Abstract
The mi transcription factor (MITF) is a basic helix-loop-helix leucine zipper (bHLH-Zip) transcription factor that is important for the development of mast cells. Mast cells ofmi/mi genotype express normal amount of abnormal MITF (mi-MITF), whereas mast cells of tg/tg genotype do not express any MITFs. Mast cells of mi/mi mice show more severe abnormalities than those of tg/tg mice, indicating that the mi-MITF possesses the inhibitory function. The MITF encoded by the mice mutant allele (ce-MITF) lacks the Zip domain. We examined the importance of the Zip domain usingmice/micemice. The amounts of c-kit, granzyme B (Gr B), and tryptophan hydroxylase (TPH) messenger RNAs decreased in mast cells ofmice/mice mice to levels comparable to those of tg/tg mice, and the amounts were intermediate between those of +/+ mice and those of mi/mi mice. Gr B mediates the cytotoxic activity of mast cells, and TPH is a rate-limiting enzyme for the synthesis of serotonin. The cytotoxic activity and serotonin content ofmice/mice mast cells were comparable to those of tg/tg mast cells and were significantly higher than those of mi/mi mast cells. The phenotype of mice/mice mast cells was similar to that of tg/tg mast cells rather than to that of mi/mi mast cells, suggesting that thece-MITF had no functions. The Zip domain of MITF appeared to be important for the development of mast cells.
Introduction
The mi locus of mice encodes a member of the basic helix-loop-helix leucine zipper (bHLH-Zip) protein family of transcription factors (hereafter called MITF, for mitranscription factor).1,2 Spontaneous chemical, radiation, and insertional mutageneses provide abundant mutant alleles at themi locus,3,4 which are useful for the analysis of the relationship between the structure and function of MITF.5-8 The mutant allele that has been studied most intensively is mi. The mi/mi mice show depletion of pigment in both hair and eyes, microphthalmia, osteopetrosis, and deficient natural killer activity.3,4 In addition, the number of mast cells decreases and their phenotype is abnormal inmi/mi mice.9-15 Although most mast cells in the skin of normal (+/+) mice are stained with berberine sulfate that binds heparin proteoglycan, few mast cells are berberine sulfate+in the skin of mi/mi mice.13,16-19 Cultured mast cells (CMCs) derived from the spleen of mi/mi mice are deficient in the expression of various genes, such as the mouse mast cell protease (MMCP)-4,20 MMCP-5,21MMCP-6,22 c-kit,23 p75 nerve growth factor receptor,24 granzyme B (Gr B),25 tryptophan hydroxylase (TPH),25integrin α4 subunit,26 and α-melanocyte–stimulating hormone receptor genes.27 MITF encoded by themi mutant allele (mi-MITF) deletes 1 of 4 consecutive arginines in the basic domain.1,6,7 Themi-MITF is defective in the DNA binding ability and the nuclear localization potential.28,29 Themi-MITF does not appear to transactivate target genes due to these abnormalities.20-27 29
The tg is another mutant allele of the milocus.1,30 The tg/tg mice possess the insertional mutation at the promoter region of mi gene and do not express any MITFs.1,31 The tg/tg andmi/mi mice share several phenotypic features, but the phenotypic abnormality of tg/tg mice is apparently mild compared with that of mi/mi mice. The transcription of c-kit, Gr B, and TPH genes was significantly reduced inmi/mi CMCs, but the reduction was moderate intg/tg CMCs.32 This indicated that the presence of mi-MITF caused more severe abnormalities than the absence of normal (+) MITF. In addition to the loss of transactivation ability, the mi-MITF possesses an inhibitory effect on the transcription of some particular genes in mast cells.32Because the tg is considered to be a null mutant allele, thetg/tg mice may be useful for evaluating the function of other mutant MITFs. When a homozygous mouse at a certain mutantmi allele shows more severe phenotype than that of thetg/tg mouse, the MITF encoded by the mutant miallele may possess an inhibitory function. When a homozygous mouse at another mutant mi allele shows a phenotype comparable to that of tg/tg mouse, the MITF encoded by the mutantmi allele may not possess any functions.
MITF encoded by the mice mutant allele (ce-MITF) lacks the Zip domain because of a stop codon between HLH and Zip.6 To our knowledge, themice is the only available mutant of genes encoding bHLH-Zip proteins lacking the Zip domain. In the present study, we compared the phenotype of mast cells ofmice/mice mice with that ofmi/mi or tg/tg mice to clarify the importance of the Zip domain of MITF for development of mast cells. The phenotype of mice/mice mast cells was similar to that of tg/tg mast cells rather than to that of mi/mi mast cells, indicating that the ce-MITF had no functions.
Materials and methods
Mice
The original stock of C57BL/6-mi/+ mice was purchased from the Jackson Laboratory (Bar Harbor, ME) and was maintained in our laboratory by consecutive backcross with our own inbred C57BL/6 colony (more than 15 generations at the time of the present experiments). The original stock of VGA-9-tg/tg mice, in which the mouse vasopressin–Escherichia coli β-galactosidase transgene was integrated at the 5′ flanking region of the mi gene, were kindly given by Dr H. Arnheiter (National Institutes of Health, Bethesda, MD).1 The integrated transgene was maintained by repeated backcrosses to our own inbred C57BL/6 colony (more than 10 generations at the time of the present experiment). The C57BL/6-mice/mivit mice were maintained in the laboratory of Dr L. Lamoreux (Texas A&M University, College Station, TX).33 The C57BL/6-mice/mivit mice were crossed to our own inbred C57BL/6 colony, and the mice/+mice were selected by sequencing of the MITF gene. Female and male heterozygous mi/+, tg/+, ormice/+ mice were crossed together, and the resulting homozygous mi/mi, tg/tg, ormice/mice mice were selected by their white coat color.3 4 C57BL/6-+/+ mice raised in our laboratory were used as a control.
Cells
Pokeweed mitogen–stimulated spleen cell–conditioned medium (PWM-SCM) was prepared according to the method described by Nakahata et al.34 Mice ofmice/mice, mi/mi, tg/tg, and control +/+ were used at 2 to 3 weeks of age to obtain CMCs. Mice were killed by decapitation after ether anesthesia, and spleens were removed. Spleen cells were cultured in α-minimal essential medium (α-MEM; ICN Biomedicals, Costa Mesa, CA) supplemented with 10% PWM-SCM and 10% fetal calf serum (FCS; Nippon Bio-supp Center, Tokyo, Japan). Half of the medium was replaced every 5 days. Cells derived from the spleen of each mutant genotype reached 1 × 107 in number within 4 weeks. More than 95% of cells contained alcian blue+ granules and were considered to be CMCs 4 weeks after initiation of the culture. The NIH3T3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Flow Laboratories, Irvine, UK) supplemented with 10% FCS. The P815 cells were maintained in α-MEM supplemented with 10% FCS.
Staining and counting of mast cells
Mice 20 days of age were killed by decapitation after ether anesthesia. Pieces of dorsal skin were removed, smoothed onto a piece of the filter paper to keep them flat, fixed in Carnoy's solution, and embedded in paraffin. Sections of skin pieces were stained with alcian blue or with berberine sulfate. The staining method with alcian blue or berberine sulfate has been described previously.11 16-18Mast cells between epithelium and panniculus carnosus were counted under the microscope, and the number was expressed as mast cells per centimeter of skin. Berberine sulfate+ mast cells were counted under the fluorescent microscope, and the proportion of berberine sulfate+ cells to alcian blue+ cells was calculated.
In situ hybridization
Skin pieces were removed from the back of 20-day-old mice, fixed in 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4), and embedded in paraffin. The technique of in situ hybridization has been described in detail.35 To obtain the MMCP-4,36 MMCP-5,37 MMCP-6,38and mast cell carboxypeptidase A (MC-CPA)39 probes, single-stranded complementary DNA (cDNA) was generated from total RNA extracted from CMCs of +/+ mouse origin by lithium chloride-urea method.40 The specific cDNA of proteases was then amplified with specific primers for each protease by polymerase chain reaction (PCR).15 The cDNAs were subcloned into theEcoRV site of pBluescript KS− plasmid (pBS; Stratagene, La Jolla, CA) that contains T3 and T7 promoters to generate probes.
After hybridization with the antisense probe, the cells possessing signals that were stronger than those obtained with the sense probe were considered to be messenger RNA+ (mRNA+). We counted the number of mRNA+ cells per centimeter of skin. In the adjacent section, the number of alcian blue+cells per centimeter of skin was counted. Then, the proportion of various protease mRNA+ cells to alcian blue+cells was calculated.
Northern blot analysis
Each RNA sample was prepared from 1 × 107 CMCs by the lithium chloride-urea method.40 Northern blot analysis was performed using c-kit,41MMCP-4,36 MMCP-5,37 MMCP-6,38MC-CPA,39 Gr B,25 TPH,25 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH)42 cDNAs labeled with α-[32P]deoxycytidine triphosphate (370 MBq/mL; NEN Life Science Products, Boston, MA) by random oligonucleotide priming. After hybridization at 42°C, blots were washed to a final stringency of 0.2 × SSC (1 × SSC is 150 mM NaCl and 15 mM trisodium citrate, pH 7.4) and subjected to autoradiography.
Cytotoxicity assay
Mast cell cytotoxicity was measured using a 51Cr release assay according to the procedure described by Bissonnete and Befus.43 As a positive control, spleen cells were freshly prepared from 8-week-old +/+ mice. The target was YAC-1 cells, which were obtained from American Type Culture Collection (Bethesda, MD). The cells were maintained in α-MEM supplemented with 10% FCS. CMCs derived from +/+, mi/mi, mice/mice, and tg/tg mice and spleen cells of +/+ mice were washed, suspended in α-MEM supplemented with 10% FCS, and distributed at different concentrations (0.5, 1.0, and 2.5 × 106 cells) in triplicate into 96-well microtiter plates with round bottoms. YAC-1 cells (5 × 106) were labeled with 3.7 MBq μCi [51Cr]Na2CrO4 (Amersham-Pharmacia Biotech, Amersham Place, UK) for 2 hours, washed 3 times, and resuspended in α-MEM supplemented with 10% FCS. Labeled YAC-1 cells (1.0 × 104) were mixed with various numbers of CMCs in a total volume of 200 μL/well. Plates were incubated at 37°C for 18 hours in a CO2 incubator and spun at 150g for 10 minutes, and the radioactivity was determined in 100 μL samples of cell-free supernatants. The radioactivity released in the well containing YAC-1 cells alone was designated spontaneous release (SR). Total 51Cr release (TR) was measured by adding 0.01% Triton X-100 to the well containing YAC-1 cells alone. The percentage of specific 51Cr release was calculated using the following formula: (cpm in the presence of CMCs − SR)/(TR − SR) × 100.
Concentration of serotonin
The concentration of serotonin was measured using high performance liquid chromatography (HPLC) with electrochemical detection.44 Briefly, CMCs were collected, washed with phosphate-buffered saline (PBS), counted, and sonicated for 20 seconds in a sonicator (Tomy, Tokyo, Japan) in 1 mL ice-cold 3% perchloric acid containing 5 mM ethylenediaminetetraacetic acid (EDTA) and 1 mM sodium metabisulfate. The homogenate was centrifuged at 10 000g for 15 minutes at 4°C, and the supernatant was applied directly to the HPLC column. The concentration of serotonin per 1.0 × 106 cells was calculated.
Electrophoretic gel mobility shift assay (EGMSA)
The production of the fusion protein containing glutathione-S-transferase (GST) and MITF was described previously.28 To examine the DNA binding ability of MITF, an oligonucleotide that is a part of MMCP-6 promoter was used as a probe.28 The sequence of the oligonucleotide is 5′-TGGTGGGGACACATGTTACATGGA (the sequence recognized by +-MITF is underlined). The oligonucleotide was labeled with α-[32P]deoxycytidine triphosphate by filling 5′ overhangs and used as probes of EGMSA. DNA-binding assays were performed in a 20 μL reaction mixture containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 75 mM KCl, 1 mM dithiothreitol (DTT), 4% Ficoll 400, 50 ng poly(dI-dC), 25 ng labeled DNA probe, and 3.5 μg GST-MITF fusion protein. After the incubation at room temperature for 15 minutes, the reaction mixture was subjected to electrophoresis at 14 V/cm at 4°C on a 5% polyacrylamide gel in 0.25 × Tris-borate-EDTA (TBE) buffer (1 × TBE is 90 mM Tris-HCl, 64.6 mM boric acid, and 2.5 mM EDTA, pH 8.3). The polyacrylamide gels were dried on Whatman 3MM chromatography paper and subjected to autoradiography.
Construction of expression plasmids and immunocytochemistry
The pBS containing the whole coding region of +-MITF,ce-MITF, or mi-MITF was constructed in our laboratory (hereafter called pBS-+-MITF, pBS-ce-MITF, and pBS-mi-MITF, respectively). To generate the Myc-tagged MITF construct, we subcloned the SmaI-HincII fragment of pBS-+-MITF, pBS-ce-MITF, or pBS-mi-MITF into the StuI site of the CS2+MT expression vector that provides 6 copies of the Myc epitope tag at the N-terminal end of the protein (a gift from Dr I. Matsumura, Osaka University, Osaka, Japan).45 The resultant chimeric gene was subcloned into pEF-BOS expression vector kindly provided by Dr S. Nagata (Osaka University, Osaka, Japan).46 The expression plasmid was transfected into NIH3T3 cells, and the overexpressed MITF protein was detected by anti-Myc antibody as described previously.29Briefly, the cells were fixed with 100% methanol, permealized by treatment with 0.2% Triton X-100 in PBS, and incubated with the mouse monoclonal anti-Myc antibody (9E10; Pharmingen, San Diego, CA). Immunoreacted cells were detected by direct immunofluorescence with goat antimouse immunoglobulin G antibody conjugated with fluorescein isothiocyanate (MBL, Nagoya, Japan).
ce-MITF cDNA containing the nuclear localization
The nuclear localization signal (NLS) of SV40 large-T antigen (PKKKRKV)47 was inserted into the N-terminus ofce-MITF by PCR. The amplified product was verified by sequencing and cloned into CS2+MT expression vector. Then, the resultant chimeric gene was subcloned into pEF-BOS expression vector. The subcellular localization of the ce-MITF with NLS of SV40 large-T antigen was examined by immunocytochemistry as described above.
Immunoblotting for the nuclear and cytoplasmic extracts
Nuclear and cytoplasmic extracts of NIH3T3 cells transfected with expression plasmid of Myc-tagged +-MITF, ce-MITF, ormi-MITF were prepared as described before.29Briefly, transfected cells were washed with PBS twice and resuspended in ice-cold buffer containing 10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, and 1 mM phenylmethylsufonyl fluoride (PMSF). Then, Nonidet P-40 was added to a final concentration of 0.1%. After vigorous vortexing, the homogenate was centrifuged at 3000 rpm for 3 minutes. The supernatant was used as the cytoplasmic fraction. The pellet was resuspended in ice-cold buffer containing 400 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 50 mM Tris-HCl (pH 7.4), and 1 mM PMSF, kept on ice for 1 hour, and centrifuged at 15 000 rpm. The supernatant was used as the nuclear fraction. Samples were separated by SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane (Immobilon P, Millipore, Bedford, MA). The blots were incubated with 5% skim milk in Tris-buffered saline (20 mM Tris-HCl [pH 7.4], 150 mM NaCl). Then, the blots were incubated with Tris-buffered saline containing 5% skim milk with the anti-Myc antibody. The membrane was incubated with peroxidase-conjugated goat antimouse immunoglobulin G antibody, and the immune complexes were visualized with Western blot chemiluminescence reagent (NEN Life Science Products).
Transient cotransfection assay
The reporter plasmid that contained Gr B promoter starting from nt −910 (+1 shows a transcription initiation site) was previously reported.25,32 As the expression plasmids, theSmaI-HincII fragment of pBS-ce-MITF, or pBS-mi-MITF, was introduced into the bluntedXbaI site of pEF-BOS, and the PCR-amplified fragment ofce-MITF with the NLS of SV40 large-T antigen was also cloned into the blunted XbaI site of pEF-BOS. A total of 10 μg of a reporter, 2 μg of an expression plasmid, and 3 μg of an expression vector containing β-galactosidase gene were cotransfected into P815 cells by electroporation. The expression vector containing β-galactosidase gene was used as an internal control. The cells were harvested 48 hours after the transfection and lysed with 0.1 M potassium phosphate buffer (pH 7.4) containing 1% Triton X-100. Soluble extracts were then assayed for luciferase activity with a luminometer LB96P (Berthold, Wildbad, Germany) and for β-galactosidase activity. The luciferase activity was normalized by the β-galactosidase activity and total protein concentration according to the method described by Yasumoto et al.48 The normalized value was expressed as the relative luciferase activity.
Results
The number of mast cells was examined in the skin ofmice/mice mice. Histologic sections of the skin pieces of mice/mice mice were stained with alcian blue. The number of mast cells in themice/mice mice decreased to one third that of the control +/+ mice and was comparable to that ofmi/mi and tg/tg mice (Table1). In +/+ mice, most of skin mast cells were berberine sulfate+, indicating that they contained heparin.16 The proportion of berberine sulfate+ mast cells in the skin ofmice/mice mice was comparable to that of +/+ mice and was also comparable to that of tg/tgmice (Table 1). In contrast, the proportion of berberine sulfate+ mast cells was 3% in the skin of mi/mimice, as reported previously.8 11
Number of mast cells and proportion of berberine sulfate+ cells to alcian blue+ cells in the skin of +/+, mi/mi,mice/mice, andtg/tg 20-day-old mice
| Genotype of mice . | No. alcian blue+ cells per cm of skin* . | Proportion of berberine sulfate+ to alcian blue+ cells, %* . |
|---|---|---|
| +/+ | 426 ± 29 | 95 ± 5 |
| mice/mice | 142 ± 26† | 83 ± 5 |
| mi/mi | 125 ± 20† | 3 ± 1† |
| tg/tg | 168 ± 31† | 84 ± 6 |
| Genotype of mice . | No. alcian blue+ cells per cm of skin* . | Proportion of berberine sulfate+ to alcian blue+ cells, %* . |
|---|---|---|
| +/+ | 426 ± 29 | 95 ± 5 |
| mice/mice | 142 ± 26† | 83 ± 5 |
| mi/mi | 125 ± 20† | 3 ± 1† |
| tg/tg | 168 ± 31† | 84 ± 6 |
Mean ± SE of 5 mice.
P < .01 compared with the value of +/+ mice byt test.
The expression of mast cell–specific protease genes was analyzed by in situ hybridization in mast cells of the skin. The proportion of MMCP-4 mRNA+ and MMCP-6 mRNA+ mast cells decreased remarkably in the skin of mice/micemice, but that of MMCP-5 mRNA+ and MC-CPA mRNA+mast cells did not (Table 2). No significant differences were detectable in the expression of mast cell proteases among skin mast cells ofmice/mice, tg/tg, andmi/mi mice.
Proportion of MMCP-4, MMCP-5, MMCP-6, and MC-CPA mRNA+ cells to alcian blue+ cells in the skin of +/+, mi/mi,mice/mice, andtg/tg 20-day-old mice
| Genotype of mice . | Proportion of cells expressing mRNA of each protease to alcian blue+ cells, %* . | |||
|---|---|---|---|---|
| MMCP-4 . | MMCP-5 . | MMCP-6 . | MC-CPA . | |
| +/+ | 55 ± 4 | 55 ± 4 | 45 ± 2 | 92 ± 4 |
| mice/mice | 5 ± 1† | 51 ± 5 | 3 ± 2† | 89 ± 3 |
| mi/mi | 6 ± 2† | 57 ± 6 | 3 ± 1† | 90 ± 1 |
| tg/tg | 5 ± 2† | 52 ± 4 | 4 ± 2† | 87 ± 4 |
| Genotype of mice . | Proportion of cells expressing mRNA of each protease to alcian blue+ cells, %* . | |||
|---|---|---|---|---|
| MMCP-4 . | MMCP-5 . | MMCP-6 . | MC-CPA . | |
| +/+ | 55 ± 4 | 55 ± 4 | 45 ± 2 | 92 ± 4 |
| mice/mice | 5 ± 1† | 51 ± 5 | 3 ± 2† | 89 ± 3 |
| mi/mi | 6 ± 2† | 57 ± 6 | 3 ± 1† | 90 ± 1 |
| tg/tg | 5 ± 2† | 52 ± 4 | 4 ± 2† | 87 ± 4 |
Mean ± SE of 5 mice.
P < .01 compared with the value of +/+ mice byt test.
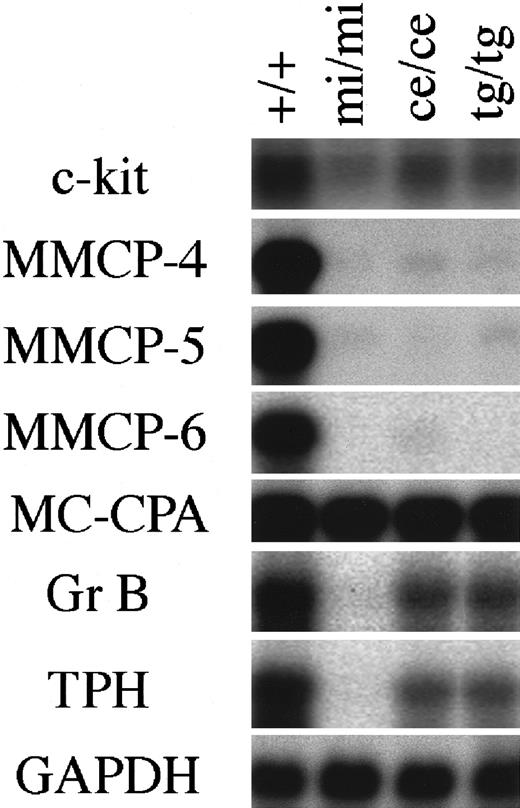
The expression of genes that had been demonstrated to be affected by MITF was examined in CMCs derived from the spleen ofmice/mice, tg/tg, andmi/mi mice using Northern blot. The amount of c-kit mRNA reduced in mi/mi CMCs, as reported previously.23 The amount of c-kit mRNA ofmice/mice CMCs was comparable to that of tg/tg CMCs and was intermediate between the amount of +/+ CMCs and that of mi/mi CMCs (Figure1).
Expression of various genes in CMCs derived from +/+,mi/mi, mice/mice, andtg/tg mice.
The blot was hybridized with 32P-labeled cDNA probe of c-kit, MMCP-4, MMCP-5, MMCP-6, MC-CPA, Gr B, TPH, or GAPDH. Three independent experiments were done, and comparable results were obtained. A representative experiment is shown.
Expression of various genes in CMCs derived from +/+,mi/mi, mice/mice, andtg/tg mice.
The blot was hybridized with 32P-labeled cDNA probe of c-kit, MMCP-4, MMCP-5, MMCP-6, MC-CPA, Gr B, TPH, or GAPDH. Three independent experiments were done, and comparable results were obtained. A representative experiment is shown.
The amounts of MMCP-4, MMCP-5, and MMCP-6 mRNAs inmice/mice CMCs reduced to the levels comparable to those of mi/mi and tg/tg CMCs (Figure 1). In contrast to the reduced expression of MMCP-5 mRNA inmice/mice CMCs, the proportion of MMCP-5 mRNA+ mast cells was not reduced in the skin ofmice/mice mice (Table 2). We previously reported that the addition of stem cell factor (SCF) significantly increased the amount of MMCP-5 mRNA in mi/miCMCs and speculated that SCF synthesized by skin fibroblasts may induce the MMCP-5 expression in mi/mi skin mast cells.21 The different pattern of MMCP-5 expression between CMCs and skin mast cells ofmice/mice, tg/tg, andmi/mi mice may be attributable to the different concentration of SCF surrounding mast cells.
The amount of Gr B mRNA reduced inmice/mice CMCs to a level comparable to that of tg/tg CMCs, but their magnitude of reduction was significantly smaller than that of mi/mi CMCs (Figure 1). We compared the cytotoxic activity of various CMCs, because Gr B mediates the cytotoxic activity of mast cells against YAC-1 cells.32 CMCs of +/+, mi/mi, mice/mice, or tg/tggenotype were cultured together with 51Cr-labeled YAC-1 cells, and the 51Cr release from YAC-1 cells was measured after 18 hours. At an effector:target (E:T) ratio of 50, neither +/+,mi/mi, mice/mice, nortg/tg CMCs showed any cytotoxic activity (Table3). At an increased E:T ratio of 100 or 250, a remarkable cytotoxic activity ofmice/mice CMCs was detected as in the case of +/+ or tg/tg CMCs, but no cytotoxic activity was observed in mi/mi CMCs (Table 3).
Cytotoxic activity of +/+, mi/mi,mice/mice, andtg/tg CMCs to YAC-1 cells
| E:T ratio . | Specific 51Cr release, %3-150 . | ||||
|---|---|---|---|---|---|
| +/+ CMCs . | Mi/MiCMCs . | mice/miceCMCs . | tg/tg CMCs . | +/+ spleen cells . | |
| 50 | < 0.1 | < 0.1 | < 0.1 | < 0.1 | 52.2 ± 3.83-151 |
| 100 | 12.8 ± 0.23-151 | < 0.1 | 10.1 ± 0.33-151 | 11.9 ± 0.63-151 | 58.3 ± 5.23-151 |
| 250 | 25.2 ± 2.43-151 | < 0.1 | 16.6 ± 2.33-151 | 19.1 ± 1.63-151 | NE |
| E:T ratio . | Specific 51Cr release, %3-150 . | ||||
|---|---|---|---|---|---|
| +/+ CMCs . | Mi/MiCMCs . | mice/miceCMCs . | tg/tg CMCs . | +/+ spleen cells . | |
| 50 | < 0.1 | < 0.1 | < 0.1 | < 0.1 | 52.2 ± 3.83-151 |
| 100 | 12.8 ± 0.23-151 | < 0.1 | 10.1 ± 0.33-151 | 11.9 ± 0.63-151 | 58.3 ± 5.23-151 |
| 250 | 25.2 ± 2.43-151 | < 0.1 | 16.6 ± 2.33-151 | 19.1 ± 1.63-151 | NE |
E:T ratio is the ratio of CMCs or spleen cells to YAC-1 cells.
NE indicates not examined.
Mean ± SE of 3 experiments.
P < .01 by t test compared with the value of mi/mi CMCs.
The amount of TPH mRNA also reduced inmice/mice CMCs to a level comparable to that of tg/tg CMCs, but the magnitude of reduction was significantly smaller than that of mi/mi CMCs (Figure 1). The serotonin content of various CMCs was compared, because TPH is the rate-limiting enzyme of the serotonin synthesis.49 The serotonin content of mice/mice CMCs was comparable to that of tg/tg CMCs. Both values were smaller than the value of +/+ CMCs but were larger than the value of mi/mi CMCs (Table4).
Concentration of serotonin in +/+,mi/mi,mice/mice, andtg/tg CMCs
| Genotype of CMCs . | Serotonin concentration4-150 (nM/106cells) . |
|---|---|
| +/+ | 8.86 ± 0.73 |
| mi/mi | 1.32 ± 0.084-151,‡ |
| mice/mice | 5.02 ± 0.354-151 |
| tg/tg | 6.24 ± 0.284-151 |
| Genotype of CMCs . | Serotonin concentration4-150 (nM/106cells) . |
|---|---|
| +/+ | 8.86 ± 0.73 |
| mi/mi | 1.32 ± 0.084-151,‡ |
| mice/mice | 5.02 ± 0.354-151 |
| tg/tg | 6.24 ± 0.284-151 |
Mean ± SE of 3 experiments.
P < .05 compared with the value of +/+ mice byt test.
P < .05 compared with the value oftg/tg mice by t test.
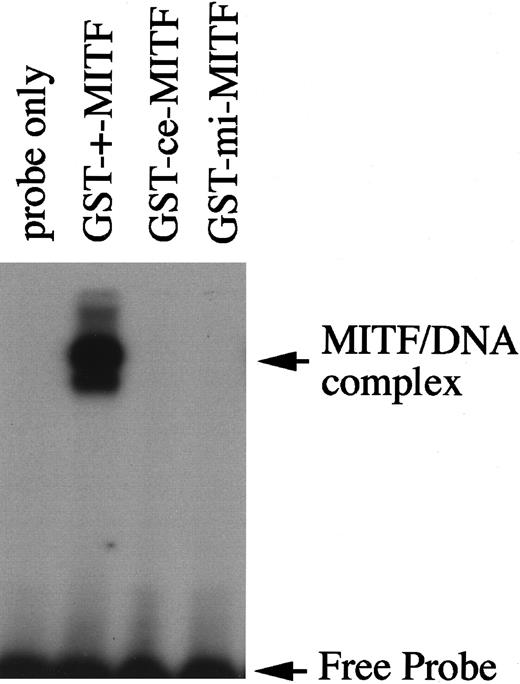
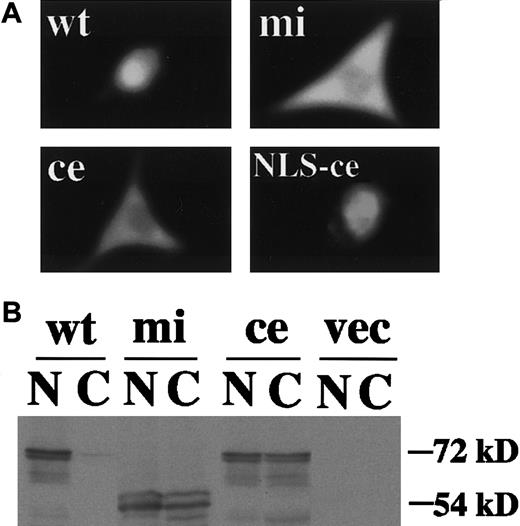
We then examined functions of ce-MITF. First, the DNA binding ability of ce-MITF was examined by EGMSA. A part of the MMCP-6 promoter containing the MITF binding motif, CACATG, was used as a probe. The specific binding of ce-MITF was not detectable as in the case of mi-MITF (Figure2). Second, the subcellular localization of ce-MITF was examined by immunocytochemistry. Because thece-MITF lacks the carboxy-terminal region that is recognized by anti-MITF antibody used in our previous studies, we detected the localization of various epitope-tagged MITFs in the present study.8 The expression vector that contained +-MITF,ce-MITF, or mi-MITF cDNA downstream from the sequence of Myc-epitope was transfected into NIH3T3 cells. The localization of MITF was detected with anti-Myc antibody. A strong signal was detected only in the nucleus of the NIH3T3 cells overexpressing Myc-tagged +-MITF (Figure3A). In contrast, signals were detected in both nucleus and cytoplasm of the NIH3T3 cells overexpressing Myc-tagged ce-MITF (Figure 3A). Signals were detected in both nucleus and cytoplasm as well in the NIH3T3 cells overexpressing Myc-tagged mi-MITF (Figure 3A). When the modifiedce-MITF possessing the NLS of SV40 large-T antigen was overexpressed, signals were detected only in the nucleus (Figure 3A).
DNA binding ability of +-MITF, ce-MITF, or mi-MITF examined by EGMSA.
The labeled 5′-TGGTGGGGACACATGTTACATGGA oligonucleotide was used as a probe (underline shows hexameric motif recognized by MITF). Each lane contains 3.5 μg GST-+-MITF, GST-ce-MITF, or GST-mi-MITF.
DNA binding ability of +-MITF, ce-MITF, or mi-MITF examined by EGMSA.
The labeled 5′-TGGTGGGGACACATGTTACATGGA oligonucleotide was used as a probe (underline shows hexameric motif recognized by MITF). Each lane contains 3.5 μg GST-+-MITF, GST-ce-MITF, or GST-mi-MITF.
Subcellular localization of various MITFs examined by immunocytochemistry or immunoblotting.
(A) NIH3T3 cells were transfected with expression vector containing Myc-tagged +-MITF, ce-MITF, mi-MITF, orce-MITF with the NLS of SV40 large-T antigen. After 48 hours of transfection, cells were stained with anti-Myc antibody. Magnification × 4000. (B) Nuclear and cytoplasmic extracts of NIH3T3 cells transfected with the above-mentioned expression plasmid were immunoblotted with anti-Myc antibody. Extracts of cells transfected with expression vector alone were used as a negative control (shown as vec).
Subcellular localization of various MITFs examined by immunocytochemistry or immunoblotting.
(A) NIH3T3 cells were transfected with expression vector containing Myc-tagged +-MITF, ce-MITF, mi-MITF, orce-MITF with the NLS of SV40 large-T antigen. After 48 hours of transfection, cells were stained with anti-Myc antibody. Magnification × 4000. (B) Nuclear and cytoplasmic extracts of NIH3T3 cells transfected with the above-mentioned expression plasmid were immunoblotted with anti-Myc antibody. Extracts of cells transfected with expression vector alone were used as a negative control (shown as vec).
Subcellular localization of +-MITF, ce-MITF, andmi-MITF was also examined by immunoblotting analysis. A strong signal was detected in the nuclear fraction prepared from the NIH3T3 cells transfected with Myc-tagged +-MITF cDNA but not in the cytoplasmic fraction. In contrast, moderate signals were detected in both nuclear and cytoplasmic fractions of the NIH3T3 cells transfected with Myc-tagged ce-MITF or Myc-tagged mi-MITF cDNA (Figure 3B). The size of the immunoreactive protein in NIH3T3 cells transfected with Myc-tagged ce-MITF cDNA was smaller than that of NIH3T3 cells transfected with Myc-tagged +-MITF cDNA or Myc-tagged mi-MITF cDNA because of the truncation of the C-terminal region of ce-MITF (Figure 3B).
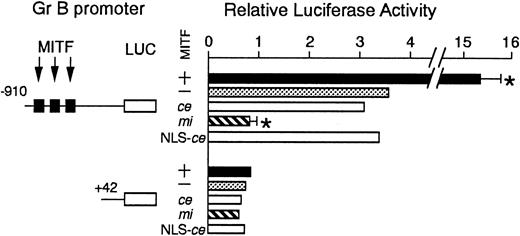
We examined the effect of ce-MITF on the transactivation of the Gr B promoter using the transient cotransfection assay (Figure4). The 5′ flanking sequence of the Gr B gene (nt −910 to +42) was cloned upstream of the luciferase gene. Three functional CANNTG motifs were present in this region.25 The luciferase construct was cotransfected into P815 cells with the expression plasmid containing no insert, +-MITF,mi-MITF, or ce-MITF cDNA. The coexpression of +-MITF significantly increased the luciferase activity, whereas that ofmi-MITF significantly reduced it. The expression ofce-MITF showed the luciferase activity comparable to the value obtained by the expression of vector alone (Figure 4). We further examined the modified ce-MITF that possessed the NLS of SV40 large-T antigen. The luciferase activity obtained by the expression of the modified ce-MITF was comparable to the value obtained by the expression of original ce-MITF (Figure 4).
The effect of coexpression of +-MITF,ce-MITF, mi-MITF, or ce-MITF with NLS of SV40 large-T antigen on the luciferase activity under the control of the Gr B promoter.
Variousreporter and effector constructs were introduced into P815 cells by electroporation. Three solid squares represent CANNTG motifs between −910 and +42, ie, CAGATG (nt −521 to −516), CACGTG (nt −530 to 525), and CATTTG (nt −521 to −516) motifs. The bars represent the mean ± SE of 3 independent experiments. In some cases, the SE was too small to be shown by the bars.P < .01 by t test when compared with the control, in which expression vector containing no insert was cotransfected.
The effect of coexpression of +-MITF,ce-MITF, mi-MITF, or ce-MITF with NLS of SV40 large-T antigen on the luciferase activity under the control of the Gr B promoter.
Variousreporter and effector constructs were introduced into P815 cells by electroporation. Three solid squares represent CANNTG motifs between −910 and +42, ie, CAGATG (nt −521 to −516), CACGTG (nt −530 to 525), and CATTTG (nt −521 to −516) motifs. The bars represent the mean ± SE of 3 independent experiments. In some cases, the SE was too small to be shown by the bars.P < .01 by t test when compared with the control, in which expression vector containing no insert was cotransfected.
Discussion
We examined the importance of Zip domain of MITF by comparing the mast cell abnormalities of mice/micemice with those of tg/tg and mi/mi mice. First, we examined the abnormalities of skin mast cells. Inmice/mice mice, the number of skin mast cells decreased to a level comparable to that of mi/miand tg/tg mice. Although the proportion of berberine sulfate+ mast cells decreased in mi/mi mice, the proportion was normal in mice/micemice as in the case of tg/tg mice. The proportions of MMCP-4 mRNA+ and MMCP-6 mRNA+ skin mast cells reduced in mice/mice mice to levels comparable to those of tg/tg and mi/mi mice. The abnormalities of skin mast cells ofmice/mice mice were similar to those of tg/tg mice rather than to those ofmi/mi mice.
Next, we compared the phenotype ofmice/mice CMCs to that oftg/tg and mi/mi CMCs. The amounts of MMCP-4, MMCP-5, and MMCP-6 mRNAs reduced inmice/mice CMCs to levels comparable to those of tg/tg and mi/mi CMCs. The amounts of c-kit, Gr B, and TPH mRNAs inmice/mice CMCs were comparable to those of tg/tg CMCs and were intermediate between the amounts of +/+ and those of mi/mi CMCs. The phenotype ofmice/mice CMCs was similar to that of tg/tg CMCs rather than to that of mi/mi CMCs.
The level of c-kit mRNA expression inmice/mice and tg/tg CMCs was higher than that of mi/mi CMCs. In contrast, numbers of mast cells in the skin of mice/miceand tg/tg mice were comparable to those ofmi/mi mice. The expression level of c-kit in CMCs was not necessarily proportional to the number of mast cells in the skin. The mechanisms remain to be clarified.
There was a possibility that the decreased mast cell number in the skin of 20-day-old mi/mi, mice/mice, and tg/tg mice might be a consequence of delayed homing or development of the mutant mast cells. If the decrease in 20-day-old mice was due to the delay of homing or development, the number of mast cells may be corrected in older mice. We examined the number of mast cells in the skin of approximately 60-day-old tg/tg mice, but the number of mast cells was comparable to that of 20-day-old tg/tg mice (unpublished data). We considered that the homing or development of mast cells was completed in 20-day-old mutant mice.
CMCs of mice/mice genotype killed YAC-1 cells as effectively as +/+ or tg/tg CMCs, but the cytotoxic activity of mi/mi CMCs was deficient. This was partly consistent with the expression level of Gr B mRNA demonstrated by the Northern analysis. Probably the expression level of Gr B observed in mice/mice andtg/tg CMCs may be enough for the cytotoxic activity. Serotonin contents of +/+,mice/mice, tg/tg, ormi/mi CMCs were well correlated with the expression levels of the TPH gene in mice of each genotype. Because we have not determined the amount of Gr B or TPH proteins, we were not able to indicate the direct correlation between mRNA and protein levels. Determination of the amounts of Gr B and TPH proteins will clarify this point.
We examined the function of ce-MITF by the luciferase assay using the Gr B promoter. As previously reported, the expression of +-MITF increased the activity of Gr B promoter significantly, whereas the expression of mi-MITF reduced it.32 The expression of ce-MITF showed a promoter activity comparable to the value obtained by the expression of vector alone, suggesting that the ce-MITF lacked not only the transactivation ability but also the inhibitory effect on transcription. This was consistent with the fact that the phenotype ofmice/mice mast cells was similar to that of tg/tg mast cells.
The mi-MITF showing the inhibitory effect possessed the mutated basic domain and the intact Zip domain. Because thece-MITF did not show such an inhibitory effect, the Zip domain may be necessary for the inhibitory effect of MITF. This was consistent with the result of Krylov et al that mutants of various bHLH-Zip proteins showing inhibitory effects possessed the abnormal basic domain and the normal Zip domain.50
The effect of Zip domain on DNA binding ability has been reported in various bHLH-Zip proteins. Mutant upstream stimulatory factor (USF) and Max lacking the Zip domain bind DNA.51,52 On the other hand, TFE3 lacking Zip domain did not bind it.53 Fisher and his colleagues reported that the ce-MITF synthesized by reticulocyte lysates did not bind DNA.7 Here, we obtained the same result using recombinant GST-ce-MITF fusion protein. The Zip domain of MITF and TFE3 appeared to be indispensable for DNA binding, whereas the Zip domain of USF and Max did not. The different attitude of Zip domain between MITF/TFE3 and USF/Max may be related to the fact that MITF forms a heterodimer with TFE3 but not with USF or Max.7
The ce-MITF was detected both in the nucleus and cytoplasm as in the case of mi-MITF.8,29 Small molecules less than 50 kd are able to pass through the nuclear pore by passive diffusion.54 Because the molecular mass of monomeric Myc-tagged ce-MITF was approximately 50 kd, the monomeric form of ce-MITF may diffuse passively through the nuclear pore. The inability to dimerize and the reduction of the molecular mass due to truncation of Zip domain might cause passive diffusion of ce-MITF. The second explanation is that the Zip domain of MITF possesses an NLS. Recently, Nagoshi et al reported that the NLS located in the Zip domain of sterol regulatory element binding protein 2 (SREBP2), another bHLH-Zip protein.55 The truncation of Zip domain abolishes the nuclear localization of SREBP2. Further biochemical studies may clarify whether the NLS is present in the Zip domain of MITF.
There was a possibility that the abnormality of ce-MITF was simply due to its inability to translocate into the nucleus. To examine this possibility, we constructed the modified ce-MITF that possessed the NLS of SV40 large-T antigen. The expression of the modified ce-MITF did not show any promoter activities as that of the original ce-MITF. This indicated that the abnormality of ce-MITF was not due to a defect in the nuclear localization.
Taken together, the Zip domain was important for the function of MITF. The mice/mice mice are useful for clarifying the function of MITFs.
The authors thank Dr S. Nagata of Osaka University for pEF-BOS and Dr I. Matsumura of Osaka University for CS2+MT.
Supported by grants from the Ministry of Education, Science and Culture; the Ministry of Health and Welfare; the Organization of Pharmaceutical Safety and Research; the Welfide Medicinal Research Foundation; and NIH grant EY-10223 to M.L.L.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Eiichi Morii, Dept of Pathology, Osaka University Medical School, Yamada-oka 2-2, Suita 565-0871, Japan; e-mail:morii@patho.med.osaka-u.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal