Abstract
This study demonstrates in both stable and inducible BCR-ABL–expressing hematopoietic cells a down-regulation of the major mammalian DNA repair protein DNA-PKcs by BCR-ABL. Similar results were found in BCR-ABL CD34+ cells from patients with chronic myelogenous leukemia (CML). DNA-PKcs down-regulation is a proteasome-dependent degradation that requires tyrosine kinase activity and is associated with a marked DNA repair deficiency along with increased sensitivity to ionizing radiation. The conjunction of a major DNA repair deficiency and a resistance to apoptosis, both induced by BCR-ABL, provides a new mechanism to explain how secondary genetic alterations can accumulate in CML, eventually leading to blast crisis. The down-regulation of DNA-PKcs was reversible in CD34+ CML cells suggesting that this approach might offer a novel and powerful therapeutic strategy in this disease, especially to delay the blast crisis.
Introduction
BCR-ABL p210, the molecular counterpart of the t(9;22) (Philadelphia chromosome, Ph1), is the pathognomonic marker of chronic myelogenous leukemia (CML). The increased tyrosine kinase (TK) activity of the BCR-ABL fusion protein is crucial for its transforming ability.1 CML is characterized also by a progression from chronic toward an acute phase (blast crisis), which is invariably fatal. This transition is generally accompanied by an increased BCR-ABL expression in leukemic cells, with evidence of additional genetic and chromosomal abnormalities, suggesting a genetic instability in Ph1 cells.2
The precise molecular mechanisms underlying this genetic instability, which has been extensively studied in both human CML as well as in experimental cell transformation systems, remain undetermined. In human CML, studies of microsatellite instabilities3,4 as well as those of the methylation of the ABL1 promoter5,6 have given conflicting results. In the experimental setting, cells expressing BCR-ABL present cytogenetic abnormalities during extensive culture7 and an enhanced mutator phenotype associated with an increased expression of the DNA polymerase beta.8 We hypothesized that the DNA-PK (DNA-dependent protein kinase) complex, a major DNA repair system in mammalian cells, could also play a role in this process because it is involved in DNA double-strand breaks (DSB) repair and V(d)J recombination, and can interact and bind with c-abl.9 The DNA-PK complex is composed of a DNA-targeting component, the heterodimer Ku (Ku80/Ku70) and a catalytic subunit designed as the DNA-PKcs. Ku and c-abl provide opposite functions with regard to DNA-PK activity because binding of Ku to DNA-PKcs is required for DNA repair, whereas c-abl dissociates the DNA-PKcs/Ku/DNA complex.10
In contrast to c-abl, it is not known whether BCR-ABL can influence the DNA-PK complex, although BCR-ABL has been reported to activate the phosphatidyl inositol 3 kinase (PI-3K) pathway and could influence the expression of genes belonging to the PI-3K family such as ATM and DNA-PKcs.11
In this study, we found both in murine and human hematopoietic cells transfected with BCR-ABL a major decrease in DNA-PKcs expression that was inversely correlated with BCR-ABL protein levels. A direct relationship between these 2 events was demonstrated by using the murine Ba/F3 cell line expressing a BCR-ABL gene under the control of a tetracycline-inducible promoter. The DNA-PKcs down-regulation was reversible by the use of proteasome inhibitors as well as TK inhibitors. Importantly, a major decrease of DNA-PKcs expression was also found in CD34+ cells from patients with CML. The decreased expression of DNA-PKcs induced by BCR-ABL was associated with a marked DNA repair deficiency, suggesting that this phenomenon could play a role in determining the genetic instability associated with the blast crisis of CML.
Materials and methods
Generation of BCR-ABL–expressing cell lines
Murine Ba/F3 cell clones stably expressing BCR-ABL p210 have been generated as previously described.23 Individual clones expressing high and low levels of BCR-ABL were used in all experiments. BCR-ABL–expressing human hematopoietic UT-7 cells were obtained using MP-ZEN or MSCV-p210 retroviral vectors.18The UT-7 clones stably expressed low (UT-7/E8-1) or high (UT-7/E8-2, UT-7/9) levels of the p210 fusion protein. The UT-7 clones were cultured in the presence of recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF; 10 ng/mL; R&D Systems, Minneapolis, MN) except for the BCR-ABL clones UT-7/9 and UT-7/E8-2 expressing high levels of BCR-ABL. The Ba/F3 cell line was cultured in the presence of 10% WEHI conditioned medium as a source of interleukin 3 (IL-3). For irradiation experiments, all clones were grown in the presence of recombinant human granulocyte-macrophage stem cell factor (rhGM-SCF) (UT-7) or WEHI (Ba/F3). All BCR-ABL–expressing Ba/F3 clones, regardless their levels of BCR-ABL expression, were IL-3 independent for their growth and survival. As previously reported,23 the clone C6 expressed the highest levels of BCR-ABL, whereas the clones C5 and C3 expressed intermediate and low levels of BCR-ABL protein, respectively.
Generation of tetracycline-inducible Ba/F3-BCR-ABL transfectants
To construct a tetracycline-sensitive promoter containing BCR-ABL vector, we have used the SIN vector possessing both the tTA and the tet-OP sequences separated by an IRES.24 The 7.2-kb BCR-ABL complementary DNA (cDNA) insert was subcloned into the SIN vector in the 5′-3′ orientation at the EcoRI site and the resulting vector was transfected in the 293 EBNA cell line to generate transient supernatants. Ba/F3 cell line was transduced with the resulting retrovirus and after 48 hours cells were deprived of IL-3 (10% WEHI-CM). After establishment of IL-3 independence, cells were amplified using IL-3 and BCR-ABL expression was monitored in the presence (BCR-ABL OFF) or absence of doxycycline (BCR-ABL ON). High-level BCR-ABL–expressing cells were then amplified in the presence of doxycycline at 0.1 μg/mL and 1 μg/mL to determine their viability. Experiments to determine the DNA-PKcs levels and the clonogenic survival of cells were performed on a polyclonal stock showing greater than 80% reduction of BCR-ABL protein levels 48 hours after addition of doxycycline (1 μg/mL) and 10% WEHI-CM. On deprivation of IL-3, 50% of the cells undergo apoptotic cell death when grown in the same concentrations of doxycycline (data not shown).
Irradiation and clonogenic assay
Irradiation was performed using a 137Cs γ-ray source at a dose rate of 1.45 Gy/min as previously described.25 Cells were harvested in exponential phase in semiliquid methyl cellulose and irradiated at room temperature (0-6 Gy). The linear-quadratic model was used for fitting the clonogenic survival curves.26 Cells were harvested in exponential phase and irradiated at room temperature. Both UT-7 and Ba/F3 clones were transplanted in methylcellulose (Stem Cell Technologies) at appropriate concentration in the presence of 10 ng/mL rhGM-SCF (UT-7) or 10% WEHI (Ba/F3). The surviving fraction was determined by measuring the viability of colony-forming unit compared with the corresponding untreated controls. Each data point represents the average and SD from 4 independent experiments performed in triplicate. Colonies were counted 10 to 14 days later.
Purification and culture of CD34+ cells
Light-density cells from a leukeopheresis sample of a child with neuroblastoma as well as from 2 patients with chronic phase CML at diagnosis were subjected to a standard CD34+ immunomagnetic beads separation using the miniMACS system following the manufacturer's guidelines (Miltenyi Biotec, Auburn, CA). The purity of CD34+ cells was more than 90%. For Western blot analyses, cryopreserved CD34+ cell samples were thawed and expanded for 6 days in long-term culture medium (Stem Cell Technologies, Vancouver, BC, Canada) containing, rhSCF (50 ng/mL, Amgen, Thousand Oaks, CA), rhIL-3 (50 ng/mL; Novartis, Basel, Switzerland), recombinant human pegylated-megakaryocyte growth and differentiation factor (rhPEG-MGDF; 100 ng/mL; Amgen) and the rhFlt3-ligand (Flt3-L; 100 ng/mL; Diaclone, Besançon, France). Amplified cells were then used in Western blot analyses.
Western immunoblotting
Whole cell extracts (70 μg) were boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and separated by 10% SDS-PAGE gel for Ku70/80, 5% for DNA-PKcs, and 7.5% for BCR-ABL. The protein transfer to polyvinylidene difluoride (PVDF) membranes was performed with a wet-blotting system and blocked for 2 hours in 5% no-fat milk solution. Thereafter, DNA-PKcs (Neomarker, Fremont, CA ) and abl (Oncogene, Cambridge, MA) primary monoclonal antibody and diluted 1:300 overnight in TBS Tween. After washing for 1 hour, the secondary antimouse antibody diluted at 1:5000 was applied to the membrane in TBS Tween solution and then rewashed. Blotting was revealed using an enhanced chemiluminescence (ECL) kit (Amersham, Uppsala, Sweden). Equivalent protein loading was controlled by Red Ponceau and β-actin expression.
Proteasome inhibitors lactacystin and LLnl (Calbiochem, La Jolla, CA) were diluted in DMSO. Lactacystin was used at the final concentration of 2 or 10 μM and LLnL at the final concentration of 50 μM. Cell lines were exposed to proteasome inhibitors 8 hours in RPMI supplemented with 10% fetal calf serum and cytokines, for the cell lines, 4 hours for the primary human cells were cultured.
AG957 (Calbiochem) diluted in DMSO was added to cells to a final concentration of 1 μM and cells were collected after 3 hours of exposure.
Northernblot
Total RNA from parental UT7 cell line and UT7-E8-1, UT7-E8-2, UT-7/9 clones was extracted by the TRIZOL reagent (Life Technologies, Rockville, MD) and the integrity was checked under UV exposure. Total RNA (10 μg) was then separated by electrophoresis through 1% agarose gel containing 7.1% formaldehyde, transferred to nylon membranes (Genescreen, NEN Life Science Products, Boston, MA) and UV cross-linked. To prepare the DNA-PKcs probe, we used a 587-bp DNA fragment amplified from UT7 cell line using DNA-PKcs primers (see below). This fragment was labeled with a 32P dCTP by using an RTS Radprime DNA Labeling System (Life Technologies, Cergy pontoise, France), purified with Nick Columns (Amersham Pharmacia Biotech, Saclay Orsay, France). The membrane was hybridized overnight at 42°C using formamide and washed with SPE 1 ×, 0.1% SDS at 55°C for 45 minutes. The hybridized bands were visualized by autoradiography.
Preparation of the DNA-PKcs probe by reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA from the parental UT7 cell line was reverse transcribed using 200 U Super SCRIPT II RT (Life Technologies, Rockville, MD) and 300 ng random primer p(dn)6(Roche Boehringer-Mannheim, Mannheim, Germany) for 1 hour at 42°C. Five hundred nanograms of cDNA was then amplified by adding 1 U Taq DNA polymerase (ATGC, Orléans, France) and 100 pmol paired primers DNA-PKcs (Genset, Paris, France): (antisense primer, positions 1634-1611): 5′-CCT GCG GGT AGT TAT CAG TGA TTT-3′ (sense primer, positions 1047-1071): 5′-CCA CTT ACA GGA TCA TAG CGA CGA ATG C-3′. The PCR reaction was performed under the following conditions: 35 cycles of 1 minute at 94°C, 1 minute at 55°C, 1 minute at 72°C, and final elongation for 7 minutes at 72°C. This reaction gives rise to an amplified fragment of 587 bp, which was eluted from the gel and quantified before labeling.
Assessment of DNA repair by chromosomal painting (fluorescence in situ hybridization)
Chromosome preparation was performed as previously described.28 Cells were incubated 24 hours after x-irradiation with phytohemagglutinin M (Gibco/BRL, Grand Island, NY), and 1% bromodesoxyuridine BrdU, Colcemid (0.1 μg/mL) was added 2 hours before harvesting; cells were given hypotonic shock (KCl at 0.075 M) for 20 minutes at 37°C and slides with chromosomes in metaphase were prepared according to the standard methanol acetic acid (3:1, v/v) procedure. The slides were stained by Fluorescent Plus Giemsa; whole chromosome 1, 3, and 4 painting was performed with a specifically fluorescent-labeled probe (Gibco/BRL). The slides were analyzed under a fluorescence microscope by visual scoring of translocations, insertions, deletions, and chromosome breaks (200 cells were counted for each condition).
Assessment of apoptosis induced by ionizing radiation
Early apoptotic changes were detected using fluorescein isothiocyanate (FITC)-annexin V, which binds to phosphatidylserine exposed on the outer leaflet of apoptotic cell membranes. Propidium iodide (PI) staining was used for the discrimination between apoptotic and necrotic cells among the annexin V+ cells. Cells (106) were washed and resuspended in 490 μL binding buffer solution (Annexin V-FITC Kit, Immunotech). Annexin V-FITC (5 μL) and 5 μL PI were then added to the cell suspension for 10 minutes followed by FACS analysis.
Results
DNA-PKcs is down-regulated by BCR-ABL
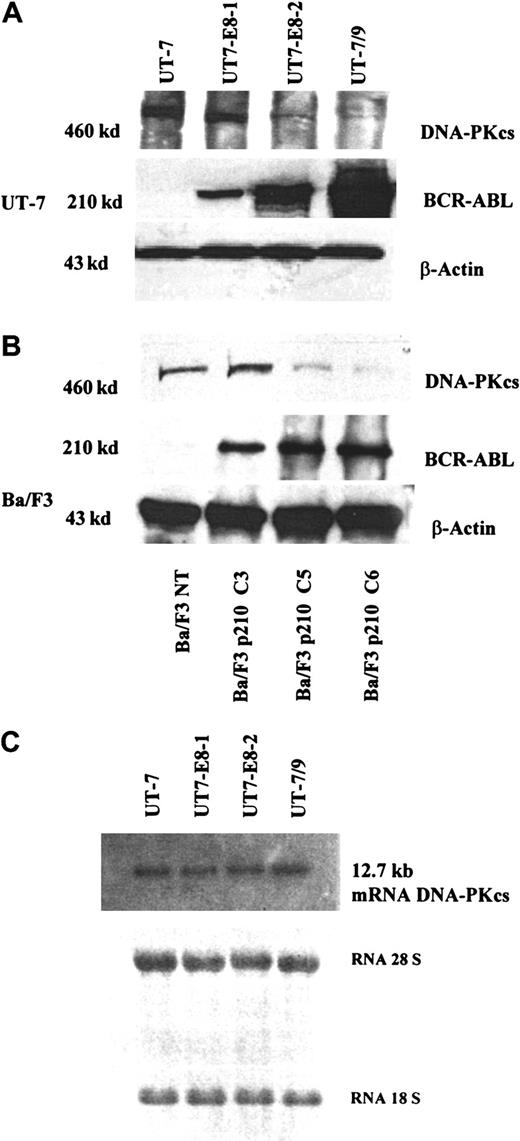
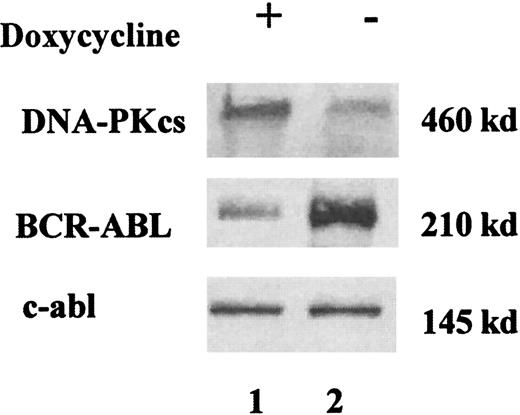
Using Western immunoblotting, a major decrease in DNA-PKcs expression was found in the human UT-7 clones expressing BCR-ABL, as compared to the parental UT-7 cells (Figure1A). The clone with the highest levels of p210 (UT-7/9, Figure 1, lane 4) had a nearly undetectable DNA-PKcs protein expression, whereas clones with low levels of BCR-ABL expressed higher levels of DNA-PKcs (Figure 1A, lanes 2 and 3). In contrast, no decrease in the expression of the other components of the DNA-PK complex (Ku70 and Ku80) was seen in both human and murine clones (data not shown), and no difference in the DNA-PKcs messenger RNA (mRNA) levels was observed in the different UT-7 clones expressing various levels of p210 (Figure 1C). Therefore this result indicates that BCR-ABL–induced down-regulation of DNA-PKcs is posttranscriptional. As shown in Figure 1B, a major decrease in DNA-PKcs expression inversely correlated with BCR-ABL protein levels was also found in 3 individual Ba/F3 murine clones expressing high (C6), intermediate (C5), and low (C3) levels of BCR-ABL, as previously described.12 To demonstrate a direct relationship between BCR-ABL and DNA-PKcs expression, we generated a Ba/F3 cell line in which a tetracycline-inducible BCR-ABL construct was expressed by retrovirus-mediated gene transfer. These cells, selected initially on the basis of their growth factor-independent behavior and high levels of BCR-ABL protein, were grown in the presence of tetracycline and 10% WEHI-CM, as a source of murine IL-3. In these conditions, there was a minimal level of BCR-ABL expression with a BCR-ABL/ABL ratio of 0.6 (Figure 2, lane 1). When the cells were deprived of tetracycline for 48 hours, there was a marked increase of BCR-ABL expression, with a BCR-ABL/ABL ratio of 5 (Figure 2, lane 2). The same protein extracts were then evaluated for the DNA-PKcs expression in the conditions of low or high BCR-ABL expression. As shown in Figure 2, the DNA-PKcs expression was reduced 4-fold when BCR-ABL protein expression was “switched on.” These results strongly suggest the regulation of the DNA-PKcs protein by BCR-ABL expression.
Western blot analysis showing DNA-PKcs and BCR-ABL expression in the human UT-7 clones, expressing variable levels of BCR-ABL.
(A) Western blot analysis showing DNA-PKcs and BCR-ABL expression in the murine Ba/F3 (B) clones, expressing increasing level of BCR-ABL (C3, low; C5, intermediate; C6, high, as previously reported12). This figure, showing a decreased expression of DNA-PK as BCR-ABL increases, is representative of 3 independent experiments. (C) Northern blot analysis showing the levels of DNA-PKcs mRNA in human UT-7 clones.
Western blot analysis showing DNA-PKcs and BCR-ABL expression in the human UT-7 clones, expressing variable levels of BCR-ABL.
(A) Western blot analysis showing DNA-PKcs and BCR-ABL expression in the murine Ba/F3 (B) clones, expressing increasing level of BCR-ABL (C3, low; C5, intermediate; C6, high, as previously reported12). This figure, showing a decreased expression of DNA-PK as BCR-ABL increases, is representative of 3 independent experiments. (C) Northern blot analysis showing the levels of DNA-PKcs mRNA in human UT-7 clones.
A representative Western blot analysis showing DNA-PKcs and BCR-ABL expressions in doxycycline-inducible Ba/F3 cell line.
Cells were grown in IL-3 (10% WEHI), with (+, lane 1) and without (−, lane 2) doxycycline (1 μg/mL) for 48 hours. Cells extracts were immunoblotted with anti-DNA-PKcs and anti-BCR-ABL antibodies. Equal protein loading was assessed by protein dosage, Ponceau staining, and the presence of an equal intensity of the endogenous c-abl. When the cells were deprived of tetracycline for 48 hours, a marked increase of BCR-ABL expression was found, along with a reduction of DNA-PKcs expression.
A representative Western blot analysis showing DNA-PKcs and BCR-ABL expressions in doxycycline-inducible Ba/F3 cell line.
Cells were grown in IL-3 (10% WEHI), with (+, lane 1) and without (−, lane 2) doxycycline (1 μg/mL) for 48 hours. Cells extracts were immunoblotted with anti-DNA-PKcs and anti-BCR-ABL antibodies. Equal protein loading was assessed by protein dosage, Ponceau staining, and the presence of an equal intensity of the endogenous c-abl. When the cells were deprived of tetracycline for 48 hours, a marked increase of BCR-ABL expression was found, along with a reduction of DNA-PKcs expression.
BCR-ABL–induced DNA-PKcs down-regulation is proteasome dependent and requires TK activity
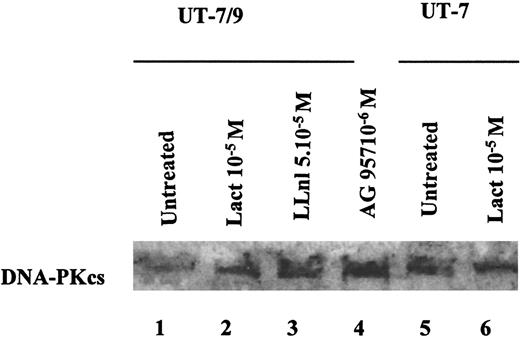
We then studied the possible mechanisms underlying this phenomenon. Preliminary experiments had shown that DNA-PKcs levels in the parental UT-7 were lower when diluted with BCR-ABL UT-7/9 cell extracts, suggesting that BCR-ABL might activate a protease activity (data not shown). We therefore explored the possible implication of the ubiquitin-proteasome pathway in the down-regulation of DNA-PKcs. In the UT-7/9 clone expressing the highest levels of BCR-ABL, incubation with the proteasome inhibitors lactacystin or LLnl was able to restore DNA-PKcs expression (Figure 3, lanes 2 and 3), whereas no effect on DNA-PKcs expression was observed in the parental cells after exposure to these proteasome inhibitors (Figure 3, lanes 5 and 6). These data suggest that BCR-ABL expression could elicit the degradation of DNA-PKcs through the ubiquitin-proteasome pathway. The degradation of DNA-PKcs also required the TK activity of BCR-ABL because the TK inhibitor AG957 was able to restore the DNA-PKcs protein levels (Figure 3, lane 4).
Representative Western blot, showing the effects of BCR-ABL TK inhibition and proteasome inhibition on DNA-PKcs expression in UT-7/9 BCR-ABL expressing cells and parental UT-7 cells.
Cells were grown in rhGM-CSF and exposed to tyrphostin AG957 1 μM for 3 hours (lane 4), proteasome inhibitors lactacystin (10 μM; lanes 2 and 6) and LLnl (50 μM; 8 hours' incubation; lane 3) diluted in DMSO, similar volume of DMSO was added for 8 hours as a control (lanes 1 and 5). The DNA-PKcs protein levels could be increased using both TK and proteasome inhibitors.
Representative Western blot, showing the effects of BCR-ABL TK inhibition and proteasome inhibition on DNA-PKcs expression in UT-7/9 BCR-ABL expressing cells and parental UT-7 cells.
Cells were grown in rhGM-CSF and exposed to tyrphostin AG957 1 μM for 3 hours (lane 4), proteasome inhibitors lactacystin (10 μM; lanes 2 and 6) and LLnl (50 μM; 8 hours' incubation; lane 3) diluted in DMSO, similar volume of DMSO was added for 8 hours as a control (lanes 1 and 5). The DNA-PKcs protein levels could be increased using both TK and proteasome inhibitors.
DNA-PKcs expression in CD34+ cells from patients with CML
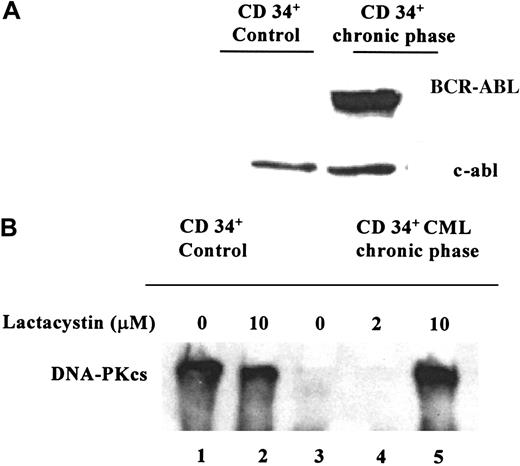
The expression of DNA-PKcs was studied in BCR-ABL+CD34+ cells from 2 patients with CML showing in both samples a very low or undetectable level of DNA-PKcs protein. In contrast, a relatively high DNA-PKcs expression was found in CD34+ cells purified from a leukapheresis sample from a patient with a neuroblastoma (Figure 4, lane 1). As shown in Figure 4, the down-regulation of DNA-PKcs was reversible in CD34+ CML cells, using 10 μM lactacystin for 4 hours. This treatment restored the DNA-PKcs expression in the CML cells, whereas, remarkably, no significant change was documented in the control CD34+ cells cultured with the same concentration of lactacystin (10 μM) indicating the specificity of the proteasome-dependent DNA-PKcs down-regulation in CML cells (Figure 4, lane 5). Interestingly, the low-dose lactacystin (2 μM) was not able to restore DNA-PKcs expression (Figure 4, lane 4).
Representative Western blot.
These blots show (A) BCR-ABL expression and (B) DNA-PKcs expression and the effects of proteasome inhibition in CD34+ cells from a patient with CML and in CD34+ control cells from a patient with a neuroblastoma. Cells were grown in the presence of rhGM-CSF for 4 hours, with 2 μM lactacystin (lane 4) or 10 μM lactacystin (lanes 2 and 5) and without lactacystin (lanes 1 and 3). Using 10 μM lactacystin, it was possible to reverse the down-regulation of DNA-PKcs in the CD34+CML cells.
Representative Western blot.
These blots show (A) BCR-ABL expression and (B) DNA-PKcs expression and the effects of proteasome inhibition in CD34+ cells from a patient with CML and in CD34+ control cells from a patient with a neuroblastoma. Cells were grown in the presence of rhGM-CSF for 4 hours, with 2 μM lactacystin (lane 4) or 10 μM lactacystin (lanes 2 and 5) and without lactacystin (lanes 1 and 3). Using 10 μM lactacystin, it was possible to reverse the down-regulation of DNA-PKcs in the CD34+CML cells.
Induction of a major DNA repair deficiency by BCR-ABL expression
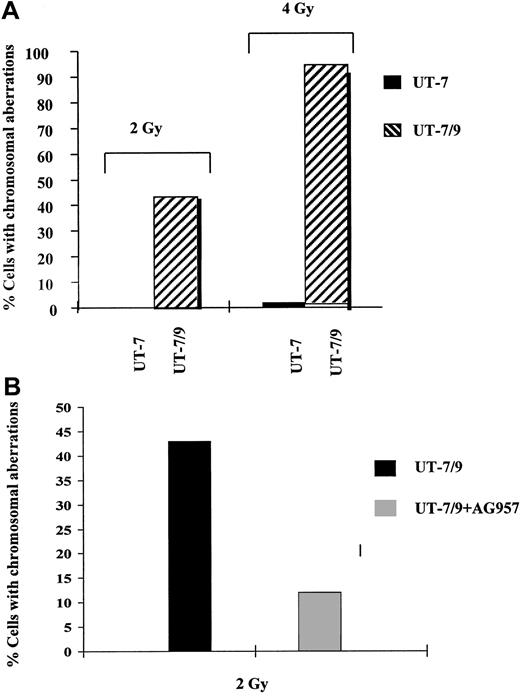
Because DNA-PKcs plays a major role in DNA DSB repair in mammalian cells,12 we then studied DNA repair after exposure to ionizing radiation, a DNA damaging agent able to induce DSB. This DNA repair function was studied by chromosome fluorescence in situ hybridisation (FISH) in UT-7 clones, showing a major DNA repair defect in the clone exhibiting the highest levels of p210 (UT-7/9, Figure5A). Interestingly, this DNA repair deficiency was markedly reduced when UT-7/9 cells were incubated with a TK inhibitor (AG957) (Figure 5B). The defect in DNA repair in the BCR-ABL–expressing cells after radiation exposure was also reflected by the number of micronuclei per cell, which was 3 times higher for the BCR-ABL–expressing UT-7/9 cells, as compared to the UT-7 parental cells (data not shown).
FISH analysis.
(A) Effect of BCR-ABL expression on DNA repair, as evaluated by FISH. Human parental UT-7 and BCR-ABL transfected UT-7/9 cells were irradiated at 2 and 4 Gy and collected 24 hours later. Histogram shows the percentage of cells with chromosomal abnormalities and SD; 200 metaphases were counted for each point. The percentage of cells with chromosomal abnormalities was extremely low in the UT-7 parental cells and dramatically increased in the BCR-ABL–expressing UT-7/9 cells. (B) Effect of BCR-ABL TK inhibition with AG957 on DNA repair as evaluated by FISH. BCR-ABL–expressing UT-7/9 cells were pretreated with AG957 1 μM for 3 hours, irradiated at 2 Gy, and collected 24 hours later. A marked decrease of the percentage of cells with chromosomal abnormalities was observed after exposure to AG957.
FISH analysis.
(A) Effect of BCR-ABL expression on DNA repair, as evaluated by FISH. Human parental UT-7 and BCR-ABL transfected UT-7/9 cells were irradiated at 2 and 4 Gy and collected 24 hours later. Histogram shows the percentage of cells with chromosomal abnormalities and SD; 200 metaphases were counted for each point. The percentage of cells with chromosomal abnormalities was extremely low in the UT-7 parental cells and dramatically increased in the BCR-ABL–expressing UT-7/9 cells. (B) Effect of BCR-ABL TK inhibition with AG957 on DNA repair as evaluated by FISH. BCR-ABL–expressing UT-7/9 cells were pretreated with AG957 1 μM for 3 hours, irradiated at 2 Gy, and collected 24 hours later. A marked decrease of the percentage of cells with chromosomal abnormalities was observed after exposure to AG957.
BCR-ABL expression confers increased sensitivity to ionizing radiation
Given this major DNA repair deficiency, which was positively correlated with BCR-ABL protein levels, we further studied the response of BCR-ABL–expressing cells to ionizing radiation. As shown in Figure6, the UT-7 and Ba/F3 clones exhibiting the highest levels of p210 were the most radiosensitive. In each condition, the clones with the highest BCR-ABL expression had the lowest clonogenic survival. The same results were obtained using the Ba/F3 cells expressing a tetracycline-inducible BCR-ABL construct, with evidence of increased radiosensitivity in the absence of doxycycline, a condition in which BCR-ABL expression was the highest (Tet-OFF) (Figure6B). However, the increased radiosensitivity of BCR-ABL–expressing cells was relatively modest, a finding that prompted us to assess the apoptotic response of BCR-ABL cells, when exposed to ionizing radiation. As shown in Figure 7, both the UT-7 parental and the low-level BCR-ABL–expressing UT-7/E8-1 clones exhibited high sensitivity to radiation-induced apoptosis, whereas the 2 clones with the highest levels of BCR-ABL (UT-7/8-2 and UT-7-9) were refractory to apoptosis. Thus, the impact on radioinduced cell death due to the major DNA repair deficiency in BCR-ABL–expressing cells was probably counterbalanced by a major resistance to radio-induced apoptosis. These 2 opposite effects ultimately resulted in a modest increase in radiosensitivity (Figure 6).
Clonogenic survival curves.
(A) Human UT-7 cells: UT-7, open squares; UT-7/E8-1, open circles; UT-7/E8-2, closed triangles; UT-7/9, closed squares. Both UT-7/E8-2 and UT-7/9 expressing high levels of BCR-ABL exhibited lower clonogenic survival (ie, increased radiosensitivity) than low BCR-ABL expressing UT-7/E8-1 and the parental UT-7 cells after 2, 4 and 6 Gy. (B) Murine BaF3 cells: Ba/F3 NT, open squares; Ba/F3 p210C3, closed triangles; Ba/F3 p210 inducible with doxycycline, open circle; Ba/F3 p210 inducible without doxycycline, closed square. The tet-inducible BCR-ABL SIN-p210 cells were grown in the presence of IL-3 (10% WEHI) with or without doxycycline (1 μg/mL). Cells were irradiated in exponential phase and plated in methylcellulose as described above, in the presence of 10% WEHI. The clonogenic survival of Ba/F3 cells decreased in the presence of BCR-ABL expression, both in stable and inducible BCR-ABL transfectants.
Clonogenic survival curves.
(A) Human UT-7 cells: UT-7, open squares; UT-7/E8-1, open circles; UT-7/E8-2, closed triangles; UT-7/9, closed squares. Both UT-7/E8-2 and UT-7/9 expressing high levels of BCR-ABL exhibited lower clonogenic survival (ie, increased radiosensitivity) than low BCR-ABL expressing UT-7/E8-1 and the parental UT-7 cells after 2, 4 and 6 Gy. (B) Murine BaF3 cells: Ba/F3 NT, open squares; Ba/F3 p210C3, closed triangles; Ba/F3 p210 inducible with doxycycline, open circle; Ba/F3 p210 inducible without doxycycline, closed square. The tet-inducible BCR-ABL SIN-p210 cells were grown in the presence of IL-3 (10% WEHI) with or without doxycycline (1 μg/mL). Cells were irradiated in exponential phase and plated in methylcellulose as described above, in the presence of 10% WEHI. The clonogenic survival of Ba/F3 cells decreased in the presence of BCR-ABL expression, both in stable and inducible BCR-ABL transfectants.
Effect of BCR-ABL expression on γ irradiation-induced apoptosis.
UT-7 and BCR-ABL transfected cells were irradiated at 0, 2, 5, and 10 Gy and collected 24 hours later. Histograms show the average and SD for at least 3 independent experiments evaluating the apoptotic fractions quantified by flow cytometric analysis of PI iodide and annexin V-FITC–stained cells. The clones with the highest levels of BCR-ABL (UT-7-8/1 and UT-7/9) were refractory to radioinduced apoptosis.
Effect of BCR-ABL expression on γ irradiation-induced apoptosis.
UT-7 and BCR-ABL transfected cells were irradiated at 0, 2, 5, and 10 Gy and collected 24 hours later. Histograms show the average and SD for at least 3 independent experiments evaluating the apoptotic fractions quantified by flow cytometric analysis of PI iodide and annexin V-FITC–stained cells. The clones with the highest levels of BCR-ABL (UT-7-8/1 and UT-7/9) were refractory to radioinduced apoptosis.
Discussion
Little is known about the possible implication of BCR-ABL in the DNA repair processes. A deficiency of the DNA repair mechanisms in CML cells has been suggested with evidence of a colocalization of BCR-ABL and the excision repair gene XPB.13 However, the demonstration of a direct regulatory role of BCR-ABL in the DNA repair process is lacking. Moreover, the XPB-related DNA repair is essentially involved in UV-induced thymidine dimer excision process and, thus, is unlikely to account for all the genetic and chromosome abnormalities accompanying the blast crisis. We therefore hypothesized that some other DNA repair genes might be involved in this process, such as the DNA-PK complex, which is one of the major systems involved in DNA repair in mammalian cells.
To address this question we used human and murine hematopoietic cell lines stably transfected with BCR-ABL and expressing variable levels of the p210 fusion protein. Convergent results were obtained both in murine and human cells showing a marked decrease in DNA-PKcs expression with increasing BCR-ABL protein levels. A direct relationship between these 2 events was demonstrated using an inducible promoter-controlled BCR-ABL expression. These findings constitute the first demonstration of a direct involvement of BCR-ABL in the regulation of one of the major DNA repair proteins. Importantly, a decreased expression of DNA-PKcs was also found in CD34+ cells from patients with CML at diagnosis, suggesting that this phenomenon constitutes an important feature of this disease.
As previously reported in various other cellular models,14the decreased expression of DNA-PKcs was associated with a major DNA repair deficiency after exposure to DNA-damaging agents (eg, ionizing radiation). It is thus possible that the down-regulation of DNA-PKcs by BCR-ABL could play a role in determining one of the main characteristics of CML, which is the chromosome and genetic instability2 leading to the blast crisis.15
One of the possible mechanisms involved in the down-regulation of DNA-PKcs could be a degradation through the ubiquitin-proteasome pathway. Indeed, exposure to proteasome inhibitors (lactacystin, LLnl) prevented DNA-PKcs degradation in BCR-ABL–expressing cells, and had no effect in the parental cells (Figure 3). Remarkably, Western blot analysis of the DNA PKcs expression in CD34+ CML cells revealed a dramatic increase of the DNA-PKcs levels after 4 hours of lactacystin exposure (Figure 4, lane 5), whereas this same treatment did not induce any significant change of DNA-PKcs levels in the control CD34+ cells (Figure 4, lane 2). These results strongly suggest a specific interaction between the proteasome pathway and BCR-ABL expression. These data are in agreement with previous experiments that showed that herpes simplex virus (HSV-1) could induce a proteasome-dependent degradation of DNA-PKcs.16 How BCR-ABL interacts with the proteasome pathway to regulate DNA-PKcs degradation remains however, to be determined.
We and others have previously reported that BCR-ABL–expressing cells exhibited resistance to drug-induced apoptosis.17,18DNA-PKcs has recently been shown to be involved in the regulation of apoptosis.19 Whether BCR-ABL–induced down-regulation of DNA-PKcs interferes with the antiapoptotic potential of BCR-ABL remains to be studied.
In our study, the decrease of DNA-PKcs induced by BCR-ABL was found to enhance the sensitivity to ionizing radiation, which is commonly used as a conditioning regimen for allogeneic bone marrow transplantation in CML. The low DNA-PKcs expression in CML cells should therefore render them more sensitive to ionizing radiation than the normal cells. Indeed, low-dose splenic irradiation has been extremely efficient to reduce the tumor mass in patients with accelerated or acute phase of CML.20 These findings are in agreement with previous works having shown hypersensitivity to ionizing radiation in DNA-PKcs–deficient cells.21 Radiation sensitivity of Bcr-Abl expressing cells is controversial; an increased radiation sensitivity in Bcr-Abl expressing cells has been previously reported in 32d BCR-ABL mouse cells,22 whereas the radiation resistance was also described in patient Bcr-Abl–expressing cells.17 It is remarkable that ionizing radition could be the only cytotoxic agent found to be more efficient as the level of BCR-ABL increases, which may have some implications to better integrate total body irradiation in the management of advanced cases of CML. In the experimental studies reported here, it should be pointed out, however, that the magnitude of the radiosensitization was relatively modest (Figure 6) and likely less than expected, given the major decrease of the DNA-PKcs protein induced by BCR-ABL. A possible explanation for this discrepancy was that the impact on radio-induced cell death due to the DNA repair deficiency could be counterbalanced by the major resistance to radio-induced apoptosis, as BCR-ABL p210 increased (Figure 7). This resistance to apoptosis as BCR-ABL expression increases has already been reported for some cytotoxic drugs.17 18 These 2 opposite effects ultimately resulted in a moderate increase in radiosensitivity (Figure 6). The results reported in this work might also have other important implications. Indeed, the fact that the CML cells are both deficient in DNA repair and refractory to apoptosis may explain that chromosomal and genetic abnormalities can accumulate in leukemic cells and why this myeloproliferative disorder constantly progresses to a blast crisis. Finally, we showed that DNA-PKcs expression could be increased in CML cells by proteasome inhibitors, suggesting that this approach might represent a novel and powerful strategy to decrease the accumulation of genetic alterations in these cells and hence to delay the occurrence of the blast crisis.
We thank H. M. Blau (Stanford University) for the gift of the retro-TET-Sin vector.
Supported by individual grants from the ARC (association de recherche contre le cancer), Ligue nationale contre le cancer et Ligue contre le cancer, section des Hauts de Seine (to J.B.) and from the ARC, comité de recherche clinique (CRC) of the Institut Gustave Roussy (to A.G.T.) and a graduate student grant (to A.D.) from the Ministère de la Recherche. E.D. and A.D. contributed equally to this work as first authors; A.G.T. and J.B. contributed equally as last authors.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jean Bourhis, UPRES EA 27-10, Institut Gustave Roussy, 39 rue Camille Desmoulins, 94805 Villejuif, France; e-mail:bourhis@igr.fr.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal