Abstract
PU.1 is a member of the Ets family of transcription factors required for the development of various lymphoid and myeloid cell lineages, but its role in natural killer (NK) cell development is not known. The study shows that PU.1 is expressed in NK cells and that, on cell transfer into alymphoid Rag2/γc−/−mice, hematopoietic progenitors of PU.1−/−fetal liver cells could generate functional NK cells but not B or T cells. Nevertheless, the numbers of bone marrow NK cell precursors and splenic mature NK cells were reduced compared to controls. Moreover,PU.1−/− NK cells displayed reduced expression of the receptors for stem cell factor and interleukin (IL)-7, suggesting a nonredundant role for PU.1 in regulating the expression of these cytokine receptor genes during NK cell development.PU.1−/− NK cells also showed defective expression of inhibitory and activating members of the Ly49 family and failed to proliferate in response to IL-2 and IL-12. Thus, despite the less stringent requirement for PU.1 in NK cell development compared to B and T cells, PU.1 regulates NK cell differentiation and homeostasis.
Introduction
Natural killer (NK) cells are a distinct subset of lymphocytes that mediate important functions in innate immunity being able to eliminate tumor cells and to produce cytokines without prior sensitization (reviewed in Trinchieri1). NK cells can distinguish cells with disparate levels of major histocompatibility complex (MHC) class I expression, killing cells that, due to viral infections or transformation, have low MHC expression, and sparing those with normal expression (reviewed in Ljunggren and Karre2). Such discrimination is mediated by self MHC-specific inhibitory receptors on NK cells that belong to one of 3 groups: killer immunoglobulin-like receptors on human NK cells, lectin-like Ly49 receptors on murine NK cells, and lectin-like NKG2/CD94 cells that are found on both human and rodent NK cells (reviewed in Lanier3). Despite our appreciation of these different NK cell functions, the developmental relationship of NK cells with other hematopoietic lineages is not clear. A rare population of common lymphoid progenitors (CLPs) that can give rise to T, B, and NK cells has been identified in mouse bone marrow (BM),4 and NK and T cells have been suggested to derive from a common progenitor during fetal life.5 6 NK and T cells also share expression of several differentiation antigens and effector functions, yet the great majority of T cells are generated in the thymus, whereas the predominant site for NK cell development is the BM.
Part of the difficulty in understanding the developmental relationship of NK cells with other hematopoietic lineages stems from our incomplete knowledge of NK cell ontogeny. Notwithstanding, it is now clear that development of NK cells is strictly dependent on cytokines that promote survival, proliferation, and differentiation. Interleukin (IL)-15 plays a pivotal role in NK cell differentiation, thus NK cells are extremely reduced or absent in mice deficient for IL-157 or for any of the IL-15 receptor subunits (IL-15Rα,8IL-2Rβ,9 γ) or its downstream signaling molecules Jak311 and Stat5.12However, IL-15 intervenes in a rather late stage of development, when the commitment to the NK cell lineage has already been made. Although “early acting” cytokines, including IL-7, stem cell factor (SCF), and Flk2L/Flt3L, are best candidates for driving the commitment to the NK cell lineage (reviewed in Williams et al13), the relative contribution of these growth factors is not fully appreciated. Therefore, the molecular mechanisms marking NK cell specification are not completely understood. It is conceivable that these cytokines activate genetic programs that use multiple transcription factors to silence or to activate lineage-specific genes. However, although the transcriptional regulation of lineage commitment during lymphopoiesis has been quite extensively studied in the context of B- and T-cell development (reviewed in Glimcher and Singh14), the transcription factors that control engagement to the NK cell lineage have only recently started to be identified. Mice deficient for interferon regulatory factor-1 fail to develop NK cells, due to impaired transcriptional activation of the Il15locus.15 The absence of CCAAT/enhancer binding protein γ (CREB-γ) impinges selectively on NK cell development and not on B or T lymphopoiesis.16 Disruption of the Id2 gene results in the block of NK cell development, whereas B- and T-cell differentiation unfolds normally.17 The further understanding of the transcriptional regulation of NK cells may help to define stages in which lineage specification takes place, thereby clarifying the developmental relationship of NK cells with B- and T-cell lineages.
The Ets (E26 transformation specific) family of oncogenic transcription factors comprises more than 20 members that are conserved throughout evolution, regulating cell fate of multiple cell types in worms, flies, birds, and mammals (reviewed in Bassuk and Leiden18). Several mammalian Ets transcription factors are expressed in the hematopoietic system. Gene-targeting experiments have helped to define the role of these transcription factors in mouse hematopoietic development. One Ets family member, PU.1 (purine rich box-1) or Spi-1 (spleen focus-forming virus integration site-1) is required for the differentiation of multiple hematopoietic lineages.19,20Although specification of erythrocytes and megakaryocytes can occur in the absence of PU.1, monocytes, mature granulocytes, myeloid-derived dendritic cells, and B lymphocytes fail to develop inPU.1−/− mice.19,20 T-cell development is also strongly affected, with few or no T cells developing in PU.1−/− mice19,20or in fetal thymic organ culture.21 In contrast, becausePU.1−/− mice die at E17.5 during embryonic life19 or just after birth,20 the role of PU.1 in NK cell differentiation has not been examined.
To overcome this limitation, we utilized a recently developed complementation system in which fetal liver (FL) cells are used as a source of hematopoietic stem cells (HSCs) to reconstitute alymphoidRag2/γc−/− mice. In these hematopoietic chimeras, all lymphoid cells are donor derived.22-24 We found that PU.1−/−Rag2/γc−/− hematopoietic chimeras were essentially devoid of B and T cells, whereas NK cells could be generated. Thus, despite sharing a close developmental relationship, B, T, and NK cells are differentially dependent on PU.1 for their generation.
Materials and methods
Mice and generation of FL hematopoietic chimeras
Mice with a null mutation in the common γ chain (γ) were from the fourth generation backcross to the C57Bl/6 background. Rag2−/− mice (10th backcross to C57Bl/6) were bred with B10.BR (H-2k), and F1 progeny were intercrossed to generate RAG-2–deficient mice on the H-2k background. These mice were then bred withRag2/γc−/− mice22 to generateRag2/γc−/− mice carrying the H-2k haplotype. PU.1−/+mice19 (C57Bl/6 × 129, H-2b) were screened by polymerase chain reaction (PCR), using genomic DNA and specific primers for PU.1 and Neo gene (5′-CGG ATG TGC TTC CCT TAT CAA AC-3′, 5′-TGA CTT TCT TCA CCT CGC CTG TC-3′, 5′-CAG AAA GCG AAG GAG CAA AGC TG-3′). PU.1+/− mice were intercrossed to generate PU.1−/− and control (PU.1+/+, or PU.1+/−, thereafter referred to as WT) embryos. The morning of the vaginal plug discovery was designated as day 0.5 of gestation. FL cells were obtained from day 15.5 embryos by passage of the tissue through a 23-gauge needle, and the genotypes were determined by fluorescence-activated cell sorter (FACS) analysis, using antibodies specific for Mac-1 (macrophages fail to develop inPU.1−/− embryos).19Rag2/γc−/− mice on H-2kbackground (> 6 weeks of age) were irradiated with 300 rads from a cobalt source and 2 hours later injected intravenously with 8, 25, or 75 × 105 FL cells as a source of hematopoietic progenitors, including HSCs, which we will refer to as FL-HSCs.
Flow cytometry analysis and cell sorting
Single cell suspensions were prepared from blood, thymus, BM, spleen, and liver. Erythrocytes were lysed in ammonium chloride, and cells were resuspended in phosphate-buffered saline with 1% bovine serum albumin and 0.01% sodium azide. Cell viability was evaluated by trypan blue exclusion. Monoclonal antibodies (mAbs) directly conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), Tricolor (TRI), allophycocyanin (APC), or biotin were used for immunofluorescence analysis, including mAbs specific for immunoglobulin M (IgM), IgD, T-cell receptor αβ (TCRαβ), TCRγδ, CD3, CD4, CD8, CD11a (LFA-1), CD11b (Mac-1), CD19, CD45R (B220), CD90 (Thy-1.2), CD117 (c-kit), CD122 (IL-2Rβ), CD161 (NK1.1), DX5, 2B4, Ly49A, Ly49C/I, Ly49D, Ly49G2, Gr-1, TER-119, and H-2k (all from Pharmingen, San Diego, CA). Biotin-conjugated mAbs were revealed by streptavidin-TRI (Caltag, Burlingame, CA). Cells (106) were first incubated with anti-FcγRII/III (hybridoma 2.4G2) for 20 minutes on ice to avoid unspecific binding to low affinity FcRs. Thereafter, cells were stained with a mixture of biotinylated and fluorochrome-labeled mAbs at saturating concentrations, washed twice, and finally incubated with streptavidin-TRI.
To evaluate rare BM progenitor populations, 20 × 106cells were incubated with purified rat mAbs for lineage (Lin) markers (CD11b, CD19, TER-119, and Gr-1) on ice for 20 minutes. After washing, cells were incubated with goat antirat IgG coupled with magnetic beads (Dynal, Oslo, Norway), and the majority of Lin+ cells were removed on a magnet. Cells were then stained with H-2k (to detect host-derived cells), washed, and incubated with mAbs specific for rat IgG, CD3, CD4, CD8, and B220 (all coupled with TRI). TRI+ cells were subsequently electronically gated, thereby excluding Lin+ and host-derived precursors.
Analysis was performed on a FACScan or FACScalibur flow cytometer, using the Cellquest software (Becton Dickinson, San Diego, CA). Dead cells were excluded by means of their forward and side scatters, and an electronic gate was set to acquire 104 lymphoid cells. Cells derived from the host (H-2k) were excluded from the analysis by electronic gating.
To purify splenic NK cells, cell suspensions were stained with mAbs specific for NK1.1, and CD3 and NK cells (CD3−NK1.1+) were sorted using a FACStar+ (Becton Dickinson). The purity of the sorted populations was reproducibility greater than 95%.
NK cell lytic activity
A standard 51Cr release assay was used to measure NK lytic activity in vitro as described.22 YAC-1 cells (mouse thymoma; H-2a) were used as target cells and were maintained in complete medium (CM; RPMI-1640 with 10% fetal calf serum, 10−5 M β-ME, 100 mg/mL streptomycin, 100 U/mL penicillin). Target cells were labeled with 100 μCi 51Cr (ICN Pharmaceutical, Costa Mesa, CA), and 2.5 to 5 × 103cells were incubated with graded numbers of effector cells in 200 μL medium for 4 hours. Effector cells were either NK cells that were isolated from splenocytes by cell sorting or IL-2–activated NK cell cultures. The radioactivity released into the cell-free supernatant was measured, and the percentage of specific lysis was calculated as follows: 100 × (experimental release − spontaneous release) / (maximum release − spontaneous release).
Reverse transcriptase–PCR and Western blotting
RNA was isolated from freshly sorted NK cell populations with RNABle (EUROBIO, Les Ulis, France) according to the manufacturer's instructions. Complementary DNA was synthesized, using reverse transcriptase (RT) from avian myeloblastosis virus (PROMEGA, Madison, WI), hexanucleotides, and oligo-dT (Amersham Pharmacia, Uppsala, Sweden). PCR was performed, using Taq Platinum polymerase (GIBCO BRL). Primer sequences were as follows: PU.1 forward 5′-GAG TTT GAG AAC TTC CCT GAG-3′ and reverse 5′-TGG TAG GTC ATC TTC TTG CGG-3′; Ets-1 forward 5′-CTA CGG TAT CGA GCA TGC TCA GTG-3′ and reverse 5′-AAG GTG TCT GTC TGG AGA GGG TCC-3′; Id2 forward 5′-TCT GAG CTT ATG TCG AAT GAT AGC-3′ and reverse 5′-CAC AGC ATT CAG TAG GCT CGT GTC-3′; IL-7Rα forward 5′-CTT TTA CGA GTG AAA TGC CTA ACT-3′ and reverse 5′-CAG GTA TGA TTC AAG AAT GCA ATA CA-3′; TCF-1 forward 5′-CTC TGC CTT CAA TCT GCT CAT-3′ and reverse 5′-TGG GTT CTG CCT GTG TTT TCA-3′; hypoxanthine phospho ribosyl transferase (HPRT) forward 5′-CAC AGG ACT AGA ACA CCT GC-3′ and reverse 5′-GCT GGT GAA AAG GAC CTC T-3′ (RNA control).
Proteins were extracted from cell lysates derived from splenocytes, thymocytes, pre-B cells (cell line 18.81), and purified IL-2–activated NK cells. Proteins were then resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Immunoreactive PU.1 protein was revealed with an affinity-purified rabbit anti-PU.1 antibody as described.25
Cell-cycle analysis
Cell-cycle analysis was performed on in vitro IL-2–activated NK cells and on circulating CD3−NK1.1+ NK cells from peripheral blood, using 7-aminoactinomycin-D (7-AAD) incorporation into saponin-permeabilized cells as described.23 Sorted splenic CD3−NK1.1+ cells were plated at 104 cells/well in round-bottom microtitre plates in 200 μL CM and activated with 1000 U/mL huIL-2 (Peprotech, Rocky Hill, NJ) for 7 days. To stimulate NK cell proliferation and to promote subsequent activation-induced cell death, 2 ng/mL mIL-12 (Peprotech) was added during the final 48 hours before the cell-cycle analysis.
Statistical analysis
Data were analyzed with the Microsoft Excel software, applying the paired Student t test. The null hypothesis was rejected, and difference was assumed significant when P < .05.
Results
Expression of PU.1 in NK cells
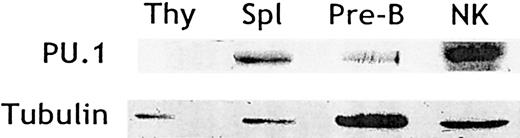
Previous studies have assessed PU.1 expression in CLPs, immature B- and T-cell precursors, and mature B cells.21,26 27However, little is known about PU.1 expression in NK cells and its putative role during NK cell differentiation. We, therefore, tested and found PU.1 protein in cell lysates derived from sorted NK cells expanded in IL-2 (Figure 1). As expected, PU.1 was also found in control lysates derived from total splenocytes and pre-B cells but not in thymocyte lysates (Figure 1). Thus, mature NK cells, like B cells and unlike T cells, maintain PU.1 expression throughout development.
NK cells express PU.1.
Cell lysates were generated from thymocytes, splenocytes, pre-B cells (cell line 18.81), and purified splenic IL-2–activated NK cells. Protein extracts were resolved by SDS-PAGE and probed by Western blotting with an affinity-purified anti-PU.1 antibody. Blots were stripped and reprobed with antitubulin antibody to control for sample loading.
NK cells express PU.1.
Cell lysates were generated from thymocytes, splenocytes, pre-B cells (cell line 18.81), and purified splenic IL-2–activated NK cells. Protein extracts were resolved by SDS-PAGE and probed by Western blotting with an affinity-purified anti-PU.1 antibody. Blots were stripped and reprobed with antitubulin antibody to control for sample loading.
Lymphoid cell development in the absence of PU.1
To address the role of PU.1 in NK cell development in vivo, we generated hematopoietic chimeras by injectingPU.1−/− or WT FL-HSCs (H-2b) into alymphoid Rag2/γc−/−(H-2k) mice. In this system,22-24 all lymphoid cells are donor derived, and any host-derived cell can be identified by virtue of their differential H-2 expression. BecausePU.1−/− FL cells are known to contain fewer hematopoietic progenitors than control cells,25 the reconstitution capacity of 3 different doses (8 × 105, 25 × 105, or 75 × 105) of FL-HSCs fromPU.1−/− embryos was analyzed and compared to WT controls. Seven to 13 weeks after the transfer, chimeras were killed, and the lymphoid cellularity in thymus, BM, spleen, and liver was evaluated. Splenic T cells (CD3+NK1.1−) were virtually absent in PU.1−/− chimeras (Table 1), and the thymi ofPU.1−/− chimeras were hypocellular, containing almost exclusively early CD4−CD8−double-negative thymocytes (Figure 2). However, in very rare PU.1−/− chimeras, we could detect the 4 populations of CD4+, CD4+CD8+, CD8+, and CD4−CD8− thymocytes (data not shown), consistent with a variable penetrance of the T-cell–deficiency phenotype.20,21 B220+IgM+ B cells were absent in the spleen (Table 1) and BM ofPU.1−/− chimeras (Figure 2), confirming the essential role of PU.1 in B-cell differentiation.19 20
Splenic cellularity (cell numbers × 105)
| FL-HSC* . | FL cells injected . | Total† . | NK cells (NK1.1+CD3−) . | T cells (NK1.1−CD3+) . | B cells (B220+IgM+) . | n . |
|---|---|---|---|---|---|---|
| WT | 8 | 518 ± 147 | 5.2 ± 1 | 75.7 ± 1.3 | 267 ± 111 | 7 |
| PU.1−/− | 8 | 47 ± 23 | 0.48 ± 0.29 | 0.21 ± 0.18 | 0.04 ± 0.02 | 8 |
| WT | 25 | 1250 ± 243 | 16.1 ± 1.2 | 206.6 ± 98 | 429 ± 99 | 4 |
| PU.1−/− | 25 | 96 ± 46 | 3.7 ± 3 | 0.2 ± 0.1 | 0.2 ± 0.1 | 8 |
| FL-HSC* . | FL cells injected . | Total† . | NK cells (NK1.1+CD3−) . | T cells (NK1.1−CD3+) . | B cells (B220+IgM+) . | n . |
|---|---|---|---|---|---|---|
| WT | 8 | 518 ± 147 | 5.2 ± 1 | 75.7 ± 1.3 | 267 ± 111 | 7 |
| PU.1−/− | 8 | 47 ± 23 | 0.48 ± 0.29 | 0.21 ± 0.18 | 0.04 ± 0.02 | 8 |
| WT | 25 | 1250 ± 243 | 16.1 ± 1.2 | 206.6 ± 98 | 429 ± 99 | 4 |
| PU.1−/− | 25 | 96 ± 46 | 3.7 ± 3 | 0.2 ± 0.1 | 0.2 ± 0.1 | 8 |
NK indicates natural killer; FL-HSC, fetal liver hematopoietic stem cell; IgM, immunoglobulin M; WT, wild type.
Genotype of FL-HSCs used to reconstitute alymphoidRag2/γc−/− mice.
Numbers of cells found in the spleen of reconstitutedRag2/γc−/− mice.
NK cells but not B and T cells can be generated in the absence of PU.1.
Seven to 13 weeks after transfer of FL-HSCs, cells were isolated from thymus, BM, and spleen and were stained respectively with mAbs specific for CD8-FITC and CD4-TRI, IgM-PE and B220-TRI, as well as CD3-APC and NK1.1-PE. An electronic gate based on morphologic criteria was set to exclude most nonlymphoid cells. An example of the gating strategy for lymphoid cell analysis in the spleen is shown. Lymphoid organs ofPU.1−/− chimeras contained virtually no T or B cells, whereas NK cells were present in all chimeras analyzed. Data are from representative of 8 independent experiments.
NK cells but not B and T cells can be generated in the absence of PU.1.
Seven to 13 weeks after transfer of FL-HSCs, cells were isolated from thymus, BM, and spleen and were stained respectively with mAbs specific for CD8-FITC and CD4-TRI, IgM-PE and B220-TRI, as well as CD3-APC and NK1.1-PE. An electronic gate based on morphologic criteria was set to exclude most nonlymphoid cells. An example of the gating strategy for lymphoid cell analysis in the spleen is shown. Lymphoid organs ofPU.1−/− chimeras contained virtually no T or B cells, whereas NK cells were present in all chimeras analyzed. Data are from representative of 8 independent experiments.
In contrast with the T- and B-cell deficiency, CD3−NK1.1+ NK cells were present in spleen, liver, and BM of all PU.1−/− chimeras analyzed (n = 16), although they were reduced in absolute numbers compared to controls (n = 11; Table 1). The kinetics of peripheral NK cell generation in the absence of PU.1 was somewhat slower than controls (data not shown), a phenomenon already described for the in vivo generation of T cells in the viablePU.1−/− strain20 and the in vitro generation of lymphoid-derived PU.1−/−dendritic cells.28 The data in Table 1 demonstrate that by injecting more PU.1−/− FL-HSCs, more NK cells could be generated, but the NK cell numbers inPU.1−/− chimeras never reached those found inWT chimeras. Thus, 10-fold lessPU.1−/− NK cells were present in the spleens of chimeras generated with 8 × 105 FL-HSCs, but only 3- to 4-fold less PU.1−/− NK cells were present in the spleens of PU.1−/− chimeras generated with 25 × 105 FL-HSCs. However, T- and B-cell numbers did not significantly increase with higher doses of FL-HSCs and remained 400- to 6000-fold reduced compared to controls. The numbers of NK cells generated with even higher doses of FL-HSCs (75 × 105) were not greater than those obtained by injecting 25 × 105 FL-HSCs (Table 1 and data not shown), suggesting that the reduction of NK cells inPU.1−/− chimeras is not only due to the reduced frequency of progenitors in the FL ofPU.1−/− embryo donors. These results clearly demonstrate that NK cell development is permissive in the absence of PU.1.
Reduced production of early NK cell precursors in the absence of PU.1
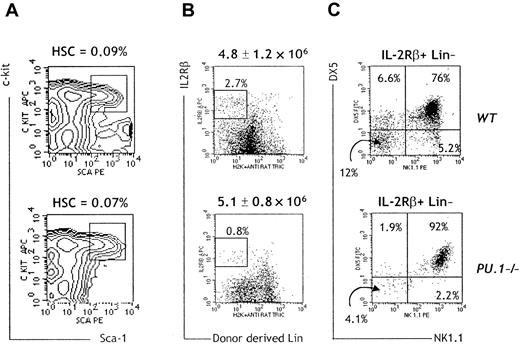
HSCs of PU.1−/− embryos fail to express VLA-4/CD49d− VLA-5/CD49e-CD11b integrins that has been hypothesized to result in defective homing to the BM.29 To directly test whether defects in engraftment ofPU.1−/− FL-HSCs in the adult BM could explain the lower numbers of NK cells in PU.1−/−chimeras, we searched for donor-derived HSCs 8 weeks post-transfer. These cells are contained in a population of BM cells that is negative for the host H-2k, does not express lineage-specific markers (including CD19, B220, CD3, CD4, CD8, Gr-1, Mac-1, TER-119, and NK1.1), and is positive for both Sca-1 and c-kit. We found HSCs (Sca-1+, c-kit+, Lin−, and H-2k−) in PU.1−/− chimeras (Figure 3A) and the presence of donor-derived CD45+ cells at 16 weeks post-transfer (data not shown). These results rule out a major defect in engraftment or homing of FL-HSCs in PU.1−/− chimeras.
Reduced numbers of donor-derived NK cell precursors inPU.1−/− chimeras.
(A) BM cells of WT and PU.1−/−chimeras were isolated, and lineage-positive cells were eliminated by a combination of magnetic bead depletion and electronic gating. HSCs (boxed cells) were identified by c-kit and Sca-1 expression as shown. Percentages were calculated on total BM cells. Data are representative of 4 independent experiments. (B) Lin-depleted BM cells were in parallel stained with mAbs specific for DX5-FITC, NK1.1-PE, and IL-2Rβ-APC. Absolute numbers ± standard deviation of Lin-depleted BM cells are indicated on top of the dot plots. Lin-IL-2Rβ+ cells were electronically gated, and their percentages are indicated. (C) The percentages of gated Lin-IL-2Rβ+ cells that were positive or negative for NK1.1 and DX5 are indicated. Data are representative of 3 independent experiments, including 8 WT and 5PU.1−/− chimeras.
Reduced numbers of donor-derived NK cell precursors inPU.1−/− chimeras.
(A) BM cells of WT and PU.1−/−chimeras were isolated, and lineage-positive cells were eliminated by a combination of magnetic bead depletion and electronic gating. HSCs (boxed cells) were identified by c-kit and Sca-1 expression as shown. Percentages were calculated on total BM cells. Data are representative of 4 independent experiments. (B) Lin-depleted BM cells were in parallel stained with mAbs specific for DX5-FITC, NK1.1-PE, and IL-2Rβ-APC. Absolute numbers ± standard deviation of Lin-depleted BM cells are indicated on top of the dot plots. Lin-IL-2Rβ+ cells were electronically gated, and their percentages are indicated. (C) The percentages of gated Lin-IL-2Rβ+ cells that were positive or negative for NK1.1 and DX5 are indicated. Data are representative of 3 independent experiments, including 8 WT and 5PU.1−/− chimeras.
Collectively, these observations suggest that PU.1 plays an intrinsic and essential role in early NK cell differentiation. We have recently identified a cell population in murine BM that appears to represent a committed NK cell precursor (NKP), having lost any potential for B, T, or myeloid differentiation (E. Rosmaraki et al, manuscript submitted). NKPs are Lin− and share IL-2Rβ+ expression with mature NK cells, but in contrast they do not express NK1.1 or DX5. Eight to 10 weeks after the transfer, we enumerated these Lin− IL-2Rβ+ NK1.1-DX5 NKPs in WTand PU.1−/− chimeras. NKPs were 10- to 12-fold reduced in the BM of PU.1−/− chimeras (Figure3B,C). This observation suggests that PU.1−/−hematopoietic progenitors can only poorly generate the early NK cell compartment. Accordingly, the numbers of NK cells in olderPU.1−/− chimeras (> 30 weeks; n = 5) decline with age to almost undetectable levels (data not shown).
Characterization of PU.1−/−NK cells
The development of hematopoietic progenitors is associated with dynamic changes in the expression of a number of transcription factors.26,27 The transcription factors Ets-1 and Id2 have been shown to be crucial for NK cell development.17 30Figure 4A shows thatPU.1−/− NK cells express Id2 transcripts and, interestingly, up-regulate expression of Ets-1 compared to control NK cells.
Characterization of
PU.1−/− NK cells. (A) RT-PCR for the indicated transcripts was performed, using RNA prepared from sorted splenic NK cells (CD3−NK1.1+) fromWT and PU.1−/− chimeras. (B) Splenic NK cells were stained with mAbs specific for the indicated surface antigens. Histogram profiles are shown for CD3−NK1.1+ gated cells. Data are representative of 8 independent experiments.
Characterization of
PU.1−/− NK cells. (A) RT-PCR for the indicated transcripts was performed, using RNA prepared from sorted splenic NK cells (CD3−NK1.1+) fromWT and PU.1−/− chimeras. (B) Splenic NK cells were stained with mAbs specific for the indicated surface antigens. Histogram profiles are shown for CD3−NK1.1+ gated cells. Data are representative of 8 independent experiments.
We further analyzed the cell surface phenotype of the splenic NK cells that developed in the absence of PU.1. A series of differentiation antigens, including NK1.1, DX5, CD2, 2B4, Mac-1, and Thy-1, were expressed at expected frequencies in PU.1−/−NK cells and at levels comparable to those of controls (Figure 4B and data not shown). The LFA-1 complex is expressed on all leukocytes and is composed of CD11a and CD18 integrins. CD11a is a putative target gene of PU.1,31,32 neverthelessPU.1−/− NK cells expressed normal levels of LFA-1 (Figure 4B). Mature NK cells express inhibitory and activating members of the Ly49 family of receptors that recognize MHC class I antigens on the surface of target cells and play a critical role in regulating NK cell cytotoxic activity (reviewed in Raulet33). PU.1−/− chimeras contained normal percentages of splenic NK cells expressing the inhibitory receptors Ly49C/I and Ly49G2 (Figure 4B). However, the NK cell fractions expressing the inhibitory receptor Ly49A and the activating receptor Ly49D were clearly underrepresented inPU.1−/− NK cells (Figure 4B). Along these lines, the Ly49a locus has been shown to contain a consensus PU.1 binding site (E. Hofer, 18th International NK Cell Workshop, Marseille, France, May 2000). Reduced Ly49A expression has been described in mice deficient for the high mobility group transcription factor TCF-1.34 Figure 4A shows that TCF-1 transcripts were normally detected in PU.1−/− NK cells, excluding the possibility that PU.1 is an essential regulator of TCF-1 expression.
Mature PU.1−/−NK cells fail to proliferate in response to IL-2 and IL-12
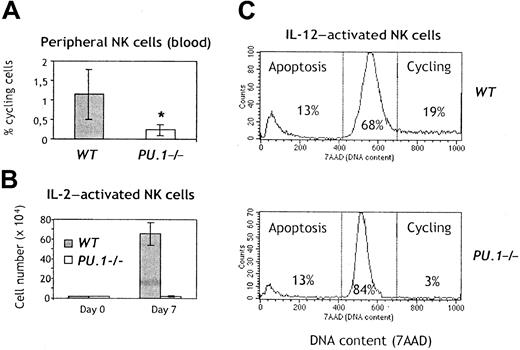
A survival and/or proliferation defect of mature NK cells could contribute to the reduced absolute numbers of NK cells inPU.1−/− chimeras. We, therefore, measured the percentages of cycling cells and hypodiploid apoptotic cells among circulating NK cells in PU.1−/− chimeras. Although there was no obvious increase in the proportion ofPU.1−/− NK cells undergoing apoptosis (data not shown), the fraction of NK cells in cycle was significantly lower inPU.1−/− NK cells, as compared with controls (Figure 5A). IL-2 mediates survival, activation, and expansion of NK cells in vitro,1 and addition of IL-12 to IL-2–stimulated NK cells promotes their activation-induced cell death. Purified splenic NK cells fromPU.1−/− chimeras remained viable throughout the culture period in IL-2 but did not appreciably expand (Figure 5B). In addition, they proliferated little in IL-12, although a normal apoptotic response to IL-2 + IL-12 was preserved (Figure 5C). Finally, we failed to generate NK cells in vitro fromPU.1−/− FL cells (data not shown), using a combination of cytokines (SCF, Flk2L/Flt3L, IL-7, and IL-2) that drives NK cell differentiation with high efficiency from WT FL cells.13 23 Together, these results indicate thatPU.1−/− NK cells have defective responses to cytokines.
Reduced proliferation in
PU.1−/− NK cells. (A) Blood cells were isolated from chimeras 4 weeks after the transfer of FL-HSCs. The figure indicates the mean and standard deviation of cycling NK cells in 6 WT and 6 PU.1−/− chimeras; *P = .008. (B) Freshly sorted NK cells from spleen of chimeras were plated at 2 × 104 cells/well and expanded in IL-2 for 7 days, and thereafter counted. (C) The same cells were replated at 105 cells/well and further stimulated with IL-12 overnight. For detection of apoptotic and proliferating NK cells, cells were stained with mAbs specific for NK1.1-PE and further stained with 7-AAD to reveal the DNA content. Proliferating cells (in G2/M) contain more DNA, whereas cells dying by apoptosis are hypodiploid.
Reduced proliferation in
PU.1−/− NK cells. (A) Blood cells were isolated from chimeras 4 weeks after the transfer of FL-HSCs. The figure indicates the mean and standard deviation of cycling NK cells in 6 WT and 6 PU.1−/− chimeras; *P = .008. (B) Freshly sorted NK cells from spleen of chimeras were plated at 2 × 104 cells/well and expanded in IL-2 for 7 days, and thereafter counted. (C) The same cells were replated at 105 cells/well and further stimulated with IL-12 overnight. For detection of apoptotic and proliferating NK cells, cells were stained with mAbs specific for NK1.1-PE and further stained with 7-AAD to reveal the DNA content. Proliferating cells (in G2/M) contain more DNA, whereas cells dying by apoptosis are hypodiploid.
Expression of growth factor receptors onPU.1−/−NK cells
PU.1 has been implicated in the regulation of cytokine receptor genes.35 Developing NK cell precursors rely on Flk2L/Flt3L, SCF, IL-7, and IL-15 to survive, proliferate, and differentiate (reviewed in Williams et al13). The receptor for IL-7 is expressed during the early lymphopoiesis, and transcripts for the IL-7Rα chain are also found in mature NK cells (Figure6A). However,PU.1−/− NK cells failed to express IL-7Rα. Interactions of SCF with its c-kit receptor are essential for expansion of NK cell precursors and full maturation of NK cells.23 A small subset of mature NK cells express the c-kit receptor, but this subset was 6- to 7-fold reduced in PU.1−/− NK cells (Figure 6B). NK cells derived from c-kit–deficient FL-HSCs also express low levels of the activation marker B220.23Consistent with the reduction in c-kit expression, the B220+ NK cell fraction was clearly underrepresented inPU.1−/− chimeras (Figure 6B). Signaling through the IL-15 receptor is crucial to drive NK cell development.7-9 This tripartite receptor is composed of the IL-15Rα, IL-2Rβ, and γc chains (the latter 2 are shared with the IL-2 receptor and can signal in response to high doses of IL-2). In line with the capacity of IL-2 to promote survival ofPU.1−/− NK cells (Figure 5B), essentially all of these cells expressed IL-2Rβ and contained RNA transcripts for IL-15Rα and γc (Figure 6B and data not shown).
Expression of growth factor receptors and activation markers on
PU.1−/− NK cells. (A) RT-PCR for the indicated transcripts was performed, using RNA prepared from sorted splenic NK cells (CD3−NK1.1+) fromWT and PU.1−/− chimeras. (B) Splenic NK cells were stained with mAbs specific for the indicated surface antigens. Histogram profiles are shown for CD3−NK1.1+ gated cells. Data are representative of 8 independent experiments.
Expression of growth factor receptors and activation markers on
PU.1−/− NK cells. (A) RT-PCR for the indicated transcripts was performed, using RNA prepared from sorted splenic NK cells (CD3−NK1.1+) fromWT and PU.1−/− chimeras. (B) Splenic NK cells were stained with mAbs specific for the indicated surface antigens. Histogram profiles are shown for CD3−NK1.1+ gated cells. Data are representative of 8 independent experiments.
Lytic activity of PU.1−/−NK cells
We have previously observed a correlation between reduced expression of B220 and defective lytic activity inc-kit−/− NK cells.23 Because B220 and c-kit expression were reduced in PU.1−/−NK cells, we assessed the capacity of freshly isolated splenicPU.1−/− NK cells to lyse YAC-1 thymoma targets in a standard 51Cr release assay. As shown in Figure7, PU.1−/− NK cells were fully competent in lysing this NK-sensitive target. In addition, IL-2–activated PU.1−/− NK cells could kill both YAC-1 and P815 targets (data not shown). These results show that differentiation of the lytic machinery for natural cytolysis does not require PU.1.
PU.1−/−NK cells are able to lyse tumor cells in vitro.
Splenic CD3−NK1.1+ NK cells were purified by sorting and were used as effectors in a classical 51Cr release assay versus YAC-1 thymoma cells. There was no significant difference in the lytic capacity of splenic NK cells purified fromPU.1−/− (○) or WT (●) chimeras. Data from 2 separate experiments are shown.
PU.1−/−NK cells are able to lyse tumor cells in vitro.
Splenic CD3−NK1.1+ NK cells were purified by sorting and were used as effectors in a classical 51Cr release assay versus YAC-1 thymoma cells. There was no significant difference in the lytic capacity of splenic NK cells purified fromPU.1−/− (○) or WT (●) chimeras. Data from 2 separate experiments are shown.
Discussion
In this report, we demonstrate that NK cells like B but not T lymphocytes express PU.1, but this transcription factor is not strictly required for the generation of functional NK cells in vivo. Our studies made use of a novel alymphoid mouse strain,Rag2/γc−/− mice, which were reconstituted with PU.1−/− FL hematopoietic progenitors. In this setting, B-cell, T-cell, and NK cell development is entirely donor derived, and little or no competition with endogenous early lymphoid precursors is observed.22-24 Moreover, the potentially lethal effects associated with defective granulocyte and macrophage development in the absence of PU.1 are avoided since these hematopoietic lineages develop normally inRag2/γc−/− mice.36 This genetic approach allowed us to assess the role of PU.1 in NK differentiation.PU.1−/− chimeras remain B and T cell deficient but generate BM precursors and peripheral functional NK cells, although both are reduced in numbers.
We show here that NK cells maintain expression of PU.1 throughout differentiation. Consistent with this finding, maturePU.1−/− NK cells displayed phenotypical abnormalities, such as reduced expression of certain surface antigens, including the receptors for IL-7 and SCF, and defective proliferative responses to potent NK cell mitogens, such as IL-2 and IL-12. However,PU.1−/− NK cells could mediate natural cytotoxicity and survive in vitro on stimulation with IL-2. Taken together, our results demonstrate a differential requirement of PU.1 for NK versus B- and T-cell lymphopoiesis.
How do we explain the partial effects of PU.1 deficiency on NK cell differentiation in the context of the known roles that this transcription factor plays during hematopoiesis? PU.1 expression appears to be regulated in a complex and dynamic fashion throughout the hematopoietic system. PU.1 is expressed in HSCs and in common myeloid progenitors and in CLPs.26 Although the precise effects of PU.1 deficiency on these hematopoietic subsets remain to be determined, PU.1 is not required for the generation of HSCs, since this cell subset could be detected after cell transfer ofPU.1−/− FL precursors intoRag2/γc−/− recipients. PU.1 is essential for myeloid differentiation,19,20 yet its absence causes divergent consequences in distinct cell subsets. Monocytes and macrophages strictly depend on PU.1 for development,37whereas neutrophils can develop in PU.1−/−mice, although they show defective effector functions.38Myeloid-derived dendritic cells also fail to develop in the absence of PU.1, whereas the requirement for PU.1 in development of lymphoid-derived dendritic cells is controversial, and the expression of PU.1 in this subset has not been documented.28,39 PU.1 expression is maintained as CLPs differentiate toward the B-cell lineage but is turned off early in T-cell development.21 27
The requirement of PU.1 for B-cell development appears absolute, as shown by the complete absence of fetal and BM-derived B lineage cells in PU.1−/− mice,19,20,25 which we confirm in this report. PU.1 regulates the expression of theIL-7Rα gene during fetal hematopoiesis.14,40PU.1−/− fetal hematopoietic progenitors do not express transcripts for the B lineage-specific transcription factors EBF and Pax-5.14,25 Thus, the profound block to B-cell development caused by the PU.1 mutation may be due to defects in both IL-7–induced proliferation as well as EBF and Pax-5–mediated differentiation of B lineage progenitors. PU.1 may directly regulate the expression of the immunoglobulin loci.41-43 Interestingly, low levels of PU.1 are essential for B-cell development, whereas high levels inhibit B-cell differentiation and instead promote macrophage development.44
In contrast, PU.1 expression is restricted to a discrete stage of T lineage differentiation. Only the earliest intrathymic progenitors (CD44+CD25−) express PU.1, which is rapidly extinguished as T cells mature.21,27 Still, T-cell development is profoundly impaired in PU.1−/−mice.19,20 However, studies using fetal thymic organ culture have shown that the few thymocytes inPU.1−/− mice that can bypass this developmental block develop into mature T cells that have normal function,21 thus PU.1 expression in T-cell differentiation is essential only during early thymopoiesis. CD44+CD25− cells of fetal thymi contain a bipotent precursor endowed with both NK and T potential,45and PU.1 deficiency may block development of these 2 closely related cell lineages. However, this bipotent precursor may not represent the main pool of NK progenitors in adult life, when NK lymphopoiesis occurs in the BM. We show here a reduction of NKP inPU.1−/− BM, whereas their frequency of HSCs was normal. This suggests a NK lineage-specific role for PU.1, which may be independent of its role in early T-cell development.
The mechanisms by which the absence of PU.1 disrupts hematopoietic development are being clarified. A major function of PU.1 is to control the transcription of growth factor receptor genes in developing blood cells.14 In the context of myeloid development, evidence supports a model in which PU.1 is required for expression of the granulocyte colony-stimulating factor, macrophage colony-stimulating factor, granulocyte-macrophage colony-stimulating factor, and c-fms receptors on early myeloid progenitors (see Held et al34and references therein). However, retroviral infection with c-fms could only restore the proliferation but not differentiation defect in PU.1-deficient myeloid precursors, suggesting additional roles for PU.1 in macrophage development.40 Thus, the absence of expression of certain cytokine receptors during early myeloid and B-cell development in PU.1 mutants can partly explain the developmental blocks.
In line with this, PU.1 deficiency impaired the expression of some cytokine and growth factor receptors on developing NK cells. Previous studies have identified 4 ligand/receptor systems that play an important role in the generation of NK cells from hematopoietic precursors: Flk2/Flt3, c-kit, IL-7, and IL-15 (reviewed in Williams et al13). Of these, we found thatPU.1−/− NK cells express IL-2Rβ and transcripts for IL-15Rα and γc. In contrast, IL-7Rα was absent, and c-kit expression was reduced on PU.1−/− NK cells. The reduction in numbers of NK precursors seen inPU.1−/− BM may well reflect a synergistic effect of the defective IL-7Rα and c-kit expression. In the absence of c-kit and IL-7R signaling, NK cell development may be sustained by Flk2L/Flt3L, IL-15, or other cytokines. A similar cumulative defect in these cytokine receptors may also explain the severe block in early T lymphopoiesis in PU.1−/− mice.46Thus, the requirement for IL-7 and c-kit in early NK lymphopoiesis may be less strict than for early thymopoiesis. Mice deficient in both IL-7 and c-kit will help to test this hypothesis.
T-cell progenitors lose the expression of PU.1 as they commit to the T lineage (CD44+CD25+), whereas NK cells express it throughout development. In line with this, maturePU.1−/− NK cells are less in cycle, fail to proliferate in response to mitogens, and do not express a normal pattern of surface antigens (including Ly49A, Ly49D, and B220) as they differentiate. Yet they are competent for natural cytolysis of lymphomas, suggesting that PU.1-independent transcriptional regulation governs this crucial NK cell effector function.
Expression of Ly49 molecules may be acquired in an ordered sequence, although expression patterns of genes within the NK cell appear to be regulated independently.47 However, the mechanisms that control this process remain ill defined. We found that PU.1 deficiency was associated with a selective reduction in the expression of Ly49A and Ly49D, whereas NK1.1, Ly49G2, and Ly49C/I were normally expressed in PU.1−/− NK cells. The significance of this observation awaits further investigation but indicates that PU.1 is involved in the fine-tuning of Ly49 expression. One possibility is that PU.1 may affect the acquisition of early (Ly49A) but not late Ly49 (Ly49C/I) members. The TCF-1 transcription factor has also been implicated in the specific regulation of Ly49A.34 However, we could exclude, on the basis of gene expression analysis, a direct effect of PU.1 on TCF-1 expression, and it is, therefore, likely that the defective Ly49 expression seen in PU.1−/−NK cells is independent of TCF-1, although TCF-1 and PU.1 may cooperate to activate Ly49A expression.
The Ets family of transcription factors comprises multiple members, and, although their expression is regulated in a dynamic way during development, overlapping expression patterns of distinct members is documented and may allow functional redundancy.27Another Ets family member expressed in developing lymphoid cells is Ets-1. Ets-1–deficient mice demonstrate T-cell survival defects, accelerated terminal differentiation of B cells,48,49 and reduced numbers of NK cells.33 We found thatPU.1−/− NK cells express higher levels of Ets-1 transcripts, and it is tempting to speculate that, in the absence of PU.1, Ets-1 may compensate to some extent for the transcriptional regulation that leads to NK cell development but not to B- and T-cell development. However, a functional redundancy between PU.1 and Ets-1 is difficult to imagine, as these 2 members share only 40% homology in their DNA-binding domain and have a different panel of target genes.18 Alternatively, they may regulate nonoverlapping functions during NK differentiation. For example, bothEts1−/− mice andPU.1−/− chimeras have reduced numbers of NK cells, but natural cytotoxicity is abolished in the absence of Ets-1 and is preserved in the absence of PU.1. Therefore, PU.1 deficiency is compatible with a normal differentiation of the lytic machinery, including expression of known targets of Ets factors such as LFA-1 and perforin,18 whereas Ets-1 may be essential for these genes. One may argue that the NK cells accumulating in the periphery ofPU.1−/− chimeras may have undergone critical alterations due to effects of the mutation at early developmental stages. As such, the role of PU.1 in mature NK cells cannot be unambiguously assessed. Conditional gene targeting may provide a solution to this problem, although no NK-cell lineage–specific transgenes have been characterized to date. Alternatively, retroviral-mediated transfer of PU.140 into maturePU.1−/− NK cells may allow for correction of their phenotypic and functional abnormalities. Such technologic improvements should allow for a finer definition of the role of PU.1 in NK cell biology. Our results suggest that there is a less restrictive requirement for PU.1 in NK cell generation, as compared to myeloid, B cells, and T cells. Nevertheless, PU.1 plays critical roles in NK cell development, during expansion of committed NK precursors, and in the homeostasis and differentiation of mature NK cells.
We would like to thank Jean-Christophe Bories for critically reading the manuscript; Jacques Roland for kindly providing the 18.81 cell line; Eleftheria Rosmaraki for the identification of bone marrow NK cell precursors; and Odile Richard, Erwan Corcuff, Géraldine Bonnefoy, and Fabien Blanchet for advice and help.
F.C. and S.I.S. contributed equally to this work.
Supported by the Association pour la Recherche sur le Cancer, Fondation pour la Recherche Médicale, Ligue Nationale Contre le Cancer and the Pasteur Institut, by Institut National de la Santé et de la Recherche Médicale (INSERM) (F.C.), and by a fellowship from the Ministère de l'Education Nationale de la Recherche et de la Technologie (S.I.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Francesco Colucci, Laboratory for Cytokines and Lymphoid Development, Department of Immunology, The Pasteur Institute, 25-28, rue Dr Roux 75015 Paris, France; e-mail: cecco@pasteur.fr.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal