Abstract
Cytotoxic activity of tumor necrosis factor–related apoptosis-inducing ligand (TRAIL/Apo-2 ligand), used alone or in different combinations with either a low (1.5 Gy) or a high (15 Gy) single dose of ionizing radiation (IR), was investigated on erythroleukemic cells (K562, HEL, Friend, primary leukemic erythroblasts) and on primary CD34+-derived normal erythroblasts. Human recombinant TRAIL alone variably affected the survival/growth of erythroleukemic cells; K562 cells were the most sensitive. Moreover, all erythroleukemic cells were radio-resistant, as demonstrated by the fact that cytotoxicity was evident only after treatment with high-dose (15 Gy) IR. Remarkably, when IR and TRAIL were used in combination, an additive effect was noticed in all erythroleukemic cells. Augmentation of TRAIL-induced cell death by IR was observed with both low and high IR doses and required the sequential treatment of IR 3 to 6 hours before the addition of TRAIL. Conversely, both TRAIL and IR showed a moderate cytotoxicity on primary CD34+-derived normal erythroblasts when used alone, but their combination did not show any additive effect. Moreover, the cytotoxicity of IR plus TRAIL observed in erythroleukemic cells was accompanied by the selective up-regulation of the surface expression of TRAIL-R1 (DR4), and it was completely blocked by the z-Val-Ala-Asp (OMe)-CH2 (z-VAD-fmk) caspase inhibitor. On the other hand, the surface expression of TRAIL-R1 in CD34+-derived normal erythroblasts was unaffected by IR, which induced the up-regulation of the decoy TRAIL-R3. These data demonstrate that treatment with IR provides an approach to selectively sensitize erythroleukemic cells, but not normal erythroblasts, to TRAIL-induced apoptosis through the functional up-regulation of TRAIL-R1.
Introduction
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL), also known as Apo-2 ligand (L), is a new member of the TNF family of cytokines.1,2 Members of this family are structurally related proteins that play important roles in regulating cell death, immune response, and inflammation.3 Like other members of the TNF family, TRAIL is a type 2 membrane protein, with an intracellular amino-terminal portion, an internal transmembrane domain, and a carboxyl terminus external to the cell. In addition, a soluble form of TRAIL has been identified,4 as previously shown for TNF-α and CD95 (Apo-1/Fas) L.5 At variance with CD95L that contains a long intracellular region of 81 amino acids, TRAIL has a short intracellular tail of 17 amino acids and appears regulated at the cell surface of different cell types by proteolytic events sensitive to cysteine protease inhibitors.4 Although these cytokines share sequence homology in their extracellular C-terminals, they do not share receptors (except for TNF-α and lymphotoxin-α)3 or lymphotoxin-β and herpes virus entry mediator ligand (HVEM-L).6 Among the TNF family members, TRAIL shares the highest amino acid identity with CD95L.1,7-10 The unique feature of TRAIL, unlike CD95L and TNF-α, is considered its ability to induce apoptosis on various continuous cell lines and primary tumor cells, including several of hematopoietic origin,5 displaying minimal toxic effects on normal tissues.11-13 It has also been shown that lymphoid and myeloid tumor cells express functional TRAIL on their surfaces.14
Although a role for this cytokine in physiological conditions has not been clearly envisioned yet, we have recently demonstrated that TRAIL shows a lineage-specific inhibitory activity on the survival/growth of CD34+-derived normal erythroblasts, showing an intermediate expression of surface glycophorin A.15 Owing to its high expression at the bone marrow level, we have proposed that TRAIL likely plays an important role in the negative regulation of normal erythropoiesis.15 Because it has been proposed that TRAIL selectively kills cancer cells with respect to their normal counterparts,7,9 16-19 we have here investigated how TRAIL, used alone or in combination with ionizing radiation (IR), modulates the survival/growth of erythroleukemic cells in comparison to normal erythroblasts. For this purpose erythroleukemic (K562, HEL, Friend) cell lines, primary blasts from 2 patients affected by acute erythroleukemia, and primary CD34+-derived glycophorin A+ erythroblasts obtained from normal donors were treated with TRAIL alone or in various combinations with IR.
Materials and methods
Reagents
Recombinant His6-tagged TRAIL and His6-tag control peptide were produced in bacteria and purified by affinity chromatography on Ni2+ affinity resin, as previously reported.19The purity of each TRAIL preparation was evaluated by loading 2 μg recombinant TRAIL in 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), containing (reducing condition) or not containing (nonreducing condition) 5% β-mercaptoethanol. The gels were then stained with the gelcode Silver SNAP Staining kit (Pierce, Boston, MA). In some cases, proteins were blotted onto nitrocellulose filters. Blotted filters were blocked for 30 minutes in a 3% suspension of dried skim milk in phosphate-buffered saline (PBS) and incubated overnight at 4°C with 1:100 dilution of anti-TRAIL rabbit antiserum (Alexis Italy, Florence, Italy). Filters were washed and further incubated for 1 hour at room temperature with 1:1000 dilution of peroxidase-conjugated antirabbit IgG (Sigma Chemical, St Louis, MO) in 0.1% bovine serum albumin (BSA, Cohn fraction V, Sigma). Specific reactions were revealed with the enhanced chemiluminescence Western blotting detection reagent (Amersham, Arlington Heights, IL).
The functional activity of each TRAIL preparation used in this study was tested on the TRAIL-sensitive Jurkat cell line (J32 clone). In previous experiments, maximal effects on Jurkat cell apoptosis were observed in the presence of 0.1 to 1 μg/mL TRAIL. In contrast, equimolar concentrations of His6-tag alone did not show any significant toxicity.20 Therefore, the concentration of 1 μg/mL TRAIL was chosen to perform the experiments in both human and murine erythroleukemic cells, as well as in primary erythroleukemic blasts and primary normal erythroblasts. In this respect, it should be noted that Walczak et al11 have previously demonstrated that human recombinant TRAIL is equally effective in inducing apoptosis on both human and murine target cells.
The broad inhibitor of caspase proteases, Cbz-Val-Ala-Asp-fluoromethyl ketone (z-VAD-fmk),21 and the peptide control Cbz-Phe-Ala-fluoromethyl ketone (z-FA-fmk), both from Enzyme Systems Products (Dublin, CA), were dissolved in dimethyl sulfoxide and stocked in aliquots at −20°C until used.
Cell lines and patient samples
Friend murine erythroleukemia, a virus-transformed cell line,22 was derivative of clone 745 from C. Friend (Mt. Sinai School of Medicine, New York, NY). K56223 and HEL24 human cell lines express erythroid markers, such as glycophorin A. However, though the HEL cell line was derivative of human erythroleukemia,24 the K562 cell line was derivative of chronic myeloid leukemia, characterized by the reciprocal chromosomal translocation 9:22 generating the Philadelphia chromosome.25 All cell lines, obtained from American Type Culture Collection (Rockville, MD), were grown in RPMI (Gibco Laboratories Life Technologies, Grand Island, NY) supplemented with 10% fetal calf serum (FCS; Gibco) at an optimal cell density of 3 × 105 to 1.5 × 106 cells/mL.
Bone marrow (BM) specimens were obtained from 2 male (16 and 56 years) patients affected by acute erythroleukemia (M6 of the French-American-British classification of acute myelogenous leukemias). We obtained their informed consent to the study, in accordance with the Helsinki Declaration of 1975. BM aspirates showing blast counts greater than 70% were collected at diagnosis before any therapy was initiated. Mononuclear cells were separated on density gradient centrifugation (Ficoll/Hypaque-1077; Sigma) and immediately frozen. Aliquots containing 40 to 45 × 106 BM mononuclear cells were thawed at the time of the study, cultured for 24 hours in RPMI + 10% FCS, subjected to density gradient centrifugation (Ficoll/Histopaque-1077) to eliminate dead cells, and treated as described below.
Purification of CD34+ cells and cultures of primary erythroid cells
Cord blood (CB) specimens, collected according to institutional guidelines, were obtained during 6 normal full-term deliveries. CB mononuclear cells were isolated by density gradient centrifugation (Ficoll/Histopaque-1077) and allowed to adhere to plastic for 1 hour at 37°C. CB CD34+ cells were then isolated from mononuclear nonadherent cells using the magnetic cell sorting program Mini-MACS and the CD34 isolation kit (Miltenyi Biotech, Auburn, CA) in accordance with the manufacturer's instructions. The purity of CD34-selected cells was determined for each isolation by Facscan (Lysis II program; Becton Dickinson, San Jose, CA), using a mAb that recognizes a separate epitope of the CD34 molecule (HPCA-2; Becton Dickinson) directly conjugated to fluorescein isothiocyanate. The purity of CD34+ cells ranged from 90% to 98%.
CD34+ cells were cultured in Ex-vivo (Biowhittaker, Walkersville, MD) serum-free medium, supplemented with nucleosides (10 μg/mL each), 0.5% BSA, 10−4 M BSA-adsorbed cholesterol, 10 μg/mL insulin, 200 μg/mL iron-saturated transferrin, and 5 × 10−5 M 2-β-mercaptoethanol (all purchased from Sigma). Cells were adjusted to an optimal cell density of 5 × 104/mL and seeded in culture in the presence of stem cell factor (SCF; 50 ng/mL) + IL-3 (0.5 ng/mL) + erythropoietin (EPO, 4 U/mL). All cytokines were purchased from Genzyme (Cambridge, MA). Every 3 to 4 days, cultures were demipopulated by removing 0.5 vol medium that was substituted with fresh medium supplemented with the erythroid-specific cytokine EPO alone. At day 10 of culture, the cells were removed, counted, stained, and analyzed by Facscan flow cytometry (Becton Dickinson). Staining was performed at 4°C for 30 minutes on 2 × 105 cells in 200 μL PBS containing 1% BSA, 5% human plasma, 0.1% sodium azide, and phycoerythrin (PE)–anti–glycophorin A (Pharmingen, San Diego, CA). After re-adjusting the cell density to 1 × 105/mL, primary glycophorin A+ erythroblasts were treated as described below.
Cell treatments
Erythroleukemic cells and normal CD34+-derived normal erythroblasts were treated with TRAIL (1 μg/mL) orrHis6-tag (0.15 μg/mL) and/or irradiated at room temperature by a Mevatron 74 Siemens (Rotterdam, Holland) linear accelerator (photonic energy, 10 MV) administering 1.5 and 15 Gy (dose rate, 3 Gy/min). All experiments were performed on exponentially growing cells, showing a viability of at least 95%. Cytotoxic effects were evaluated by counting viable cells by trypan blue dye exclusion and measuring cell cycle and apoptosis as described below.
Evaluation of cell cycle and apoptosis
Samples containing 2 to 5 × 105 cells were harvested by centrifugation at 200g for 10 minutes at 4°C, fixed with cold 70% ethanol for at least 1 hour at 4°C, and treated as previously detailed.26 Briefly, samples were pelleted, treated with 0.5 μg RNAse (type I-A) (Sigma) and resuspended in PBS containing 50 μg/mL propidium iodide (PI; Sigma). Analysis of PI fluorescence was performed by FACScan with the FL2 detector in a linear mode using the Lysis II software (Becton Dickinson). For each sample, 10 000 to 20 000 events were collected. For cell-cycle analysis, only the inferred gap 1 (G1), synthesis (S) and gap 2 plus mitosis (G2 + M) peaks were considered. The proportions of cells in the G1, S, and G2 + M phases of the cell cycle were calculated as described.27 For simplicity, the G1 and S/G2 + M values have been provided. For quantitative evaluation of apoptosis, the subdiploid (less than 2n) DNA content was calculated as described27and expressed as percentage of apoptotic versus nonapoptotic cells, regardless of the specific cell-cycle phase.
Ultrastructural analysis of erythroleukemic cells was performed by transmission electron microscopy. Briefly, cell pellets were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2, for 30 minutes at 4°C, rinsed in the same buffer, and processed for conventional Spurr embedding.
Western blotting of TRAIL receptors
For the analysis of TRAIL-R1 and TRAIL-R2 expression, 40 × 106 cells were resuspended in freshly made ice-cold lysis buffer (50 mM Tris, pH 7.5, 1% Triton X-114, 150 mM NaCl, 10 μg/mL leupeptin, 10 μg/mL aprotinin, 1 mM PMSF) for 15 minutes. Lysates were centrifuged at 15 000 rpm for 10 minutes at 4°C. After discharging the pellets (mainly containing cell nuclei and debris), the proteins contained in the aqueous and membrane-associated phases were separated by adding 1% Triton X-114 at 37°C for 3 minutes. After centrifugation at 15 000 rpm, the upper aqueous phase was discharged, and the lower phase containing membrane-associated proteins was resuspended in lysis buffer (without Triton X-114). One hundred micrograms membrane-associated proteins for each sample was migrated in 12% SDS-PAGE and blotted onto nitrocellulose filters. Blotted filters were blocked for 30 minutes in a 3% suspension of dried skim milk in PBS and incubated overnight at 4°C with 1 μg anti–TRAIL-R1 or anti–TRAIL-R2 goat IgG. Filters were washed and further incubated for 1 hour at room temperature with 1:1000 dilution of peroxidase-conjugated antigoat IgG (Sigma) in 0.1% BSA. Specific reactions were revealed with the enhanced chemiluminescence Western blotting detection reagent (Amersham).
Phenotypic analysis of surface TRAIL and TRAIL receptors
At various culture time intervals after irradiation, HEL, K562, and primary cells were analyzed for the surface expression of TRAIL, TRAIL-receptor (R)1, TRAIL-R2, TRAIL-R3, and TRAIL-R4 by indirect staining with primary goat antihuman TRAIL, TRAIL-R1, TRAIL-R2, TRAIL-R3, and TRAIL-R4 IgG (all from R&D System, Oxon, United Kingdom), followed by phycoerythrin (PE)–conjugated rabbit antigoat IgG secondary antibody (Sigma). Briefly, staining was performed on 5 × 105 cells in 200 μL PBS containing 1% FCS and 5 μL each primary antibody on ice for 30 minutes. Cells were washed twice, supplemented with 3 μL PE-conjugated rabbit antigoat IgG on ice for 30 minutes, washed twice with PBS, and analyzed by FACScan. Aspecific fluorescence was assessed by using normal goat IgG and then by a second layer, as above.
Samples were assayed in duplicate, and gates containing viable cells were used to collect 10 000 events. Data are presented as either percentage of positive cells or mean fluorescence intensity (MFI) values.
Statistical analysis
Data were analyzed using the 2-tailed, 2-sample ttest (statistical analysis software; Minitab, State College, PA).P < .05 was considered significant. Interactions between TRAIL and IR were classified by the fractional inhibition method as follows: when expressed as the fractional inhibition of cell viability, additive inhibition produced by both inhibitors occurred when i1,2 = i1 + i2; synergism occurred when i1,2 > i1 + i2; and antagonism occurred when i1,2 < i1 + i2.28
Results
Low susceptibility of erythroleukemic cells to the cytotoxic activity of TRAIL or IR, used alone
Because it has been demonstrated that the homotrimerization of TRAIL is critical for its tumoricidal activity,29 each His6-tagged recombinant TRAIL preparation was analyzed by SDS-PAGE analysis performed in both reducing and nonreducing conditions and visualized by silver staining. As shown in Figure1A, a single band of approximately 32 kd, corresponding to monomeric TRAIL, was recovered when SDS-PAGE was performed under reducing conditions. On the other hand, bands of approximately 60 kd and 90 kd, reflecting multimeric forms of TRAIL were observed in nonreducing conditions. Thus, human recombinant TRAIL protein used in this study is predominantly trimeric in solution.
Analysis of TRAIL protein.
(A) Silver staining of SDS-PAGE analysis of purified recombinant TRAIL, performed in reducing and nonreducing conditions. Sizes of the molecular mass markers are indicated in kilodaltons (kd) on the left. Sizes of TRAIL monomers, dimers, and trimers are indicated on the right. The gels shown are representative of 5 purification experiments. (B) Evaluation of the cytotoxic activity of human recombinant TRAIL (1 μg/mL) on erythroleukemic cell lines. Viable cells were counted by trypan blue dye exclusion at 6 and 24 hours after the addition of either TRAIL or His6-tag control peptide in culture. Data are expressed as percentage control (His6-tag peptide-treated) cells and represent the mean ± SD of 5 independent experiments performed in duplicate.
Analysis of TRAIL protein.
(A) Silver staining of SDS-PAGE analysis of purified recombinant TRAIL, performed in reducing and nonreducing conditions. Sizes of the molecular mass markers are indicated in kilodaltons (kd) on the left. Sizes of TRAIL monomers, dimers, and trimers are indicated on the right. The gels shown are representative of 5 purification experiments. (B) Evaluation of the cytotoxic activity of human recombinant TRAIL (1 μg/mL) on erythroleukemic cell lines. Viable cells were counted by trypan blue dye exclusion at 6 and 24 hours after the addition of either TRAIL or His6-tag control peptide in culture. Data are expressed as percentage control (His6-tag peptide-treated) cells and represent the mean ± SD of 5 independent experiments performed in duplicate.
The cytotoxic effect of TRAIL protein was evaluated by 2 independent methods: count of viable cells by trypan blue dye exclusion and analysis of cell cycle and apoptosis by flow cytometry after PI staining. At both 6 and 24 hours after the addition of TRAIL (1 μg/mL), erythroleukemic cell lines showed distinct patterns of response to TRAIL from the relatively high (P < .01) sensitivity of K562 (up to 45% decrease with respect to control cells treated with His6-tag peptide) to the moderate (P < .05) sensitivity of Friend (28% decrease) and the modest sensitivity of HEL (19% decrease) (Figure 1B). These data indicate that erythroleukemic cells show a variable response to TRAIL and that the peak of cytotoxicity was at 24 hours.
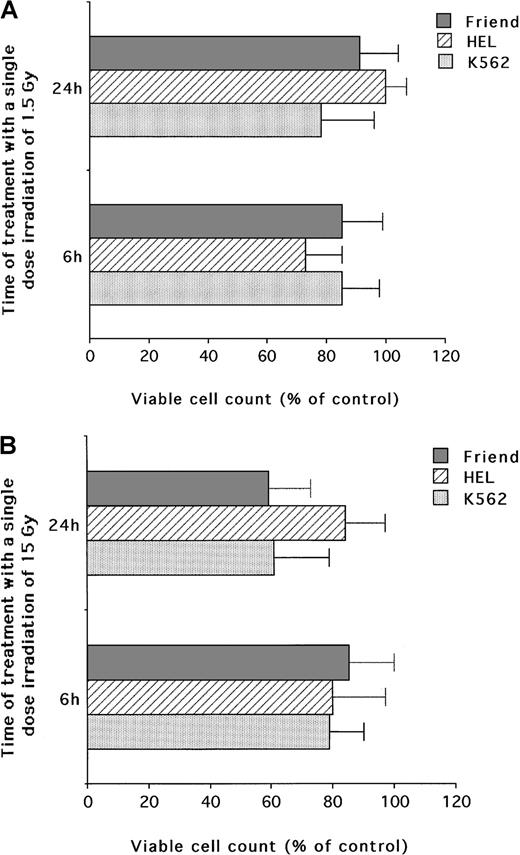
In parallel experiments, K562, HEL, and Friend cells were treated with IR. The choice of 1.5 and 15 Gy IR doses was made on the basis of the standard radiotherapeutic schemes used for the management of various human cancers.30,31 In fact, a daily fraction of 1.5 Gy or more is commonly delivered in human tumor radiotherapy,30whereas 15 Gy can be reached at the end of a therapeutic protocol32 or occasionally used as a single fraction for palliative treatment. As shown in Figure2A, relatively modest cytotoxic effects (less than 30% decrease in the number of viable cells in comparison to untreated control cells) were observed in the presence of low IR doses (1.5 Gy) in all cell lines. After 24 hours, in the presence of higher IR doses (15 Gy), the cytotoxicity ranged from a 15% decrease (for HEL) to a 40% decrease for both Friend (P < .01) and K562 (P < .01) cells (Figure 2B).
Evaluation of the cytotoxic activity of low and high single-dose IR on erythroleukemic cell lines.
(A) Low (1.5 Gy) single-dose IR. (B) High (15 Gy) single-dose IR. Viable cells were counted by trypan blue dye exclusion at 6 and 24 hours after irradiation. Data are expressed as percentage of control cells and represent the mean ± SD of 5 independent experiments performed in duplicate.
Evaluation of the cytotoxic activity of low and high single-dose IR on erythroleukemic cell lines.
(A) Low (1.5 Gy) single-dose IR. (B) High (15 Gy) single-dose IR. Viable cells were counted by trypan blue dye exclusion at 6 and 24 hours after irradiation. Data are expressed as percentage of control cells and represent the mean ± SD of 5 independent experiments performed in duplicate.
Augmentation of the TRAIL-mediated cytotoxicity on erythroleukemic cells by pretreatment with IR
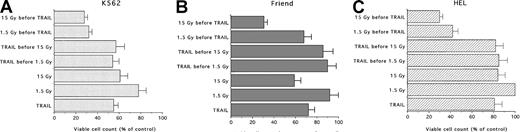
In the following experiments, the effects of different combinations of TRAIL and IR were investigated. The results are summarized in Figure 3. When cells were pretreated with TRAIL for 6, 18, or 24 hours before IR (1.5-15 Gy), no additive or synergistic effects were noticed between the 2 treatments, irrespective of the IR doses used in the cell lines examined (Figure 3A-C). On the other hand, when the cells were pretreated with IR for 3 to 6 hours before TRAIL, a potentiation of TRAIL-mediated cytotoxicity was observed (Figure 3A-C). As previously shown for the 2 treatments used alone, the behavior of the 3 erythroleukemic cell lines was not uniform. In fact, in K562, the combination of either 1.5 Gy (20% decrease in the number of viable cells with respect to control cells) or 15 Gy (40% decrease) plus TRAIL (40%-45% decrease) resulted in an additive cytotoxic effect (68%-72% decrease) (Figure 3A). Similar additive effects (approximately 70% decrease) were noticed in Friend cells treated with 15 Gy (41% decrease) followed by TRAIL (28% decrease) (Figure 3B). Of note, HEL cells showed the maximal response to this combination of treatments. In fact, TRAIL (19% decrease alone) showed a synergistic cytotoxic effect when used with either 1.5 Gy (0% decrease alone versus 58% decrease in association with TRAIL) or 15 Gy (16% decrease alone versus 70% decrease in association with TRAIL) (Figure3C).
Evaluation of the combined cytotoxic activity of TRAIL (1 μg/mL) plus low (1.5 Gy) and high (15 Gy) single-dose IR on erythroleukemic cell lines.
(A) K562, (B) Friend, and (C) HEL erythroleukemic cell lines. Cells were treated with 1 μg/mL TRAIL (for 6, 18, or 24 hours) before IR or IR (3-6 hours) before the addition of 1 μg/mL TRAIL. Viable cells were counted by trypan blue dye exclusion 24 hours after irradiation. Data are expressed as percentage of control (His6-tag peptide-treated) cells and represent the mean ± SD of 5 independent experiments performed in duplicate.
Evaluation of the combined cytotoxic activity of TRAIL (1 μg/mL) plus low (1.5 Gy) and high (15 Gy) single-dose IR on erythroleukemic cell lines.
(A) K562, (B) Friend, and (C) HEL erythroleukemic cell lines. Cells were treated with 1 μg/mL TRAIL (for 6, 18, or 24 hours) before IR or IR (3-6 hours) before the addition of 1 μg/mL TRAIL. Viable cells were counted by trypan blue dye exclusion 24 hours after irradiation. Data are expressed as percentage of control (His6-tag peptide-treated) cells and represent the mean ± SD of 5 independent experiments performed in duplicate.
Selective induction of apoptosis in erythroleukemic cells but not in normal glycophorin A+ erythroblasts by the combination of IR followed by TRAIL
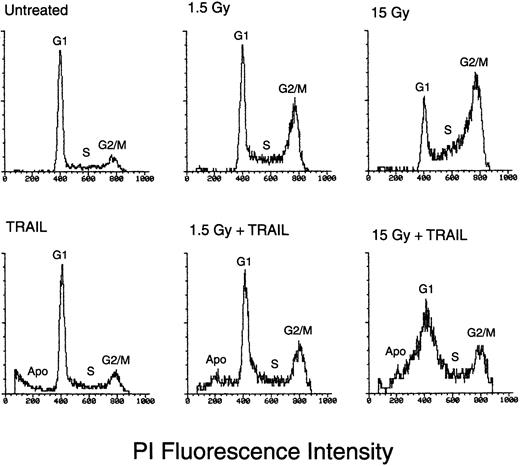
The cytotoxic mechanism of IR plus TRAIL on erythroleukemic cell lines was next investigated by analyzing the cell-cycle profile and the degree of apoptosis after PI staining and flow cytometry examination. As shown in Figure 4 and Table1, IR by itself produced profound perturbations of the cell cycle, characterized by a dose-dependent accumulation of the cells in the S/G2-M phases of the cell cycle. On the other hand, TRAIL by itself induced modest changes in the cell cycle of the cells examined (Table 1). Rather it induced a moderate increase (P < .01) of apoptosis in K562 and HEL, but not in Friend, cells (Figure 4, Table2). In cells treated with the combination of IR followed by TRAIL, the percentage of cells distributed in the different cell-cycle phases was unchanged in comparison to cells treated with IR alone (Table 1). On the other hand, the percentage of apoptosis was significantly (P < .05) increased in erythroleukemic cells treated with IR plus TRAIL with respect to those treated with TRAIL or IR alone and in control cells treated with His6-tag peptide (Table 2). Thus, although at the time points examined the cytotoxic effects of IR on erythroleukemic cells were mainly due to cell-cycle arrest, IR sensitized these cells to TRAIL-mediated apoptosis. Ultrastructural examination confirmed the presence of apoptosis in cell lines treated with IR plus TRAIL, displaying initial aspects of chromatin clumping and condensation followed by the formation of micronuclei (Figure5).
Flow cytometric evaluation of the cell-cycle profile and of apoptosis in HEL cells left untreated or treated with IR (1.5 and 15 Gy) alone, TRAIL (1 μg/mL) alone, or a combination of IR followed after 6 hours by TRAIL.
Analysis was performed 24 hours after irradiation. The x-axis shows the DNA content, determined based on fluorescence from PI staining, and the y-axis reflects the relative number of cells. Percentage cells in apoptosis (Apo) or in the G1, S, and G2 + M phases of the cell cycle for each experimental point are reported in Tables 1 and 2. These results are representative of 4 experiments performed in duplicate.
Flow cytometric evaluation of the cell-cycle profile and of apoptosis in HEL cells left untreated or treated with IR (1.5 and 15 Gy) alone, TRAIL (1 μg/mL) alone, or a combination of IR followed after 6 hours by TRAIL.
Analysis was performed 24 hours after irradiation. The x-axis shows the DNA content, determined based on fluorescence from PI staining, and the y-axis reflects the relative number of cells. Percentage cells in apoptosis (Apo) or in the G1, S, and G2 + M phases of the cell cycle for each experimental point are reported in Tables 1 and 2. These results are representative of 4 experiments performed in duplicate.
Cell-cycle analysis performed by flow cytometry after treatment with ionizing radiation alone or in combination (6 hours before) with tumor necrosis factor–related apoptosis-inducing ligand (1 μg/mL). Analysis was performed 24 hours after irradiation
| . | HEL . | K562 . | Friend . | Erythroleukemic blasts . | Normal erythroblasts . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| G1 . | S/G2-M . | G1 . | S/G2-M . | G1 . | S/G2-M . | G1 . | S/G2-M . | G1 . | S/G2-M . | |
| Control | 65 | 35 | 58 | 42 | 57 | 43 | 58 | 42 | 54 | 46 |
| 1.5 Gy | 37 | 63 | 39 | 61 | 45 | 55 | 46 | 54 | 44 | 56 |
| 15 Gy | 17 | 83 | 33 | 67 | 34 | 66 | 33 | 67 | 42 | 58 |
| TRAIL | 57 | 43 | 58 | 42 | 55 | 45 | 54 | 46 | 53 | 47 |
| 1.5 Gy before TRAIL | 38 | 62 | 39 | 61 | 44 | 56 | 45 | 55 | 43 | 57 |
| 15 Gy before TRAIL | 15 | 85 | 30 | 70 | 35 | 65 | 36 | 64 | 40 | 60 |
| . | HEL . | K562 . | Friend . | Erythroleukemic blasts . | Normal erythroblasts . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| G1 . | S/G2-M . | G1 . | S/G2-M . | G1 . | S/G2-M . | G1 . | S/G2-M . | G1 . | S/G2-M . | |
| Control | 65 | 35 | 58 | 42 | 57 | 43 | 58 | 42 | 54 | 46 |
| 1.5 Gy | 37 | 63 | 39 | 61 | 45 | 55 | 46 | 54 | 44 | 56 |
| 15 Gy | 17 | 83 | 33 | 67 | 34 | 66 | 33 | 67 | 42 | 58 |
| TRAIL | 57 | 43 | 58 | 42 | 55 | 45 | 54 | 46 | 53 | 47 |
| 1.5 Gy before TRAIL | 38 | 62 | 39 | 61 | 44 | 56 | 45 | 55 | 43 | 57 |
| 15 Gy before TRAIL | 15 | 85 | 30 | 70 | 35 | 65 | 36 | 64 | 40 | 60 |
Data represent the means of 2 (erythroleukemic blasts), 4 (HEL, K562, and Friend cell lines) and 5 (normal erythroblasts) separate experiments, performed in duplicate. Fourteen percent of the observations fell within one SD of the means.
TRAIL indicates tumor necrosis factor–related apoptosis-inducing ligand.
Quantitative evaluation of apoptosis, performed by flow cytometry 24 hours after treatment with ionizing radiation alone or in combination with (6 hours before) tumor necrosis factor–related apoptosis-inducing ligand (1 μg/mL)
| . | HEL . | K562 . | Friend . | Erythroleukemic blasts . | Normal erythroblasts . | |
|---|---|---|---|---|---|---|
| No. 1 . | No. 2 . | |||||
| Control | 4 ± 2.0 | 6 ± 2.0 | 1 ± 0.5 | 10 | 14 | 14 ± 5.0 |
| 1.5 Gy | 5 ± 1.8 | 5 ± 2.0 | 2 ± 1.0 | 15 | 19 | 25 ± 5.5 |
| 15 Gy | 6 ± 2.0 | 6 ± 2.5 | 3 ± 1.5 | 17 | 18 | 28 ± 6.0 |
| TRAIL | 18 ± 2.0 | 12 ± 3.0 | 3 ± 1.0 | 17 | 20 | 27 ± 3.5 |
| 1.5 Gy before TRAIL | 45 ± 5.0 | 17 ± 3.0 | 10 ± 3.0 | 28 | 31 | 29 ± 5.0 |
| 15 Gy before TRAIL | 51 ± 5.0 | 20 ± 5.0 | 12 ± 4.0 | 31 | 38 | 30 ± 6.5 |
| . | HEL . | K562 . | Friend . | Erythroleukemic blasts . | Normal erythroblasts . | |
|---|---|---|---|---|---|---|
| No. 1 . | No. 2 . | |||||
| Control | 4 ± 2.0 | 6 ± 2.0 | 1 ± 0.5 | 10 | 14 | 14 ± 5.0 |
| 1.5 Gy | 5 ± 1.8 | 5 ± 2.0 | 2 ± 1.0 | 15 | 19 | 25 ± 5.5 |
| 15 Gy | 6 ± 2.0 | 6 ± 2.5 | 3 ± 1.5 | 17 | 18 | 28 ± 6.0 |
| TRAIL | 18 ± 2.0 | 12 ± 3.0 | 3 ± 1.0 | 17 | 20 | 27 ± 3.5 |
| 1.5 Gy before TRAIL | 45 ± 5.0 | 17 ± 3.0 | 10 ± 3.0 | 28 | 31 | 29 ± 5.0 |
| 15 Gy before TRAIL | 51 ± 5.0 | 20 ± 5.0 | 12 ± 4.0 | 31 | 38 | 30 ± 6.5 |
Data represent the mean ± SD of 4 (HEL, K562, and Friend cell lines) or 5 (normal erythroblasts) separate experiments, performed in duplicate. Data of experiments performed in duplicate with erythroleukemic blasts obtained from 2 different patients (nos. 1 and 2) are reported individually.
TRAIL indicates tumor necrosis factor–related apoptosis-inducing ligand.
Ultrastructural examination of HEL cells treated with His6-tag control peptide or with the combination of IR (6 hours before) plus TRAIL (1 μg/mL).
Note that in comparison with the normal morphology shown in the left panel, several HEL cells, on treatment with IR plus TRAIL, showed a characteristic chromatin cap (right upper panel) and the formation of micronuclei (right lower panel) typical of apoptosis. Representative results from 1 of 7 separate experiments are shown.
Ultrastructural examination of HEL cells treated with His6-tag control peptide or with the combination of IR (6 hours before) plus TRAIL (1 μg/mL).
Note that in comparison with the normal morphology shown in the left panel, several HEL cells, on treatment with IR plus TRAIL, showed a characteristic chromatin cap (right upper panel) and the formation of micronuclei (right lower panel) typical of apoptosis. Representative results from 1 of 7 separate experiments are shown.
We were also able to examine the cytotoxic activity of TRAIL alone or in combination with IR on primary blasts obtained at diagnosis from 2 patients affected by acute erythroleukemia and on primary CD34+-derived normal erythroblasts. As shown in Table 1, both primary erythroleukemic blasts and glycophorin A+normal erythroblasts (Figure 6) showed an accumulation of the cells in the S/G2-M phases of the cell cycle after treatment with IR. On the other hand, TRAIL—used either alone or in combination with IR—did not substantially modify the cell-cycle profile of primary erythroleukemic and normal cells. Of interest, analogous with the data obtained with erythroleukemic cell lines, the percentage of apoptosis in primary erythroleukemic blasts increased in the presence of the combined treatment with IR followed by TRAIL (Table 2). On the contrary, though both TRAIL and IR were able to induce a moderate increase of apoptosis in primary normal erythroblasts when used alone, they did not show any additive cytotoxic effect when used in combination (Table 2).
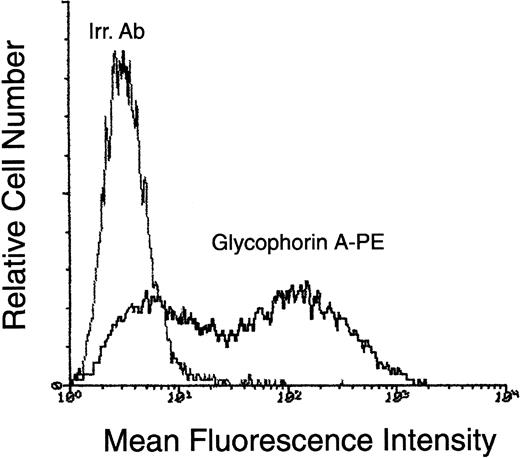
Surface phenotype of CD34+-derived erythroblasts at day 10 of liquid culture.
At day 10, cells obtained in liquid cultures supplemented with the SCF + IL-3 + EPO were phenotypically characterized by staining with PE-conjugated anti–glycophorin A. Negative controls are represented by cells stained with isotype-matched PE-conjugated irrelevant monoclonal antibodies (Irr. Ab). Representative results from 1 of 6 separate experiments are shown.
Surface phenotype of CD34+-derived erythroblasts at day 10 of liquid culture.
At day 10, cells obtained in liquid cultures supplemented with the SCF + IL-3 + EPO were phenotypically characterized by staining with PE-conjugated anti–glycophorin A. Negative controls are represented by cells stained with isotype-matched PE-conjugated irrelevant monoclonal antibodies (Irr. Ab). Representative results from 1 of 6 separate experiments are shown.
Selective up-regulation of TRAIL-R1 (DR4) in human erythroleukemic cells but not in normal erythroblasts in response to IR
It has been previously demonstrated that genotoxic agents are able to up-regulate TRAIL-R2 (DR5) and TRAIL-R3 in different cell types through p53-dependent and p53-independent pathways.15,32,33 Therefore, we next investigated whether IR treatment was able to modulate the surface expression of the different TRAIL receptors and of surface TRAIL.12 Because of the low number of primary blasts in the specimens obtained from 2 patients affected by acute erythroleukemia, these experiments could only be performed using human erythroleukemic cell lines and primary CD34+-derived normal erythroblasts.
Although untreated K562 cells showed clearly detectable levels of TRAIL-R1 (DR4) and TRAIL-R2 (DR5) on Western blot (Figure7A) and flow cytometry (Figure 7B) analyses, HEL cells were positive for TRAIL-R1 but negative for TRAIL-R2 expression (Figure 7A-C). No cross-reactivity was noticed on Western blot analysis between anti–TRAIL-R1 and anti–TRAIL-R2 polyclonal antibodies, which recognized a single band of approximately 60 and 45 kd, respectively, in agreement with the findings of other authors.32 Both cell lines showed either absent or dim expression of decoy (TRAIL-R3 and TRAIL-R4) receptors (Figure 7B-C) and of surface TRAIL (data not shown). Interestingly, exposure to either 1.5 or 15 Gy IR induced the selective up-regulation of TRAIL-R1 (DR4) after 6 hours in HEL cells (Figure 7C) and after 18 hours in K562 cells (Figure 7B). The MFI of 3 independent experiments was 18 ± 3.5 in untreated HEL cells versus 27.5 ± 4 in HEL cells irradiated for 6 hours with 1.5 Gy (P < .05), and it was 12.3 ± 2.5 in untreated K562 cells versus 22.5 ± 3 in K562 cells irradiated for 18 hours with 1.5 Gy (P < .05). No other effects on the expression of TRAIL-R2-4 or surface TRAIL could be observed up to 18 to 24 hours after irradiation in these erythroleukemic cell lines.
Evaluation of the expression of TRAIL receptors.
(A) Membrane proteins were obtained from exponentially growing HEL (H) and K562 (K) cells. After SDS/PAGE, samples were immunoblotted by using polyclonal goat antibody directed toward TRAIL-R1 and TRAIL-R2. Sizes of the molecular mass markers are indicated in kilodaltons (kd) on the left. Surface expression of TRAIL-R1, -R2, -R3, and -R4 was evaluated by flow cytometry in K562 (B), HEL (C) cell lines, and primary normal erythroblasts (D) at 6 (HEL) and 18 (K562, primary normal erythroblasts) hours after IR. Shadowed histograms represent irradiated cells stained with anti–TRAIL-RI, -R2, -R3, and -R4 antibody. Unshadowed histograms represent nonirradiated (control) cells. Negative controls are represented by cells stained with isotype-matched irrelevant goat IgG (Irr. Ab). Representative results from 1 of 4 separate experiments are shown.
Evaluation of the expression of TRAIL receptors.
(A) Membrane proteins were obtained from exponentially growing HEL (H) and K562 (K) cells. After SDS/PAGE, samples were immunoblotted by using polyclonal goat antibody directed toward TRAIL-R1 and TRAIL-R2. Sizes of the molecular mass markers are indicated in kilodaltons (kd) on the left. Surface expression of TRAIL-R1, -R2, -R3, and -R4 was evaluated by flow cytometry in K562 (B), HEL (C) cell lines, and primary normal erythroblasts (D) at 6 (HEL) and 18 (K562, primary normal erythroblasts) hours after IR. Shadowed histograms represent irradiated cells stained with anti–TRAIL-RI, -R2, -R3, and -R4 antibody. Unshadowed histograms represent nonirradiated (control) cells. Negative controls are represented by cells stained with isotype-matched irrelevant goat IgG (Irr. Ab). Representative results from 1 of 4 separate experiments are shown.
The pattern of surface expression of TRAIL receptors in primary glycophorin A+ normal erythroblasts (Figure 7D) was similar to that observed in HEL cells, in which the only surface receptor clearly detectable was TRAIL-R1 (Figure 7B). However, at variance with erythroleukemic cells, the exposure of glycophorin A+erythroblasts to IR did not modify the MFI of TRAIL-R1 after either 6 or 18 hours. On the other hand, exposure to IR induced a clear-cut up-regulation of the decoy TRAIL-R3 in primary normal erythroblasts (Figure 7D). The MFI of 3 independent experiments was 16 ± 2.8 in untreated cells versus 29.5 ± 5.5 in cells irradiated for 18 hours with 1.5 Gy (P < .05).
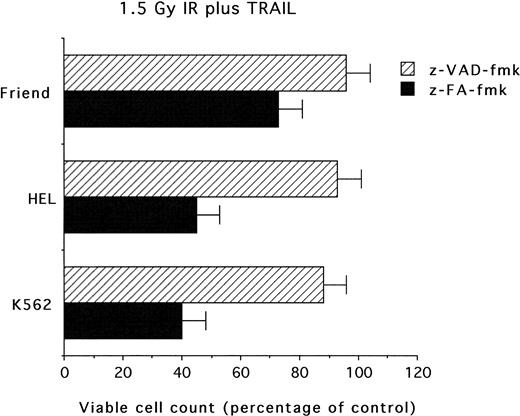
Block of the cytotoxic effects mediated by combination of IR and TRAIL by the z-VAD-fmk caspase inhibitor
We then sought to investigate whether the cytotoxicity observed in erythroleukemic cells treated with the combination of IR and TRAIL was susceptible to the pharmacologic activity of caspase inhibitors.34 Preincubation of cultures with z-VAD-fmk tripeptide at 20 μM before treatment with IR plus TRAIL completely blocked the cytotoxic activity of the combination of the 2 agents (Figure 8). The control z-FA-fmk peptide (20 μM) did not show any toxic or protective effect on erythroleukemic cells. Specificity of the z-VAD-fmk peptide inhibitor was confirmed by its ability to suppress apoptosis also in the presence of anti-CD95 agonistic antibody (data not shown), which is known to rapidly activate the caspase cascade.35
Evaluation of caspase inhibitors on the combined cytotoxicity of IR (1.5 Gy) 6 hours before the addition of TRAIL (1 μg/mL).
Cells were pretreated with z-VAD-fmk or z-FA-fmk peptides (20 μM each), irradiated with 1.5 Gy and, after 6 hours, supplemented with TRAIL (1 μg/mL). Viable cells were counted by trypan blue dye exclusion 24 hours after irradiation. Data are expressed as percentage of control (His6-tag peptide-treated) cells and represent the mean ± SD of 4 independent experiments performed in duplicate.
Evaluation of caspase inhibitors on the combined cytotoxicity of IR (1.5 Gy) 6 hours before the addition of TRAIL (1 μg/mL).
Cells were pretreated with z-VAD-fmk or z-FA-fmk peptides (20 μM each), irradiated with 1.5 Gy and, after 6 hours, supplemented with TRAIL (1 μg/mL). Viable cells were counted by trypan blue dye exclusion 24 hours after irradiation. Data are expressed as percentage of control (His6-tag peptide-treated) cells and represent the mean ± SD of 4 independent experiments performed in duplicate.
Discussion
In this study, we have demonstrated for the first time that, though IR alone exerted a variable but overall relatively modest cytotoxicity on erythroleukemic cell lines and primary malignant erythroblasts, the sequential combination of irradiation and TRAIL resulted in a synergistic cytotoxicity on these cells. The low sensitivity of erythroleukemic cells in response to radiotherapy36-39 was likely due to the high intracellular iron content of these cells compared to radio-susceptible tumor cells.36 Therefore, in spite of their radio-resistance, erythroleukemic cells are sensitized by IR to TRAIL-mediated cytotoxicity. It is also noteworthy that the additive or synergistic cytotoxic effects were observed when cells were pretreated with IR, corresponding to either a daily fraction of 1.5 Gy or a whole cycle of 15 Gy. These findings are particularly interesting because fractionated radiation plays an integral role in the treatment of human cancer. In fact, it was determined empirically that radiation delivered in small fractionated doses would produce less damage to normal tissue, providing greater tumor control than radiation delivered in large single doses.40 More important, the combination of IR plus TRAIL did not show any additive cytotoxic activity on CD34+-derived primary normal erythroblasts.
A major limitation of radiation therapy and of chemotherapy is that they require function of the p53 tumor-suppressor gene for antitumor activity,41 but more than half the human tumors acquire inactivating p53 mutations, making them resistant to conventional anticancer therapy. On the other hand, TRAIL, like other death-inducing ligands such as TNF-α and CD95L, promotes apoptosis, cell-cycle arrest, or both independently of p53 status42 and may offer a complementary approach to conventional cancer therapy. Despite the ability of TNF-α and CD95L to induce apoptosis in cancer cells, severe toxic side effects preclude the use of both these death-inducing ligands in systemic anticancer therapy. TNF-α infusion causes a lethal inflammatory response that resembles septic shock; this effect is mediated primarily by TNF activation of the proinflammatory transcription factor NF-kB in vascular endothelial cells and macrophages.42 Moreover, infusion of agonistic anti-CD95 antibody causes lethal liver damage, which is mediated by the induction of CD95-dependent apoptosis in hepatocytes that express high levels of Fas/CD95.43
Although we and others have recently shown that recombinant human TRAIL can exert cytotoxicity on normal tissues and cells, including hematopoietic progenitor cells committed to the erythroid lineage15 and hepatocytes,44 it still represents the most promising candidate among the death-inducing ligands for systemic anticancer therapy. In this respect, our data showing the ability of IR to augment TRAIL-mediated cytotoxicity in erythroleukemic cells—also at low (1.5 Gy) doses—might disclose new therapeutic perspectives. It has been shown that TRAIL can mediate the biologic activity of chemotherapeutic agents.18,35,45,46Furthermore, the use of TRAIL for anticancer treatment in vivo has been proposed on the basis of a preferential sensitivity of cancer cells rather than normal cells to TRAIL-induced apoptosis. Consistently, only mild anemia was reported by Ashkenazi et al12 after systemic in vivo administration of human recombinant TRAIL to nonhuman primates, and this has been ascribed to the frequent drawing of blood from the animals during the study. Moreover, Walczak et al11 have demonstrated that the administration of human and murine recombinant TRAIL was not toxic to normal tissues of mice and that repeated treatments with human TRAIL actively suppressed growth of the TRAIL-sensitive human mammary adenocarcinoma cell line, MDA-231, in CB.17 (SCID) mice. Similarly, it has been shown that TRAIL, used alone or in combination with camptothecin, shows significant cytotoxicity to human colon carcinoma xenografts.47
We have also addressed the mechanisms underlining the synergistic effect of IR and TRAIL on erythroleukemic cells by demonstrating that IR selectively up-regulates TRAIL-R1 (DR4) in neoplastic cells but not in normal erythroblasts. In this respect, it has been shown that TRAIL-R1 represents the major determinant of TRAIL sensitivity in cancer cells.48 Remarkably, TRAIL-R1 was also expressed on the surfaces of primary normal erythroblasts, but its expression did not change on exposure to IR. On the other hand, IR was able to substantially up-regulate TRAIL-R3 in primary normal cells. These findings strongly suggest that the differential sensitivity of erythroleukemic cells and normal erythroblasts to the combined treatment with IR plus TRAIL results from a differential modulation of TRAIL receptors by IR. In this respect, previous studies have demonstrated an extreme complexity of the expression and function of TRAIL receptors in various cell types.13
At least 5 TRAIL receptors belonging to the apoptosis-inducing TNF-receptor (R) family have been described so far. TRAIL-R1 (DR4) and TRAIL-R2 (DR5) transduce apoptotic signals on the binding of TRAIL, whereas TRAIL-R3 (DcR1), TRAIL-R4 (DcR2), and osteoprotegerin are homologous to DR4 and DR5 in their cysteine-rich extracellular domains, but they lack intracellular death domains and apoptosis-inducing capability. Although the expression of TRAIL-R3 and TRAIL-R4 do not appear to be key factors in determining the resistance or sensitivity of tumor target cells to the effects of TRAIL,45,49 they have been proposed to function as decoy receptors, protecting normal cells from apoptosis.13 Consistently, we found that TRAIL-R3 and TRAIL-R4 were either not expressed or showed dim expression in K562 and HEL cell lines and that they were not affected by IR treatment. TRAIL-R3, however, was significantly up-regulated in normal erythroblasts after IR treatment.
It should be emphasized that the ability of IR to selectively up-regulate TRAIL-R1 in erythroleukemic cells, but not in normal hematopoietic cells, is a completely novel finding. In fact, previous studies have shown that chemotherapeutic genotoxic agents can preferentially up-regulate the expression of TRAIL-R2 in glioblastoma cells, breast cancer cells, and T-lymphoma cells.16,31,50 51 Taken together, these studies and our present findings suggest that the modulation of TRAIL-R1 and TRAIL-R2, which play a major role in determining the cytotoxic response of cancer cells to TRAIL, is lineage specific and that functional TRAIL-R1 can be selectively up-regulated in erythroleukemic cells by IR.
Supported by local funds from the Universities of Chieti, Ferrara, and Bologna, and by MURST Cofin (G.Z., S.M.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Giorgio Zauli, Institute of Morphology, “G. D'Annunzio” University of Chieti, Via dei Vestini 6, 66100 Chieti Scalo (CH), Italy; e-mail: g.zauli@morpho.unich.it.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal