracellular signaling through the STAT6 pathway.
Materials and methods
Recombinant cytokines and toxins
Recombinant human IL-131,5 was produced and purified to homogeneity in our laboratory (B. H. Joshi and R.K.P., unpublished results).28 Recombinant IL13-PE38QQR was also produced and purified in our laboratory (Joshi et al, unpublished results). The cpIL4-toxin IL438-37-PE38KDEL, containing the circularly permuted IL-4 mutant in which amino acids 38-129 were linked to amino acids 1-37 via a GGNGG linker and then fused to truncated toxin PE38KDEL, consisting of amino acids 253-364 and 381-608 of PE followed by KDEL, was expressed in Escherichia coli and purified as described previously.29 30
Cell lines
CHO-K1 and human glioblastoma multiforma cell lines (T98G) were purchased from the American Type Culture Collection (Rockville, MD). CHO-K1 cells were cultured in a modified Eagle minimum essential medium (AMEM), and T98G cells were cultured in Eagle minimum essential medium (EMEM) containing 10% fetal bovine serum (Biowhittaker, Walkersville, MD), 1 mmol HEPES, 1 mmol L-glutamine, 100 μg/mL penicillin, and 100 μg/mL streptomycin (Biowhittaker).
Plasmids, mutagenesis, and transient transfection of DNA
Complementary DNAs (cDNAs) of human IL-4Rα chain,13 IL-13Rα2 chain,19 and IL-13Rα1 chain14 were cloned into pME18S mammalian expression vector [pIL4Rα, pIL13Rα (α2), and pIL13Rα′ (α1)]. The IL-13Rα deletion mutants Δ335L, Δ338R, or Δ343Y were constructed by the polymerase chain reaction (PCR), using Taq DNA polymerase with the primer 5′-CCGCTCGAGATGGCTTTCGTTTGCTTGGCTATCGG-3′ and 3′-GCTCTAGATCAACCGGTTACAAATATAACTAATATTAAG-5′ (Δ335L) or 3′-GCTCTAGATCACAAAAGCAGACCGGTTACAAATATAAC-5′ (Δ338R) or 3′-GCTCTAGATCAGGTGTTTGGCTTACGCAAAAG-5′ (Δ343Y), each containing an in-frame stop codon. The cDNAs were subcloned into the expression vector pCIneo, using the XhoI and XbaI sites. Individual plasmid DNA or a combination of multiple plasmid DNAs (6 μg/60-mm dish or 12 μg/100-mm culture dish) were transfected into semiconfluent cells, using GenePORTER transfection reagent (Gene Therapy Systems, San Diego, CA) according to the manufacturer's instructions. Briefly, cells (1 × 106/60-mm dish or 3 × 106/100-mm dish) were incubated with the DNA-GenePORTER mixture for 5 hours in Dulbecco modified Eagle medium (DMEM; Biowhittaker). Then DMEM containing 20% fetal bovine serum (FBS) was added, and incubation was continued. Twenty-four hours after transfection, the medium was changed to AMEM (CHO-K1) or EMEM (T98G) with 10% FBS, and cells were incubated for and an additional 24 hours.
Radioreceptor binding assay
Recombinant human IL-13 was labeled with 125I (Amersham, Arlington Heights, IL), using IODO-GEN reagent (Pierce, Rockford, IL) as previously described.8 The specific activity of the radiolabeled IL-13 was estimated to be 12.7 μCi/μg protein. For binding experiments, 5 × 105 cells in 100 μL binding buffer (RPMI 1640 containing 0.2% human serum albumin and 10 mmol HEPES) were incubated with 200 pmol 125I–IL-13 with or without 40 nmol unlabeled IL-13 at 4°C for 2 hours. Cell-bound 125I–IL-13 was separated from unbound by centrifugation through a phthalate oil gradient, and radioactivity was determined with a gamma counter (Wallac, Gaithersburg, MD). For the displacement assay, T98G cells were incubated with125I–IL-13 (200 pmol) with or without increasing concentration (up to 100 nmol) of IL-13, as described above.
IL-13R internalization
Internalization assays were performed as described before.21 CHO-K1 or T98G cells with or without transfected IL-13Rα chain were incubated in binding buffer containing 0.2 nmol chloroquine at 37°C for 5 minutes to prevent degradation of internalized 125I–IL-13. The cells were then washed, and 2 × 107 cells were incubated with 0.5 nmol125I–IL-13 at 4°C for 2 hours. At various time intervals, 2 duplicate sets of 50 μL aliquots were taken. One set was incubated with glycine buffer (final pH = 2.0) for 10 minutes on ice. The suspension was then centrifuged through a mixture of phthalate oils, and the radioactivity in the cell pellet (acid-resistant or internalized) and in the supernatant (surface-bound plus dissociated) was determined with a gamma counter. The other set of 50-μL aliquots was directly centrifuged through phthalate oils, and the radioactivity observed in the supernatants was used for dissociated125I–IL-13 values. Surface-bound 125I–IL-13 was determined by subtracting dissociated 125I–IL-13 values from surface-bound plus dissociated values.
Protein synthesis inhibition assay
The cytotoxic activity of IL-13 toxin or IL-4 toxin was tested as previously described.27 Typically, 104cells were cultured in leucine-free medium with or without various concentrations of IL13-PE38QQR or IL438-37-PE38KDEL for 20 to 22 hours at 37°C. Then 1 μCi of [3H]leucine (NEN Research Products, Boston, MA) was added to each well and incubated for an additional 4 hours. Cells were harvested, and radioactivity incorporated into cells was measured by a beta plate counter (Wallac).
Electrophoretic mobility shift assay
After incubation with IL-13 (250 ng/mL) for 10 minutes, cells were washed with cold extraction buffer (1 mg/mL leupeptin, 5 mg/mL pepstatin A, 2 mg/mL aprotinin, 20 mmol HEPES pH 7.0, 10 mmol KCl, 300 mmol NaCl, 0.5 mmol dithiothreitol [DTT], 0.1% NP-40, 1 mmol phenylmethylsulfonyl fluoride, 1 mmol Na3VO4, and 20% glycerol). DNA protein interactions were assessed by electrophoretic mobility shift assay (EMSA), using a Bandshift kit (Pharmacia, Piscataway, NJ). Briefly, 50 μg sample proteins was incubated in 20 μL binding buffer [10 mmol Tris-HCl (pH 7.5), 50 mmol NaCl, 0.5 mmol DTT, 10% glycerol, 0.05% NP-40, 0.05 mg/mL poly (dI-dC)2] for 20 minutes at room temperature with 1 ng32P-labeled double-stranded oligonucleotide probe SBE1. SBE1 is a STAT-binding element (5′-gatcGCTCTTCTTCCCAGGAACTCAATG-3′; 3′-CGAGAAG AAGGGTCCTTGAGTTACagct-5′) that is from the region flanking the transcription start site of the human sIL-1R antagonist gene that is necessary for response to IL-13.31 Ten times concentrated loading dye (2 μL) was added to samples that were then applied to a 4% nonreducing polyacrylamide gel and run at 150 V for 2.5 hours. Gels were dried for 2 hours and autoradiographed overnight at room temperature.
For the supershift assay, antihuman STAT6 (S-20) rabbit polyclonal immunoglobulin G (Santa Cruz Biotechnology, Santa Cruz, CA) was included in the reaction mixture before electrophoresis.
Results
125I–IL-13 binding on CHO-K1 or T98G cells transfected with IL-13Rα2, IL-13Rα1, and IL-4Rα chains
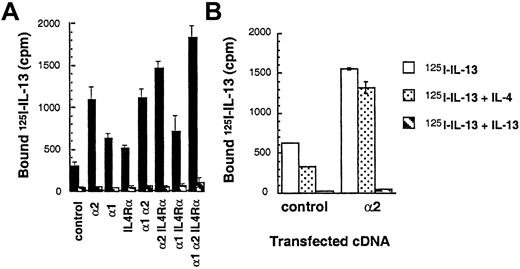
To demonstrate ligand binding and directly examine the subunit structure of IL-13R, cDNAs of various human IL-13R chains were introduced into the CHO-K1 cell line, and binding studies were performed. As shown in Figure 1A,125I–IL-13 was bound at a higher level to IL-13Rα2–transfected CHO-K1 cells as compared to cells transfected with IL-13α1 or IL-4Rα chains. A 200-fold mole per liter excess of IL-13 but not IL-4 inhibited this binding completely, indicating specific 125I–IL-13 binding (Figure 1A,B). IL-13Rα1 or IL-4Rα single-chain transfectants showed a slight increase in binding of radiolabeled ligand over vector-transfected controls. The binding of125I–IL-13 in control cells was also inhibited although only partially by IL-4 (Figure 1B). Cotransfection of IL-13Rα2 chain with IL-4Rα or IL-13Rα1 chain did not induce a significant increase in 125I–IL-13 binding. In contrast, cotransfection of IL-13Rα2 chain along with IL-13Rα1 and IL-4Rα chains resulted in125I–IL-13 binding that was significantly increased to maximum levels. These data suggest that IL-13Rα2 bind IL-13 with the greatest avidity and that this chain may not directly interact with IL-13Rα1 and IL-4Rα with respect to ligand binding.
125I–IL-13 binding to CHO-K1 and T98G cells transfected with the IL-13Rα2, IL-13Rα1, IL-4Rα, and γc chains.
cDNA for various receptor chains (6 μg/chain) was transfected into cells (1 × 106) by using GenePORTER reagent for 48 hours. For the IL-13 binding assay, 1 × 106 transfected cells were incubated with 200 pmol/L of 125I–IL-13 with or without a 200-fold molar excess of unlabeled IL-13 (A) or control CHO-K1 and IL-13Rα2–transfected cells with IL-13 or IL-4 (B). Binding assays were performed in 2 separate experiments. Cell bound radioactivity was determined as described in “Materials and methods.”
125I–IL-13 binding to CHO-K1 and T98G cells transfected with the IL-13Rα2, IL-13Rα1, IL-4Rα, and γc chains.
cDNA for various receptor chains (6 μg/chain) was transfected into cells (1 × 106) by using GenePORTER reagent for 48 hours. For the IL-13 binding assay, 1 × 106 transfected cells were incubated with 200 pmol/L of 125I–IL-13 with or without a 200-fold molar excess of unlabeled IL-13 (A) or control CHO-K1 and IL-13Rα2–transfected cells with IL-13 or IL-4 (B). Binding assays were performed in 2 separate experiments. Cell bound radioactivity was determined as described in “Materials and methods.”
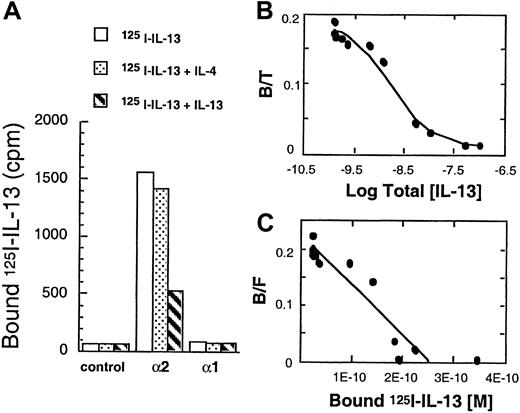
The binding characteristics and affinity of IL-13R on T98G cells transfected with IL-13Rα1 and α2 chain was also analyzed. As shown in Figure 2A, unlike control CHO-K1 cells (Figure 1A,B), vector-transfected T98G cells exhibited little125I–IL-13 binding; however, cells transfected with IL-13Rα2 chain showed more than 15-fold higher IL-13 binding when compared to the control cells. Similar to CHO-K1 cells, the binding of125I–IL-13 was inhibited by excess of IL-13 but not by IL-4. T98G cells transfected with IL-13Rα1 did not show appreciable binding to 125I–IL-13. To determine the binding affinity of 125I–IL-13 to its receptors in IL-13Rα2–transfected T98G cells, Scatchard analysis was performed, using the LIGAND program. This program allowed fitting of the data to a one-site model. As shown in Figure 2B and C, IL-13 bound to IL-13Rα2–transfected T98G cells with strong affinity (Kd = 1.12 nmol/L), and the receptor number was estimated to be approximately 30 580 sites/cell. These results suggest that IL-13Rα2 chain binds IL-13 with high affinity.
125I–IL-13 binding to T98G cells transfected with IL-13Rα1 or IL-13Rα2 chains.
cDNA for receptor chains (6 μg/chain) was transfected into T98G cells (1 × 106) by using GenePORTER reagent for 48 hours. For the IL-13 binding assay, 1 × 106 cells were incubated with 200 pmol/L of 125I–IL-13 with or without a 200-fold molar excess of unlabeled IL-4 or IL-13 (A). Displacement curve (B) and Scatchard data (C) with T98G cells transfected with IL-13Rα2 chain were analyzed by the LIGAND program.
125I–IL-13 binding to T98G cells transfected with IL-13Rα1 or IL-13Rα2 chains.
cDNA for receptor chains (6 μg/chain) was transfected into T98G cells (1 × 106) by using GenePORTER reagent for 48 hours. For the IL-13 binding assay, 1 × 106 cells were incubated with 200 pmol/L of 125I–IL-13 with or without a 200-fold molar excess of unlabeled IL-4 or IL-13 (A). Displacement curve (B) and Scatchard data (C) with T98G cells transfected with IL-13Rα2 chain were analyzed by the LIGAND program.
Internalization of IL-13R on CHO-K1 and T98G cells transfected with the IL-13Rα2 chain
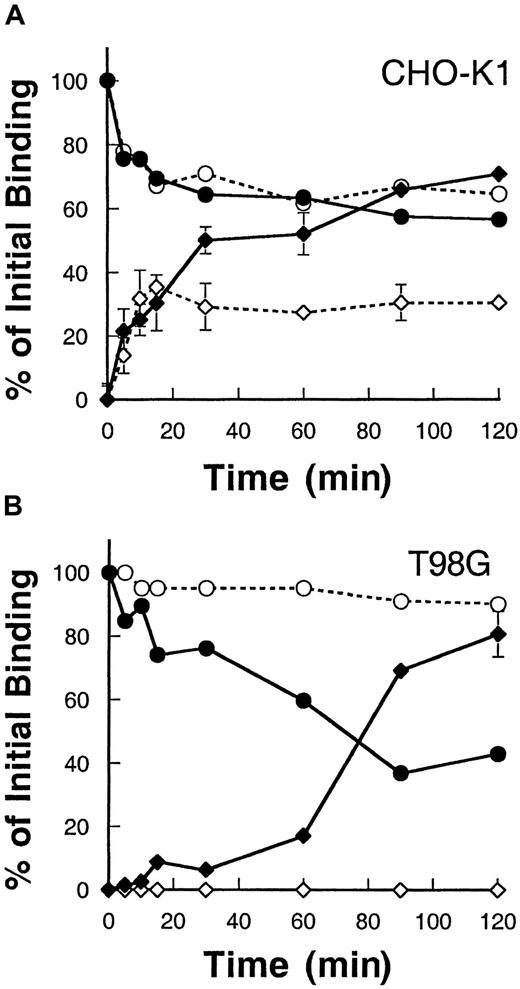
To determine whether the IL-13Rα2 chain when expressed alone can be internalized after binding to its ligand, IL-13, we examined the rate of internalization and disappearance of surface-bound IL-13 using125I–IL-13. As shown in Figure3, CHO-K1 and T98G cells transfected with IL-13Rα2 chain bound IL-13, and the level of surface-bound125I–IL-13 steadily declined with time in both CHO-K1 cells and T98G cells. Concurrently, the level of125I–IL-13 internalized increased in these cells. In CHO-K1 cells transfected with the IL-13Rα2 chain, the level of internalization was higher compared to control vector-transfected cells. IL-13Rα2 chain–transfected cells showed an approximately 35% higher internalization level at 90 minutes when compared to control CHO-K1 cells (Figure 3A). Interestingly, control CHO-K1 cells also showed internalization of IL-13, indicating that these cells express functional IL-13R and that hamster cells bind human IL-13. However, in T98G cells, vector-transfected control cells showed no internalization, and 100% of the IL-13 remained surface bound even after 2 hours of incubation at 37°C. However, IL-13Rα2 chain transfectants showed a remarkable level of internalization that reached to maximum of approximately 80% of total bound ligand at 120 minutes (Figure 3B). These results suggest that, on binding to ligand125I–IL-13, the IL-13Rα2 chain is internalized at a high rate and IL-13Rα2 is required for internalization.
Internalization of 125I–IL-13 in CHO-K1 and T98G cells.
CHO-K1 (A) and T98G (B) cells were incubated with 0.5 nmol125I–IL-13 at 4°C for 2 hours. Then the temperature was raised to 37°C, and, at various time intervals, 2 duplicate sets of 50 μL aliquots were taken. One set was incubated with glycine buffer (pH = 2.0) for 10 minutes. The mixture was then centrifuged through phthalate oils, and the radioactivity in the cell pellet (internalized) and in the supernatant (surface-bound plus dissociated) was determined with a gamma counter. The other set of 50 μL aliquots was directly centrifuged through phthalate oils, and the radioactivity observed in the supernatant was used for dissociated 125I–IL-13 values. Surface-bound 125I–IL-13 was determined by subtracting dissociated 125I–IL-13 values from (surface-bound plus dissociated) values. Data are expressed as a percentage of total IL-13 bound at time 0. Open circles, surface IL-13 bound on control cells; closed circles, surface-bound on IL-13Rα2–transfected cells; open diamonds, internalization in control cells; and closed diamonds, internalization in IL-13Rα2–transfected cells. Values are the mean of 2 independent experiments. When not shown, standard deviations are smaller than the symbol.
Internalization of 125I–IL-13 in CHO-K1 and T98G cells.
CHO-K1 (A) and T98G (B) cells were incubated with 0.5 nmol125I–IL-13 at 4°C for 2 hours. Then the temperature was raised to 37°C, and, at various time intervals, 2 duplicate sets of 50 μL aliquots were taken. One set was incubated with glycine buffer (pH = 2.0) for 10 minutes. The mixture was then centrifuged through phthalate oils, and the radioactivity in the cell pellet (internalized) and in the supernatant (surface-bound plus dissociated) was determined with a gamma counter. The other set of 50 μL aliquots was directly centrifuged through phthalate oils, and the radioactivity observed in the supernatant was used for dissociated 125I–IL-13 values. Surface-bound 125I–IL-13 was determined by subtracting dissociated 125I–IL-13 values from (surface-bound plus dissociated) values. Data are expressed as a percentage of total IL-13 bound at time 0. Open circles, surface IL-13 bound on control cells; closed circles, surface-bound on IL-13Rα2–transfected cells; open diamonds, internalization in control cells; and closed diamonds, internalization in IL-13Rα2–transfected cells. Values are the mean of 2 independent experiments. When not shown, standard deviations are smaller than the symbol.
Cytotoxicity of IL13-PE38QQR on CHO-K1 cells transfected with the IL-13Rα2, IL-13Rα1, and IL-4Rα chains
To further confirm whether the IL-13R chain is internalized after binding to ligand, the cytotoxicity of a recombinant IL13-PE38QQR targeting IL-13R was assessed. IL13-PE38QQR binds to IL-13R and is internalized by endocytosis, subsequently causing apoptotic and/or necrotic cell death through the inhibition of new protein synthesis.32 33 Thus, cytotoxicity observed in transfected cells indicates receptor internalization. CHO-K1 cells were transfected with the IL-13Rα2 chain in the absence or presence of other IL-13R chains, and sensitivity to IL13-PE38QQR was determined. As shown in Figure 4A, when CHO-K1 cells were transfected with IL-13Rα2 alone, IL13-PE38QQR was highly cytotoxic to these cells. The 50% inhibitory concentration (IC50; IL-13 toxin concentration causing 50% inhibition of protein synthesis) ranged between 7 and 10 ng/mL. When CHO-K1 cells were transfected with the IL-13Rα1 chain alone, no cytotoxicity was observed even when 1000 ng/mL IL-13 toxin was added (data not shown). However, when CHO-K1 cells were transfected with the IL-4Rα chain, 25% cytotoxicity was seen at 100 ng/mL IL-13 toxin (Figure 4A). Similar to binding data (Figure 1), when the α2 chain was cotransfected with the IL-13Rα1 or IL-4Rα chains, the cytotoxicity was similar to that seen with α2 chain transfection alone (Figure 4B). However, when all 3 chains (IL-4Rα, IL-13Rα, and α2) were cotransfected, cytotoxic activity was slightly increased compared to α2α1 or α2 IL-4Rα transfectants (IC50 of 10 ng/mL compared to 30 ng/mL in α1α2 IL-4Rα or α1α2 transfectants, respectively). Cells transfected with the α1 and IL-4Rα chains showed little sensitivity to IL13-PE38QQR.
Cytotoxicity of IL-13 toxin and IL-4 toxin to CHO-K1 cells transfected with various chains of IL-13R.
CHO-K1 cells were transfected with various receptor chains and then IL-13 toxin– or IL-4 toxin–mediated cytotoxicity was determined by protein synthesis inhibition assay. CHO-K1 transfected with IL-13Rα2 (closed circles) or IL-4Rα (open circles) chains (A); cells were transfected with various combinations of receptor chains, control (closed circles), α1 α2 (open circles), α2 IL4Rα (closed diamonds), α1 IL4Rα (open diamonds), α1α2 IL4Rα (closed triangles) (B); cells transfected with increasing concentrations of DNA for IL-13Rα2 chain, 0 μg (closed circles), 1 μg (open circles), 2 μg (closed diamonds), 3 μg (open diamonds), 4 μg (closed triangles), 5 μg (open triangles), 6 μg (closed squares) (C); transfected with 6 μg cDNA of IL-13α2 chain and incubated with various concentrations of IL13-PE38QQR (closed circles) or IL438-37-PE38KDEL (open circles) (D); or transfected with control (open circles), IL4Rα (closed triangles), γc(open triangles), IL4Rαγc (closed squares). The results are represented as means ± SD of quadruplicate determinations, and the assay was repeated several times.
Cytotoxicity of IL-13 toxin and IL-4 toxin to CHO-K1 cells transfected with various chains of IL-13R.
CHO-K1 cells were transfected with various receptor chains and then IL-13 toxin– or IL-4 toxin–mediated cytotoxicity was determined by protein synthesis inhibition assay. CHO-K1 transfected with IL-13Rα2 (closed circles) or IL-4Rα (open circles) chains (A); cells were transfected with various combinations of receptor chains, control (closed circles), α1 α2 (open circles), α2 IL4Rα (closed diamonds), α1 IL4Rα (open diamonds), α1α2 IL4Rα (closed triangles) (B); cells transfected with increasing concentrations of DNA for IL-13Rα2 chain, 0 μg (closed circles), 1 μg (open circles), 2 μg (closed diamonds), 3 μg (open diamonds), 4 μg (closed triangles), 5 μg (open triangles), 6 μg (closed squares) (C); transfected with 6 μg cDNA of IL-13α2 chain and incubated with various concentrations of IL13-PE38QQR (closed circles) or IL438-37-PE38KDEL (open circles) (D); or transfected with control (open circles), IL4Rα (closed triangles), γc(open triangles), IL4Rαγc (closed squares). The results are represented as means ± SD of quadruplicate determinations, and the assay was repeated several times.
We next examined whether sensitivity to IL-13 toxin was IL-13Rα2 concentration dependent. To address this point, we transfected CHO-K1 cells with various amounts of IL-13Rα2 cDNA and examined the cytotoxicity of IL13-PE38QQR. As shown in Figure 4C, increasing protein synthesis inhibition was observed as the amounts of cDNA used for transfection increased. When more than 3 μg IL-13Rα2 cDNA was transfected in CHO-K1 cells, the protein synthesis was inhibited by 50% with less than 100 ng/mL IL13-PE38QQR. At the highest concentration of DNA (6 μg), maximum sensitivity to IL13-PE38QQR was observed (IC50 = 4 ng/mL).
To confirm whether the cytotoxicity of IL13-PE38QQR was mediated through IL-13R, IL-13Rα2–transfected cells were tested for sensitivity to IL-4 toxin, IL438-37-PE38KDEL as well as IL13-PE38QQR. As shown in Figure 4D, CHO-K1 cells transfected with IL-13Rα2 chain were highly sensitive to IL-13 toxin but not to IL438-37-PE38KDEL even when up to 1000 ng/mL recombinant toxin was used. These results correspond to our previous findings, indicating that the IL-4R system does not utilize the IL-13Rα2 chain.9-11 15 IL-4 toxin was cytotoxic to CHO-K1 cells when transfected with IL-4Rα alone or in combination with γc chain. However, control cells and cells transfected with γc chain alone were not killed by IL-4 toxin (Figure4E).
Role of intracellular domain of IL-13Rα2 in ligand internalization
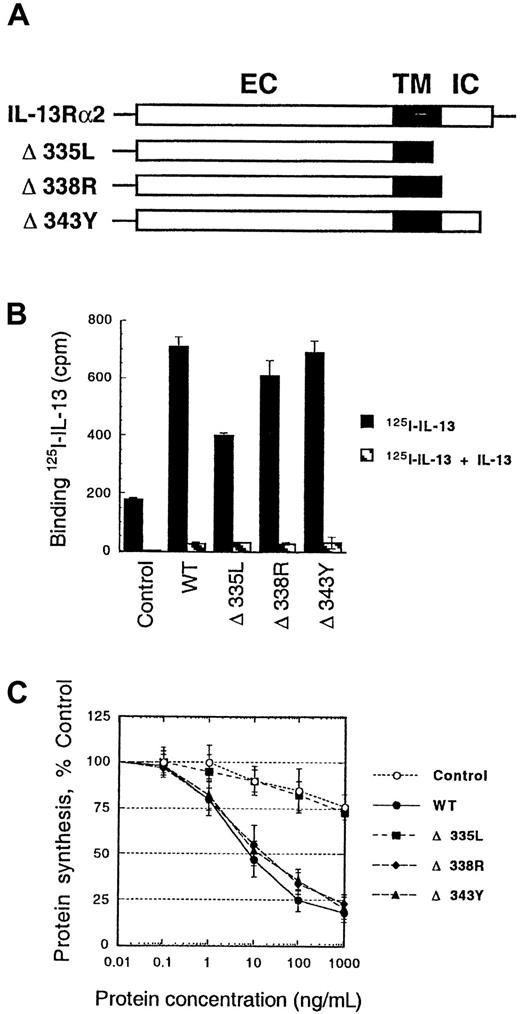
To examine the role of the intracellular domain of the IL-13Rα2 chain in the internalization process, we made deletions in the intracellular domain of α2 chain (Figure5A). After transient transfection of these deletion mutant genes in CHO-K1 cells, we performed IL-13 binding and internalization studies. As shown in Figure 5B, deletion of the part of transmembrane domain and whole intracellular domain (Δ335L mutant) caused a significant decrease in IL-13 binding. In addition, this deletion mutant showed no sensitivity to IL-13-PE38QQR similar to control cells (Figure 5C). These results indicate that the part of transmembrane domain and/or all of intracellular domain of IL-13Rα2 are required for IL-13 internalization. However, deletions of part or complete intracellular domain alone (Δ338R or Δ343Y) showed no change in IL-13 binding or sensitivity to IL-13 toxin. Therefore, it was concluded that amino acids between L335 and Y343 in the IL-13Rα2 chain might be involved in IL-13 binding and/or internalization.
125I–IL-13 binding and cytotoxicity of IL-13 toxin to CHO-K1 cells transfected with IL-13Rα2 deletion mutants.
Schematic representation of the wild-type and mutant IL-13Rα2 chains. EC, extracellular domain; TM, transmembrane domain; IC, intracellular domain (A). cDNA for various mutant receptor chains (6 μg/chain) was transfected into cells (5 × 105) by using GenePORTER reagent for 48 hours. For IL-13 binding assay, 5 × 105cells were incubated with 200 pmol/L 125I–IL-13 with or without a 200-fold molar excess of unlabeled IL-13. Binding assays were performed in 2 separate experiments. Cell-bound radioactivity was determined as described in “Materials and methods” (B). CHO-K1 cells were transfected with IL-13Rα2 and its mutants, and then IL13-PE38QQR cytotoxicity was determined by protein synthesis inhibition assay. The results are represented as means ± SD of quadruplicate determinations (C).
125I–IL-13 binding and cytotoxicity of IL-13 toxin to CHO-K1 cells transfected with IL-13Rα2 deletion mutants.
Schematic representation of the wild-type and mutant IL-13Rα2 chains. EC, extracellular domain; TM, transmembrane domain; IC, intracellular domain (A). cDNA for various mutant receptor chains (6 μg/chain) was transfected into cells (5 × 105) by using GenePORTER reagent for 48 hours. For IL-13 binding assay, 5 × 105cells were incubated with 200 pmol/L 125I–IL-13 with or without a 200-fold molar excess of unlabeled IL-13. Binding assays were performed in 2 separate experiments. Cell-bound radioactivity was determined as described in “Materials and methods” (B). CHO-K1 cells were transfected with IL-13Rα2 and its mutants, and then IL13-PE38QQR cytotoxicity was determined by protein synthesis inhibition assay. The results are represented as means ± SD of quadruplicate determinations (C).
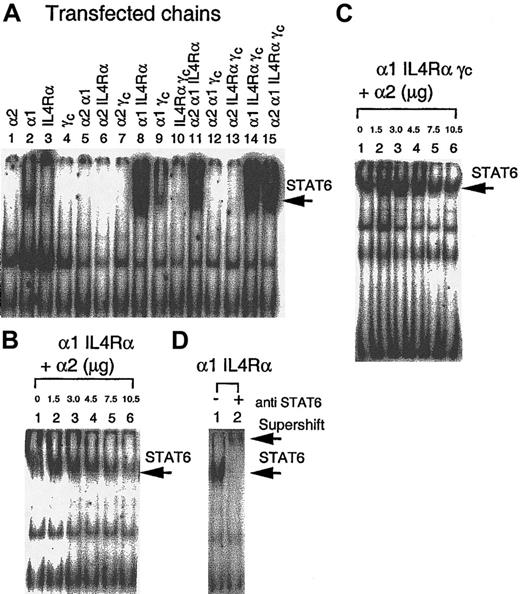
Activation of STAT6 in response to IL-13 in IL-13R chain transfectants
To demonstrate whether the IL-13Rα2 chain signals after binding IL-13 and whether IL-13Rα1 and IL-4Rα by themselves or heterodimers with the IL-13Rα2 chain are biologically functional, we analyzed STAT6 activation in response to IL-13 in various IL-13R chain transfectants. As shown in Figure 6A (lane 1), no STAT6 activation was observed in CHO-K1 cells transfected with the IL-13Rα2 chain. However, when cells were transfected with α1 or IL-4Rα chains, low-level activation was observed (Figure 6A, lanes 2 and 3). When the α1 chain was cotransfected with the IL-4Rα chain, the STAT6 activation was induced to its highest level (Figure6A, lane 8). Cotransfection of the α2 chain with the IL-4Rα chain or α1 chain did not induce STAT6 activation. In fact, α2 chain transfection abrogated IL-4Rα or α1-induced STAT6 activation (Figure 6A, lanes 5 and 6). In addition, when α1, IL-4Rα, α1γc, and IL-4Rαγc were cotransfected with the α2 chain, the STAT6 activation was decreased (Figure 6A, lanes 11-13). CHO-K1 cells transfected with all 4 chains (α2α1IL-4Rαγc) appeared to show similar STAT6 activation compared to α1 IL-4Rα transfectants (Figure 6A, lane 15).
Modulation of STAT6 activation by IL-13 and the IL-13Rα2 chain.
CHO-K1 cells were transfected with various receptor chains and incubated with IL-13 for 10 minutes, solubilized with cold whole-cell extraction buffer, and 50 μg sample protein was incubated for 20 minutes with 1 ng 32P-labeled SBE1 probe in binding buffer. DNA-protein interaction was analyzed by SDS-PAGE analysis (A). CHO-K1 cells were transfected with increasing amounts (0 to 10.5 μg) of IL-13Rα2 cDNA along with fixed amounts of DNA for α1 IL4Rα (6 μg) chains (B) or α1 IL4Rαγc (C). For supershift assay, protein extract from α1 IL4Rα–transfected CHO-K1 cells was incubated with antihuman STAT6 rabbit polyclonal immunoglobulin G before electrophoresis (D).
Modulation of STAT6 activation by IL-13 and the IL-13Rα2 chain.
CHO-K1 cells were transfected with various receptor chains and incubated with IL-13 for 10 minutes, solubilized with cold whole-cell extraction buffer, and 50 μg sample protein was incubated for 20 minutes with 1 ng 32P-labeled SBE1 probe in binding buffer. DNA-protein interaction was analyzed by SDS-PAGE analysis (A). CHO-K1 cells were transfected with increasing amounts (0 to 10.5 μg) of IL-13Rα2 cDNA along with fixed amounts of DNA for α1 IL4Rα (6 μg) chains (B) or α1 IL4Rαγc (C). For supershift assay, protein extract from α1 IL4Rα–transfected CHO-K1 cells was incubated with antihuman STAT6 rabbit polyclonal immunoglobulin G before electrophoresis (D).
To further study the impact of the IL-13Rα2 chain on STAT6 activation, we transfected CHO-K1 cells with the α1 and IL-4Rα chains along with various amounts of α2 chain cDNA. As shown in Figure 6B, the extent of STAT6 activation in response to IL-13 was gradually decreased as the amount of α2 chain cDNA transfected increased. We also assessed the effect of the IL-13Rα2 chain in IL-13–induced STAT6 activation in CHO-K1 cells transfected with IL-4Rα, IL-13Rα1, and γc chains. As shown in Figure6C, introduction of the IL-13Rα2 chain at very high concentration can still inhibit IL-13–induced STAT6 activation as seen in IL-4Rα and IL-13Rα1 transfectants. The presence of γc chain had no effect on this inhibition.
To confirm whether the IL-13–induced SBE-1–binding complex contains STAT6, an antibody supershift assay was performed. The STAT6 activation that was induced by IL-13 in α1 IL-4Rα heterodimer transfectants showed a substantial supershift by anti-STAT6 antibody (Figure 6D, lane 2). These data confirm that SBE-1 binding complexes induced by IL-13 contain the STAT6 molecule.
Discussion
The major goal of our study was to investigate whether the IL-13Rα2 chain can internalize IL-13 ligand by itself and whether this chain can induce signal transduction through STAT pathways. In this report, we demonstrate that the IL-13Rα2 chain can bind with high affinity to IL-13 and can promote internalization of ligand, but it cannot induce signaling via the STAT6 pathway. Although we cannot rule out the possibility that IL-13 may signal through different signaling cascades, our results suggest an interesting dissociation between internalization and signal transduction by IL-13R. This phenomenon is similar to that observed for the IL-6 receptor system in which IL-6 signal transducer gp130 is internalized without inducing signal transduction through the JAK/STAT pathway.34 35 The IL-13Rα2 chain affects a decrease in the level of IL-13–induced STAT6 activation in cells cotransfected with the IL-13Rα1 and IL-4Rα chains or IL-13Rα1, IL-4Rα, and γc chain. This decrease in signaling may be due in part to the “stealing” of IL-13 from other components of the receptor, thus making less IL-13 available for binding and signaling through type II IL-13 receptors.
The internalization of ligand on IL-13Rα2 transfection alone was also confirmed by cytotoxicity assays that utilize IL-13 cytotoxin. CHO-K1 cells transfected with the IL-13Rα2 chain were very sensitive to recombinant IL13-PE38QQR cytotoxin in a dose- and IL-13Rα2 concentration–dependent manner. However, IL-4 toxin was not cytotoxic, indicating specificity of the internalization. When IL-13Rα2 chain was cotransfected with IL-13Rα1 and IL-4Rα chains, a slightly higher cytotoxicity was observed. These results agree with the binding studies and further indicate that IL-13 toxin is internalized through the IL-13Rα2 chain that, when combined with the IL-13Rα1 and IL-4Rα chains, produces an additive effect.
It is of interest that about 25% protein synthesis inhibition occurs at 100 ng/mL and higher concentrations of IL13-PE38QQR in CHO-K1 cells transfected with the IL-4Rα chain alone. Because CHO-K1 cells express low levels of specific IL-13R, transfection of the IL-4Rα chain may form functional type II IL-13 complex, resulting in internalization of IL13-PE38QQR.
It has been demonstrated that a di-leucine motif in the intracellular domain of type I cytokine receptor systems (eg, IL-6R,35,36 granulocyte colony-stimulating factor receptor,37 epidermal growth factor receptor,38 growth hormone receptor,39 and human insulin receptor40) plays an essential role in the internalization of ligand. In the IL-13Rα2 chain, there is no intracellular domain L-L motif; however, 3 residues, L335, L336, and L337, are present at the carboxy terminus of the transmembrane domain.19 Whether these leucine residues interact with other amino acids and contribute to the internalization of IL13–IL-13R complex is not known. To study this, we produced 3 deletion mutants in the intracellular domain of the IL-13Rα2 chain, and the significance of different intracellular mutants was examined by binding and internalization studies. From these studies, it was concluded that amino acids between L335 and Y343 play a role in IL-13 binding and/or internalization.
We found that when CHO-K1 cells were transduced with IL-13Rα1 or IL-4Rα chain cDNA, a modest activation of STAT6 was observed. It is possible that CHO-K1 cells naturally express IL-4Rα and IL-13Rα1 chains and on introduction of either of these chains form a functional IL-13R complex. To address this issue, we performed reverse transcriptase (RT)-PCR analysis of these chains. Although we did not find expression of these chains by sensitive RT-PCR assay (data not shown), we do not rule out the possibility of their expression because (1) CHO-K1 cells specifically bind IL-13 and (2) primers used in our study were designed to amplify human receptor chain RNA. The specific primers for Chinese hamster may be needed to reverse transcribe and detect hamster receptor chains. Alternatively, the IL-13Rα1 and IL-4Rα chains can homodimerize on binding to IL-13 albeit at low levels and can activate STAT6. Homodimerization of IL-4Rα has been shown to activate STAT6 on stimulation with IL-4.15
In conclusion, we have reconstituted a functional IL-13R by transfecting various components of the IL-13R system. For the first time we provide experimental evidence that the IL-13Rα2 chain is a functional component of an IL-13R system that promotes binding and internalization of ligand. In addition, the IL-13Rα2 chain may interact with the α1 or IL-4Rα chains, or both, as it inhibits the effect of IL-13 on STAT6 activation in α1- or IL-4Rα–transfected cells. Since IL-13Rα2 is not involved in type II or type III IL-13R systems commonly expressed on normal immune and some nonimmune cells, our observations indicate that the IL-13Rα2 chain can be a potential target for receptor-directed immunotherapy or gene therapy for cancer or inflammatory diseases in which the IL-13R is involved. In addition, transfer of the IL-13Rα2 chain gene may make cells more susceptible to the cytotoxic effect of IL-13 toxin.
We thank Drs S. Rafat Husain and Bharat H. Joshi for labeling IL-13 and for providing IL13-PE38QQR, and Dr Yasuo Oshima and Ms Pamela Dover for helpful suggestions. We also thank Drs Elizabeth Shores and Donald Fink for critical reading of this manuscript.
K.K. and J.T. contributed equally to this paper.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Raj K. Puri, Laboratory of Molecular Tumor Biology, Division of Cellular and Gene Therapies, Center for Biologics Evaluation and Research, Food and Drug Administration, National Institutes of Health, Bldg 29B, Rm 2NN10, 29 Lincoln Dr MSC 4555, Bethesda, MD 20892; e-mail: puri@cber.fda.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal