Abstract
Delta-mediated Notch signaling controls cell fate decisions during invertebrate and murine development. However, in the human, functional roles for Delta have yet to be described. This study reports the characterization of Delta-1 and Delta-4 in the human. Human Delta-4 was found to be expressed in a wide range of adult and fetal tissues, including sites of hematopoiesis. Subsets of immature hematopoietic cells, along with stromal and endothelial cells that support hematopoiesis, were shown to express Notch and both Delta-1 and Delta-4. Soluble forms of human Delta-1 (hDelta-1) and hDelta-4 proteins were able to augment the proliferation of primitive human hematopoietic progenitors in vitro. Intravenous transplantation of treated cultures into immune-deficient mice revealed that hDelta-1 is capable of expanding pluripotent human hematopoietic repopulating cells detected in vivo. This study provides the first evidence for a role of Delta ligands as a mitogenic regulator of primitive hematopoietic cells in the human.
Introduction
The Notch signaling pathway has been well conserved throughout evolution and has been shown to play a crucial role in embryonic development and cell fate determination in a wide range of organisms.1,2 Activation of the Notch pathway is thought to be mediated by interactions of bordering cells via cell-to-cell contact of the Notch receptor and its membrane-associated ligands.3 Notch receptors represent a single-pass transmembrane protein composed of a large extracellular domain containing 36 epidermal growth factor (EGF)–like repeats in tandem and 3 cysteine-rich Notch/LIN-12 repeats2,4 and an intracellular domain consisting of 6 tandem cdc10/ankyrin repeats, a glutamine-rich domain (opa), and a PEST sequence critical to downstream signaling.5 Similar to the Notch receptor, putative Notch ligands are transmembrane proteins possessing EGF repeats in the extracellular domain, in addition to a unique cysteine-rich N-terminal region referred to as the Delta:Serrate:LAG2 (DSL) domain.6 The DSL domain of Notch ligands interacts stably with EGF repeats 11 and 12 of the Notch receptor in a calcium ion (Ca++)–dependent manner, leading to Notch activation.7,8 In mammals, 4 Notch genes have been described (Notch-1, Notch-2, Notch-3, and Notch-4),2,9which interact with Notch ligands, such as Delta.8Mammalian forms of Delta have recently been characterized and include Delta-1, Delta-like–1, and Delta-like–3.10 Using genetic and biochemical approaches, Artavanis-Tsakonas's group has previously shown that Delta ligand can be proteolytically processed by Kuzbanian, a member of the ADAM family of metalloproteases,11 thereby allowing a diffusible form of Delta to be secreted.
A potential role for Notch-mediated signals in hematopoiesis was introduced by the identification of a mutant form of the human homologue of the Notch-1 receptor expressed within a subset of human T-cell leukemias.12 Leukemic blasts were shown to contain a translocation between hNotch-1 and T-cell receptor β genes that results in the production of a chimeric oncoprotein.13 The Notch-1 receptor was subsequently shown to be expressed in heterogeneous populations of primitive adult human bone marrow (BM) cells expressing the cell surface marker CD34.14 These findings prompted additional studies in the mouse that indicated that members of the Notch family are involved in maturation of murine hematopoietic cells, including lineage restriction of lymphoid cells.15 More recently, a fetal stromal cell line selected for its ability to support the repopulating function of de novo isolated murine cells was shown to express Delta-like ligands, further suggesting a role of Notch regulation in uncommitted hematopoietic progenitors.16 These observations were recently supported with additional evidence indicating that Notch ligands were able to delay the acquisition of cell surface maturation markers of murine hematopoietic cells and sustain myeloid progenitors detected in vitro.10 Despite these studies in the mouse, evidence for the role of Notch signals in human hematopoietic progenitors has not been shown.
In the prevailing model of Notch signal transduction, activation of the pathway leads to transcriptional suppression of lineage-specific genes that cause inhibition of differentiation, resulting in the maintenance of cells in an uncommitted state.1,15,17-19 This role of Notch activation has led to the hypothesis that Notch ligands may be capable of modulating tissue-specific stem cell function.20 21 Here, we demonstrate that in addition to the Notch receptor, both human Delta-1 (hDelta-1) and hDelta-4, along with the Delta-processing metalloprotease Kuzbanian, are expressed in purified subsets of immature human hematopoietic cells and cells composing their putative micro-environments. Soluble forms of hDelta-1 and hDelta-4 protein were produced to investigate the functional role of these ligands in the regulation of human hematopoietic progenitor proliferation and differentiation. We show that Delta ligands are capable of expanding human clonogenic progenitors and pluripotent human repopulating cells detected in vivo. Our results indicate that Delta ligands represent hematopoietic growth factors capable of regulating primitive human hematopoietic cells. On the basis of these studies, we suggest that Delta ligands may provide a novel approach for enhanced ex vivo expansion and retroviral-mediated gene transfer of human repopulating cells.
Materials and methods
Cloning and expression of human Delta ligands and Notch receptors
Human Delta-1 complementary DNA (cDNA) was isolated by plaque hybridization from a human placenta cDNA library (γgt11) (Clontech, Palo Alto, CA). A probe was generated by degenerative polymerase chain reaction (PCR) from human fetal cDNA (Clontech). Primers were sense: 5′-TGGCARTGYAAYTGYCARGA-3′; and antisense: 5′-ATYTTYTTYTCRCARTTRAA-3′. Human Delta-4 cDNA was isolated by cross-hybridization of hDelta-1 cDNA from a human fetal lung cDNA library (γgt10, Clontech). Sequences were determined by means of ABI DNA sequencer Model 377 (Foster City, CA). Partial cDNA encoding the entire extracellular domains of Delta ligands were fused in frame to a sequence of human immunoglobulin (Ig)–G1Fc or of FLAG octapeptide. These genes were inserted into the expression vector pcDNA3 (Invitrogen, San Diego, CA) or pMKITneo (kindly provided by Dr K. Maruyama, Tokyo Medical and Dental University). These expression vectors were introduced into Chinese hamster ovary (CHO) cells by electroporation. Cells were selected in the presence of G418 and isolated. Expressions of soluble proteins of Delta ligands in culture supernatants were verified by Western blotting with anti–human IgG (Amersham, Buckinghamshire, United Kingdom) or anti-FLAG (M2) (Sigma, St Louis, MO) antibody. Human IgG1Fc chimera proteins and FLAG chimera proteins were purified from conditioned medium by affinity chromatography according to methods previously described.22 To determine the expression of other Notch signaling components, reverse transcription (RT)–PCR reactions for Notch receptors and ligands were performed in a Perkin-Elmer 9700 cycler (Foster City, CA) for a total of 40 cycles. Reactions were optimized for each set of primers by means of the following sequences: Notch-1-F 5′-GATGCCAACATCCAGGACAACATGGG-3′ and Notch-1-R 5′-GGCAGGCGGTCCATATGATCCGTGAT-3′; Notch-2-F 5′-ACATCATCACAGAC TTGGTC-3′ and Notch-2-R 5′-CATTATTGACAGCAGCTGCC-3′; Delta-1-F 5′-TGCAGGAGT- TCGTCAACAAG-3′ and Delta-1-R 5′-TCCGTAGTAGTGTTCGTCAC- 3′; Delta-4-F 5′-GCAAACCAGCACCCTCACAA-3′ and Delta-4-R 5′-TTCTTGCATGGGGAGTGGTG- 3′; Delta-3-F 5′-TGAATGCCGATGCCTAGAGG-3′ and Delta-3-R 5′-CTTCTCACAGTTGGAGCCTTGG-3′; Kuzbanian-F 5′-CATCTGACCCTAAACCAAAC-3′ and Kuzbanian-R 5′-TTAAAGTGCCTGGAAGTGGT-3′. To serve as an internal control, β-glucuronidase was amplified from all cDNA templates generated, similarly to that done previously.23,24 PCR amplification of the β-glucoronidase gene was used to provide a measure of quality control for cDNA reversed transcribed from messenger RNA (mRNA) extracted from low numbers of cells. In addition, β-glucuronidase serves as the lower limit of detection for representative cDNA since this gene is ubiquitously expressed in all cell types in as few as 1 to 2 copies per cell.23 Northern blot analysis was performed with a 32P-labeled hDelta-4 cDNA fragment (412 base pairs [bp]) prepared by PCR and hybridized to the multiple tissue Northern Blot human, human II, human III, and human fetal II (Clontech).
Purification and isolation of primary human cells
Samples of full-term human umbilical cord blood were diluted (1 to 3) in alpha-MEM (Gibco BRL, Burlington, ON, Canada), and mononuclear cells were collected by centrifugation on Ficoll-paque (Pharmacia, Piscataway, NJ) as shown previously.24,25 Cells expressing lineage-commitment markers were removed by means of an immunomagnetic separation protocol26 27 to provide primitive lineage-depleted (Lin−) cells. Lin− cells were stained with CD34 and CD38 conjugated to fluorescein isothiocyanate (FITC) and allophycocyanin (APC) (BD, San Jose, CA) and sorted on a Vantage SE (BD) to isolate subfractions. Mature populations of myeloid, T, and B cells were isolated with the use of human-specific CD33, CD3, and CD19, respectively. Sorting gates were established by means of cells stained with IgG1 conjugated to the appropriate fluorochromes (BD).
Ex vivo culture of purified primitive hematopoietic cells
Isolated CD34+CD38−Lin−populations were seeded in serum-free liquid cultures previously designed and shown to sustain human repopulating stem cells.24 Serum-free cultures contained 9500 BIT media (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with 10−4 M β-mercaptoethanol, 2 mM L-glutamine (Gibco BRL), and the following growth factors: 300 ng/mL rhu-Flt-3; 10 ng/mL rhu–interleukin(IL)–3; 10 ng/mL rhu-IL-6, (R&D Systems, Thousand Oaks, CA); 300 ng/mL rhu–stem cell factor (SCF); and 50 ng/mL rhu-G–colony stimulating factor (CSF) (Amgen, CA). Cultured subfractions were incubated with cytokine cocktail and the media conditions listed above for indicated periods at 37°C and 5% CO2 in the presence of recombinant IgG1 protein (control treated) or 10 μg/mL hDelta-1–IgG1 or hDelta-4–IgG1. Recombinant IgG1 proteins were added to cultures treated with cytokine cocktail alone, in the absence of hDelta ligands, to control for nonspecific effects of IgG1 protein. Addition of recombinant IgG1proteins had no effect when compared with conditions with hematopoietic cytokines cocktail alone. Shorter-term cultures analyzed at 4 to 12 days were done independently of longer-term cultures analyzed at 15 to 25 days.
Assays for human hematopoietic progenitors
Human clonogenic progenitor assays were performed by plating equal numbers of hDelta-1– or hDelta-4–treated or control-treated CD34+CD38−Lin− cells with the use of Methocult H4434 (Stem Cell Technologies) containing 50 ng/mL rhu-SCF, 10 ng/mL rhu-GM-CSF and rhu-IL-3, and 3 U/mL rhu-erythropoietin. Differential colony counts were assessed following incubation for 10 to 14 days at 37°C and 5% CO2 in a humidified atmosphere as shown previously.27
Transplantation of ex vivo–cultured human hematopoietic cells into nonobese diabetic/severe combined immune-deficient mice
We derived 8-week-old NOD/LtSz-scid/scid (nonobese diabetic/severe combined immune-deficient [NOD/SCID]) mice from breeding pairs originally obtained from Jackson Laboratories (Bar Harbor, ME) and maintained them under defined flora at the animal facility at the Robarts Research Institute at the University of Western Ontario. All animals were handled under sterile conditions and maintained in microisolators. Mice were sublethally irradiated at 355 cGy by means of a 137-Cs γ-irradiator prior to intravenous injection of cultured purified human cells. Mice were cotransplanted with 100 000 Lin+ irradiated (1500 rads) accessory cells.28
Analysis of human stem cell engraftment
Genomic DNA was extracted from cells harvested from the BM of the femurs, tibiae, and iliac crests of transplanted mice. For analysis, 1 to 2 μg DNA was digested with EcoRI, run on agarose gels, transferred, and probed on a Southern blot with the use of a human chromosome-17–specific α-satellite probe (p17H8).27 29Quantitation of levels of engraftment was performed by comparing the 2.7-kilobase band with standards of known mouse and human DNA mixtures; this was done with a level of resolution equal to 0.05% human DNA. In addition, BM cells from each mouse were stained with fluorochrome-conjugated antibody specific to human CD45 (a pan-leukocyte marker) and CD38 (Becton Dickinson, Immunocytometry Systems [BDIS], San Jose, CA) and analyzed by flow cytometry by means of a FACScalibur and Cell Quest software (BD). Murine BM cells from engrafted animals were further stained with CD45 APC (Pharmingen Canada) and gated to analyze human cells only in combination with either CD20 FITC and CD19 phycoerythrin (PE); CD33 FITC (BDIS) and CD15 PE (Immunotech, Marseilles, France); or CD34 FITC and CD38 PE (BDIS) monoclonal antibodies for multilineage analysis.
Results
Characterization of hDelta-4
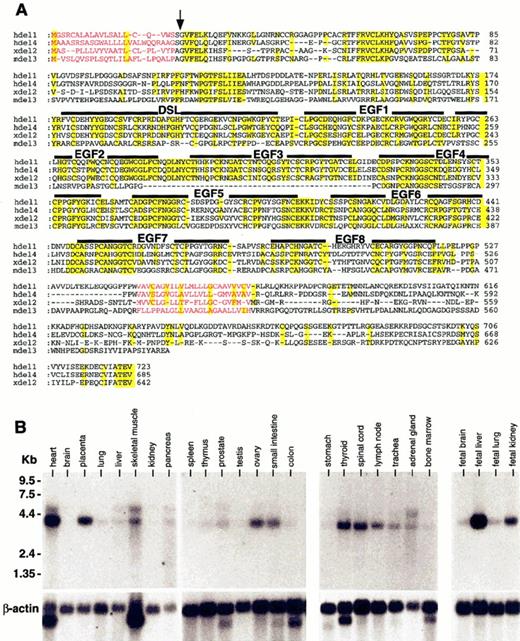
The Delta family of Notch ligands was first described in Drosophila and has since been shown to be conserved in mammals.2,8 Using low-stringency hybridization and hDelta-1 cDNA as a probe to screen a human fetal lung library, we have isolated an additional member of the Delta family, termed human Delta-4. Using a different strategy for identification and isolation, Shutter et al30 have also reported the human Delta-4 sequence. This group has characterized the expression of the mouse homologue of Delta-4 in murine embryonic development and provided evidence for the role of murine Delta-4 in endothelial cell regulation. Figure 1A illustrates an alignment of the hDelta-4 amino acid sequence, comparing hDelta-1 (hdel1),8Xenopus Delta-2 (xdel2),31 and murine Delta-3 (mdel3).32 Similar to that shown by Shutter et al,30 hDelta-4 encodes a transmembrane protein that has 8 EGF-like repeats and the DSL domain (Figure 1A). Although a second invertebrate Delta gene was reported in Xenopus as X-Delta-2,31 hDelta-4 appears to be unique among mammals and shares a low similarity (49.7%) with X-Delta-2, which is much lower than the homology of 76.2% shared by the hDelta-1 and X-Delta-1 orthologues. The intracellular domain of hDelta-4 is distinct and shares only low similarities with X-Delta-1 and X-Delta-2; these are 29.1% and 26.3%, respectively. On the basis of these comparisons, we suggest that hDelta-4 does not represent an orthologue of other mammalian Deltas.
Characterization of the human Notch ligand, Delta-4.
(A) Alignment of amino acids sequence of Delta-related proteins. The DSL domains and EGF-like repeats are marked by a black bar above alignment. The putative signal sequence (indicated with an arrow) and transmembrane domain are indicated as colored letters (red). The identical amino acids are shaded (yellow). Human Delta-1 (Gray et al, 1999) (hdel1; Genbank accession number AF003522); human Delta-4 (submitted to Genbank as human Delta-2, hdel2; Genbank accession number AF253468); X-Delta-2 (Jen et al, 1997) (xdel2; Genbank accession number U70843); and mouse D113 (Dunwoodie et al, 1997) (mdel3; Genbank accession number NM007866) represent Delta-related proteins. Human Delta-4 cDNA was isolated by cross-hybridization of hDelta-1 cDNA from fetal lung cDNA library. (B) Expression profile of hDelta-4 mRNA in tissues. As positive control, β-actin was used. For Northern blot analysis, a 32P-labeled human Delta-4 cDNA (412 bp) was used to hybridize to cDNA from various tissues of human origin.
Characterization of the human Notch ligand, Delta-4.
(A) Alignment of amino acids sequence of Delta-related proteins. The DSL domains and EGF-like repeats are marked by a black bar above alignment. The putative signal sequence (indicated with an arrow) and transmembrane domain are indicated as colored letters (red). The identical amino acids are shaded (yellow). Human Delta-1 (Gray et al, 1999) (hdel1; Genbank accession number AF003522); human Delta-4 (submitted to Genbank as human Delta-2, hdel2; Genbank accession number AF253468); X-Delta-2 (Jen et al, 1997) (xdel2; Genbank accession number U70843); and mouse D113 (Dunwoodie et al, 1997) (mdel3; Genbank accession number NM007866) represent Delta-related proteins. Human Delta-4 cDNA was isolated by cross-hybridization of hDelta-1 cDNA from fetal lung cDNA library. (B) Expression profile of hDelta-4 mRNA in tissues. As positive control, β-actin was used. For Northern blot analysis, a 32P-labeled human Delta-4 cDNA (412 bp) was used to hybridize to cDNA from various tissues of human origin.
The expression of hDelta-4 in human tissues was evaluated by hybridization to human mRNA Northern blots containing a variety of adult and fetal human tissues (Figure 1B). Variable levels of expression were detected in most adult human tissues, including sites of hematopoiesis such as BM and lymph nodes (Figure 1B). In early human development, hDelta-4 was ubiquitously expressed and was detected in all fetal tissues examined, including brain, lung, and kidney. In addition, hDelta-4 was highly expressed in the liver, which serves as a primary site of human fetal hematopoiesis (Figure 1B). Taken together, our results show that this previously uncharacterized form of human Delta is expressed in a variety of tissues composing both fetal and post-natal stages of human development.
Both Notch and Delta ligands are expressed among human hematopoietic cells and cells composing the hematopoietic micro-environment
Previous reports have shown that primitive human CD34+blood cells express Notch-1 and Notch-2,14,33 suggesting that Notch signaling may play a role in modulating hematopoietic progenitors in the human. Activation of the Notch receptor expressed on hematopoietic cells has been postulated to be derived from Notch ligands expressed among cells composing the micro-environment of active hematopoiesis, such as BM stromal or human umbilical vein endothelial cells (HUVECs). Consistent with this notion, adult BM stromal cells and HUVECs were shown to express hDelta-1 and hDelta-4, whereas hDelta-3 was not detectable in either of these tissues (Figure2A). In addition, Notch-1 and Notch-2 were shown to be expressed in both BM stromal cells and HUVECs (Figure2A). On the basis of the expression of hDelta-1 and hDelta-4 in these tissues, we investigated whether Kuzbanian, the metalloprotease responsible for cleavage of Delta ligands,11 would also be expressed in human BM stromal cells and HUVECs. Human Kuzbanian was not expressed in adult stromal cells but could be detected in HUVECs (Figure 2A) and in stromal cells isolated from human fetal liver (data not shown).
Human hematopoietic cells and their putative micro-environments express components of the Notch signaling pathway.
RT-PCR reactions were performed on cells from hematopoietic micro-environment or purified cell fractions (levels of purity greater than 99%) from a minimum of 3 independent samples. (A) Expression of human Notch receptors 1 and 2, hDelta-1, hDelta-4, hDelta-3, and hKuzbanian by putative hematopoietic micro-environment cells of adult BM stromal cells and HUVECs (n = 3). (B) Expression of hNotch-1, hNotch-2, hDelta-1, hDelta-4, hDelta-3, and hKuzbanian by primitive Lin− cell subsets (CD34+CD38−Lin− and CD34+CD38+Lin−) and mature (CD33+ myeloid cells, CD3+ T cells, and CD19+ B cells) from full-term human cord blood samples (n = 4). RT-PCR reactions were performed on fetal cDNA as a positive control while single transcript copy β-glucuronidase was used to assess the quality and integrity of cDNA templates and provide a lower limit of detection.
Human hematopoietic cells and their putative micro-environments express components of the Notch signaling pathway.
RT-PCR reactions were performed on cells from hematopoietic micro-environment or purified cell fractions (levels of purity greater than 99%) from a minimum of 3 independent samples. (A) Expression of human Notch receptors 1 and 2, hDelta-1, hDelta-4, hDelta-3, and hKuzbanian by putative hematopoietic micro-environment cells of adult BM stromal cells and HUVECs (n = 3). (B) Expression of hNotch-1, hNotch-2, hDelta-1, hDelta-4, hDelta-3, and hKuzbanian by primitive Lin− cell subsets (CD34+CD38−Lin− and CD34+CD38+Lin−) and mature (CD33+ myeloid cells, CD3+ T cells, and CD19+ B cells) from full-term human cord blood samples (n = 4). RT-PCR reactions were performed on fetal cDNA as a positive control while single transcript copy β-glucuronidase was used to assess the quality and integrity of cDNA templates and provide a lower limit of detection.
To investigate the expression of Notch and the family of Delta ligands in hematopoietic cells, highly purified subsets of myeloid, T- and B-lymphoid cells were isolated from human hematopoietic tissue by means of flow cytometric sorting (purity greater than 99%; data not shown). In addition to mature subsets, rare primitive cells depleted of lineage-committed cells (Lin−) were fractionated into CD34+CD38−Lin− or CD34+CD38+Lin− populations. While both the CD34+ subfractions contain a high frequency of hematopoietic progenitors, the CD34+CD38−Lin− subset is also enriched for human blood cells capable of pluripotent repopulating capacity in immune-deficient mice. These human hematopoietic cells are functionally defined as SCID repopulating cells (SRCs).34,35 Notch-1 and Notch-2 were expressed in primitive and mature cells within the human hematopoietic hierarchy (Figure 2B). In addition, hDelta-1 and hDelta-4, but not hDelta-3, were also expressed by mature and primitive human hematopoietic subsets (Figure 2B), along with the human homologue of the metalloprotease Kuzbanian (Figure 2B). Our analysis demonstrates that both Notch-1 and Notch-2 and at least 2 forms of Delta ligands 1 and 4 are more widely expressed among primitive and mature hematopoietic cells than was previously appreciated.17
Human Delta-1 and hDelta-4 proteins are capable of interacting with primitive human hematopoietic cells
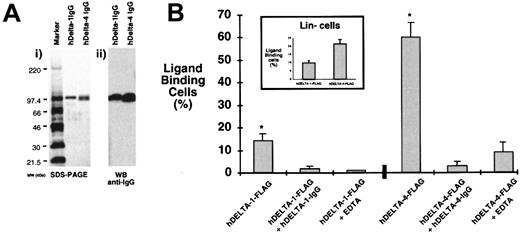
Previous studies examining the functional role of Notch ligands have used coculture or conditioned supernatants from cell lines transfected with cDNA expressing the Notch ligands.36,37However, these studies are complicated by the potential presence of unknown factors resulting from the expression of the specific ligand in the transfected cell line. Accordingly, to examine the role of Delta in human hematopoiesis, soluble forms of hDelta-1 and hDelta-4 were produced in transfected cell lines and purified by affinity chromatography.22 Since the expression of hDelta-3 was not detected in human hematopoietic subsets or stromal cells, we focused our study on the functional role of hDelta-1 and hDelta-4 in human hematopoietic development. Purified hDelta-1 and hDelta-4, expressed as chimeric proteins of either human IgG1Fc or FLAG peptide sequences, were evaluated by means of sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels and visualized by Coomassie blue staining (Figure3A, i). The specificity of Delta-1 and Delta-4 proteins was verified by Western blotting with the use of anti–human IgG1 or FLAG antibodies (Figure 3A, ii). To determine whether soluble ligands of hDelta-1 and hDelta-4 were capable of interacting with human hematopoietic cells, full-term gestation umbilical cord blood (CB) mononuclear cells and primitive CB Lin− fractions were examined. Cells were treated with purified hDelta-1– and hDelta-4–FLAG chimeric proteins complexed to anti-FLAG monoclonal antibody fluorochrome conjugates and then analyzed by flow cytometry (Figure 3B). Soluble hDelta-1 was capable of binding 15% of mononuclear cells while cells with binding affinity to hDelta-4 were considerably higher, at approximately 60% (Figure 3B). Human Delta-1 and hDelta-4 were capable of interacting with 10% and 20% of primitive CB Lin− hematopoietic cells, respectively (Figure 3B, inset). The specificity of hDelta-1 and hDelta-4–FLAG chimeric protein interactions with human hematopoietic cells was verified by pretreating cells with hDelta-1– and hDelta-4–IgG chimeric proteins (Figure 3B). Pretreatment with either hDelta-1– or hDelta-4–IgG blocked the binding of the respective Delta-FLAG chimeric ligand (Figure 3B). In addition, binding of both hDelta-1 and hDelta-4 could be abrogated by co-incubation of the ligand with 10 mM EDTA, which prevents Ca++-mediated Delta-Notch interactions via DSL-EGF domains (Figure 3B).7 Treatment with EDTA or the IgG chimeric ligands did not affect the efficiency of binding of other cell-surface–targeted fluorochrome-conjugated antibodies. Taken together, these results provide evidence for the ability of purified forms of soluble hDelta-1 and hDelta-4 to interact with subsets of primary primitive human hematopoietic cells.
Production and analysis of soluble forms of hDelta-1 and hDelta-4.
Genes encoding the extracellular domain of human Delta ligands were inserted into the expression vector pcDNA and introduced into CHO cells by electroporation. Expression of soluble proteins of Delta ligands in culture supernatants was analyzed by SDS-PAGE analysis and verified by Western blotting. (A) (i) Analysis of soluble forms of hDelta-1 and hDelta-4 by Coomassie-blue–stained SDS-PAGE gels. (ii) Western blot analysis to verify the production and purification of hDelta-1– and hDelta-4–IgG chimeras using secondary, goat anti–human IgG1 antibodies. (B) Binding and competition studies using soluble hDelta-1– and hDelta-4–FLAG–tagged chimeras on human cord blood mononuclear and Lin− cells. Results show the mean percentages ± SEM of cells detected that bind to hDelta-1– and hDelta-4–FLAG–tagged ligands. *Significant difference;P < .001. The concentration of soluble hDelta-1 or hDelta-4–FLAG was titrated, and complete saturation of cell binding was found to occur at 10 μg/mL. Prior incubation of IgG1 chimeric Delta proteins or co-incubation of 10 mM EDTA significantly inhibited detectable binding of Delta-FLAG ligands (greater than 90% inhibition). (Inset) Binding of hDelta-1 and hDelta-4 to CB Lin− cells (mean ± SEM) (n = 4). Functional binding of soluble ligands to hematopoietic cells was tested by means of FLAG tag chimera by employing secondary, fluorochrome-conjugated anti-FLAG antibody complexes as described in “Materials and methods.”
Production and analysis of soluble forms of hDelta-1 and hDelta-4.
Genes encoding the extracellular domain of human Delta ligands were inserted into the expression vector pcDNA and introduced into CHO cells by electroporation. Expression of soluble proteins of Delta ligands in culture supernatants was analyzed by SDS-PAGE analysis and verified by Western blotting. (A) (i) Analysis of soluble forms of hDelta-1 and hDelta-4 by Coomassie-blue–stained SDS-PAGE gels. (ii) Western blot analysis to verify the production and purification of hDelta-1– and hDelta-4–IgG chimeras using secondary, goat anti–human IgG1 antibodies. (B) Binding and competition studies using soluble hDelta-1– and hDelta-4–FLAG–tagged chimeras on human cord blood mononuclear and Lin− cells. Results show the mean percentages ± SEM of cells detected that bind to hDelta-1– and hDelta-4–FLAG–tagged ligands. *Significant difference;P < .001. The concentration of soluble hDelta-1 or hDelta-4–FLAG was titrated, and complete saturation of cell binding was found to occur at 10 μg/mL. Prior incubation of IgG1 chimeric Delta proteins or co-incubation of 10 mM EDTA significantly inhibited detectable binding of Delta-FLAG ligands (greater than 90% inhibition). (Inset) Binding of hDelta-1 and hDelta-4 to CB Lin− cells (mean ± SEM) (n = 4). Functional binding of soluble ligands to hematopoietic cells was tested by means of FLAG tag chimera by employing secondary, fluorochrome-conjugated anti-FLAG antibody complexes as described in “Materials and methods.”
Human Delta-1 and hDelta-4 modulate the development of primitive human hematopoietic cells in vitro
To investigate the role of human Delta ligands in human hematopoiesis, highly purified CD34+CD38−Lin− cells were cultured in serum-free media containing hematopoietic cytokines25 38 and compared with cultures where soluble forms of hDelta-1 or hDelta-4 were added. Recombinant IgG1protein was added to cultures treated with cytokine (control treated) in the absence of hDelta-1–IgG or hDelta-4–IgG to account for any nonspecific effects of IgG1. Addition of IgG1 had no effect in comparison with cells cultured with hematopoietic cytokines alone (data not shown). CD34+CD38−Lin− cells were harvested at indicated times, and changes in total cell number, primitive CD34+CD38− subsets, and hematopoietic progenitors were compared (Figure4). Treatment with hDelta-1 or hDelta-4 had no significant effect on total cell number (Figure 4A). However, in contrast to the bulk culture of total cells, both hDelta-1 and hDelta-4 treatment were able to expand the subfraction of phenotypically primitive CD34+CD38− cells (Figure 4B). Human Delta-1 was able to expand the total number of CD34+CD38− cells in both short- and long-term cultures as compared with control-treated cells (Figure 4B), whereas the proliferative effects of hDelta-4 was more pronounced after extended culture periods beyond 15 days (Figure 4B).
Functional and phenotypic analysis of human CD34+CD38−Lin− cells cultured in serum-free conditions containing soluble forms of hDelta-1 and hDelta-4.
Highly purified CD34+CD38−Lin−cells (purity greater than 99%; data not shown) were seeded in 96-well culture plates containing serum-free BIT medium and growth factors as described in “Materials and methods.” Cells were harvested at the indicated times for proliferative and differentiative analysis by fluorescence-activated cell sorting (FACS) or assayed for functional progenitors in methylcellulose containing human cytokines. (A) The fold increase in total cell number relative to cells seeded on Day 0. Cells from individual wells were counted, and the mean fold increase in total cell number was calculated. (B) Changes in the total number of primitive CD34+CD38− cells in culture. Total number of primitive CD34+CD38− cells was determined for each well from the frequencies of CD34+CD38− cells examined by FACS analysis. Mean frequencies of CD34+CD38− cells expressed as a percentage of the total cells in the culture is indicated for each treatment. (C) Effect of hDelta-1 and hDelta-4 treatment on the total number of clonogenic progenitors. Aliquots of cells were plated in methylcellulose cultures as described in “Materials and methods.” Hematopoietic progenitors (colony-forming units [CFUs]) were scored, and the frequency of progenitors was determined from the input cell number. The frequency of CFUs and the total number of cells harvested from each well were used to calculate the total number of clonogenic progenitors. Values shown are the mean ± SEM (n = 6). *Significant difference; P < .01.
Functional and phenotypic analysis of human CD34+CD38−Lin− cells cultured in serum-free conditions containing soluble forms of hDelta-1 and hDelta-4.
Highly purified CD34+CD38−Lin−cells (purity greater than 99%; data not shown) were seeded in 96-well culture plates containing serum-free BIT medium and growth factors as described in “Materials and methods.” Cells were harvested at the indicated times for proliferative and differentiative analysis by fluorescence-activated cell sorting (FACS) or assayed for functional progenitors in methylcellulose containing human cytokines. (A) The fold increase in total cell number relative to cells seeded on Day 0. Cells from individual wells were counted, and the mean fold increase in total cell number was calculated. (B) Changes in the total number of primitive CD34+CD38− cells in culture. Total number of primitive CD34+CD38− cells was determined for each well from the frequencies of CD34+CD38− cells examined by FACS analysis. Mean frequencies of CD34+CD38− cells expressed as a percentage of the total cells in the culture is indicated for each treatment. (C) Effect of hDelta-1 and hDelta-4 treatment on the total number of clonogenic progenitors. Aliquots of cells were plated in methylcellulose cultures as described in “Materials and methods.” Hematopoietic progenitors (colony-forming units [CFUs]) were scored, and the frequency of progenitors was determined from the input cell number. The frequency of CFUs and the total number of cells harvested from each well were used to calculate the total number of clonogenic progenitors. Values shown are the mean ± SEM (n = 6). *Significant difference; P < .01.
Human hematopoietic progenitor function was examined by quantitative analysis of the colony-forming unit (CFU) capacity of CD34+CD38−Lin− cells cultured with or without hDelta-1 or hDelta-4. Short-term cultures containing hDelta-1 or hDelta-4 appeared to reduce the number of progenitors expanded as compared with hematopoietic cytokine-treated cultures (Figure 4C). However, continued ligand treatment beyond 12 days increased the total number of clonogenic progenitors, with the greatest increase occurring between days 15 and 25 (Figure 4C). Since Notch activation has been associated with the cell fate determination and lineage restriction,1 we investigated whether hDelta treatment altered the differentiation program of expanded progenitors. The proportion of myeloid progenitors was similar in both control-treated and hDelta-1– or hDelta-4–treated cultures, with the exception of cultures treated with hDelta-1 for longer than 15 days, which demonstrated a modest increase in the composition of erythroid progenitors (data not shown).
Effects of hDelta-1 and hDelta-4 on human hematopoietic cells capable of pluripotent repopulating ability in NOD/SCID mice (SRCs)
To explore the potential effect of hDelta-1 and hDelta-4 in regulating primitive human repopulating cells, CD34+CD38−Lin− cells cultured with or without soluble Delta ligands were transplanted into NOD/SCID mice. This human-mouse xenotransplant model has been established as a reliable measure of human hematopoietic repopulating cell function and serves as a quantitative surrogate in vivo assay amenable to limiting dilution analysis.24,39 40 To quantitatively examine the role of hDelta ligands on SRCs, individual wells seeded with 2500 highly purified CD34+CD38−Lin− cells were treated and transplanted after 4 and 6 days into NOD/SCID mice. The BM of animals transplanted with cultured cells was analyzed 8 weeks post-transplant by flow cytometry and Southern blot analysis to determine the presence of human chimerism. A summary of the frequency of human chimeric NOD/SCID mice transplanted with control-treated, hDelta-1–treated, or hDelta-4–treated cells is shown in Table1. Primitive CD34+CD38−Lin− cells cultured for 4 days in the presence of hDelta-1 resulted in an increase in the number of human SRCs (frequency of 53%; n = 17), compared with control-treated cultures containing hematopoietic cytokines (frequency of 24%; n = 17) (Table 1). However, hDelta-1 had no effect on SRC function after 6 days of culture (Table 1). Unlike hDelta-1–treated cells, CD34+CD38−Lin− cells cultured for 4 days in the presence hDelta-4 (n = 12) demonstrated the identical frequency of chimeric NOD/SCID mice as the group of mice transplanted with control-treated cultures (Table 1). An additional 2 days of culture in the presence of hDelta-4 resulted in the loss of repopulating function (Table 1).
Quantitative analysis of human repopulating cells after 4 and 6 days of ex vivo culture in the presence or absence of soluble hDelta-1 or hDelta-4
| . | Day 4 . | Day 6 . | ||||
|---|---|---|---|---|---|---|
| No. positive . | No. negative . | No. positive out of total transplanted (% engrafted) . | No. positive . | No. negative . | No. positive out of total transplanted (% engrafted) . | |
| Control-treated | 4 | 13 | 4/17 (23%) | 4 | 5 | 4/9 (44%) |
| Delta-1 | 9 | 8 | 9/17* (53%) | 4 | 6 | 4/10 (40%) |
| Control-treated | 2 | 10 | 2/12 (17%) | 1 | 5 | 1/6 (17%) |
| Delta-4 | 2 | 10 | 2/12 (17%) | 0 | 5 | 0/5 (0%) |
| . | Day 4 . | Day 6 . | ||||
|---|---|---|---|---|---|---|
| No. positive . | No. negative . | No. positive out of total transplanted (% engrafted) . | No. positive . | No. negative . | No. positive out of total transplanted (% engrafted) . | |
| Control-treated | 4 | 13 | 4/17 (23%) | 4 | 5 | 4/9 (44%) |
| Delta-1 | 9 | 8 | 9/17* (53%) | 4 | 6 | 4/10 (40%) |
| Control-treated | 2 | 10 | 2/12 (17%) | 1 | 5 | 1/6 (17%) |
| Delta-4 | 2 | 10 | 2/12 (17%) | 0 | 5 | 0/5 (0%) |
Single wells containing 500 to 2500 highly purified CD34+CD38−Lin− human hematopoietic cells were isolated by flow cytometric sorting and cultured for 4 and 6 days in the presence of optimized combinations of hematopoietic growth factors previously shown to sustain repopulating cells. Mice were transplanted with cells harvested at the indicated time points, and the number of mice containing human hematopoietic cells (positive) was compared.
Significant difference, P < .01.
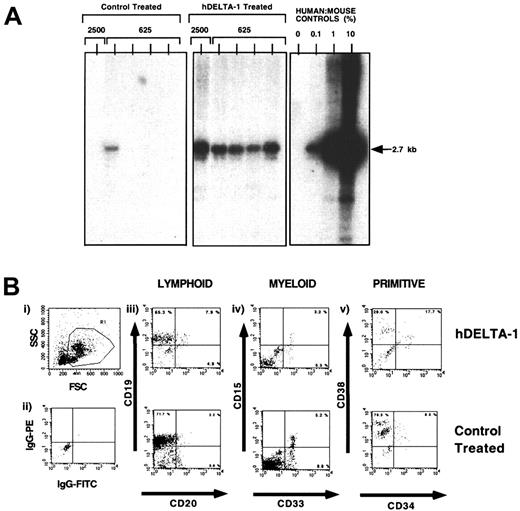
Since addition of soluble hDelta-1 increased the frequency of SRCs after 4 days, we examined whether hDelta-1 was capable of expanding SRCs in these short-term cultures. Individual wells containing hDelta-1 were seeded with 2500 CD34+ CD38−Lin− cells containing approximately 2 SRCs as demonstrated by initial limiting dilution analysis of de novo (Day 0) isolated CD34+CD38−Lin− cells (data not shown). Cells were harvested after 4 days of ex vivo culture, and either the cells were divided into 4 equal aliquots for transplantation into 4 individual NOD/SCID mice or the entire contents were transplanted into a single NOD/SCID mouse. With the use of a human-DNA–specific probe, the representative Southern blot shown in Figure 5A illustrates that mice transplanted with hDelta-1–treated cells resulted in a greater frequency of detectable SRCs as compared with engrafted mice transplanted with control-treated cultures containing hematopoietic cytokines alone (Figure 5A). Since the contents of a single well seeded with only 2 SRCs was able to engraft up to 4 individual mice after ex vivo culture, we suggest that hDelta-1 is capable of increasing the number of human SRCs detectable. To determine whether hDelta ligand addition to human CD34+CD38−Lin−cells alters the developmental program or hematopoietic lineage specification of repopulating cells, mice engrafted with hDelta-1–treated SRCs were analyzed for multilineage reconstitution by means of multiparameter flow cytometry. A representative analysis is shown in Figure 5B. Similar to control-treated SRCs, mice transplanted with hDelta-1–treated SRCs were capable of lymphoid and myeloid differentiative capacity in vivo (Figure 5B). In addition, primitive human CD34+ cells were present in transplanted mice, indicating that human SRCs treated with hDelta-1 maintain the ability to repopulate the murine BM with immature human hematopoietic subsets. No significant differences in the human hematopoietic graft composition was seen between control-treated and hDelta-1–treated stem cells (n = 8; data not shown). On the basis of these results, we suggest that addition of hDelta-1 is capable of maintaining and expanding human SRCs in ex vivo cultures.
Detection of human repopulating cells in NOD/SCID mice transplanted with CD34+CD38−Lin−cells cultured in the presence of hDelta-1.
(A) Representative Southern blot analysis of the genomic DNA extracted from the BM of individual mice transplanted with 4 aliquots of 625 cells or 2500 CD34+CD38−Lin−cells seeded into individual wells and cultured for 4 days in the absence or presence of hDelta-1. Genomic DNA extracted from BM of mice was digested with EcoR1, separated on agarose gels, and probed with human chromosome 17–specific α-satellite probe as shown previously.54 Human:mouse DNA mixture controls shown were used to measure the level of engraftment. (B) Multilineage differentiation of human repopulating cells engrafting the BM of NOD/SCID mice following hDelta-1 treatment and control-treated repopulating cells. Cells from the BM of positively engrafted mice were stained with various human-specific monoclonal antibodies and analyzed by flow cytometry. (i) Forward- and side-scatter properties were used to gate live cells in R1 for analysis. (ii) Isotype control for nonspecific IgG staining for PE and FITC fluorescence. Cells expressing the human pan-leukocyte marker CD45 were gated (not shown) and analyzed for expression of multiple-lineage human hematopoietic markers. (iii) Pan–B-cell markers CD19 and CD20 for cells of the lymphoid lineage. (iv) Myeloid markers CD15 and CD33 for immature subsets. (v) Cell surface markers CD34 and CD38 for immature subsets.
Detection of human repopulating cells in NOD/SCID mice transplanted with CD34+CD38−Lin−cells cultured in the presence of hDelta-1.
(A) Representative Southern blot analysis of the genomic DNA extracted from the BM of individual mice transplanted with 4 aliquots of 625 cells or 2500 CD34+CD38−Lin−cells seeded into individual wells and cultured for 4 days in the absence or presence of hDelta-1. Genomic DNA extracted from BM of mice was digested with EcoR1, separated on agarose gels, and probed with human chromosome 17–specific α-satellite probe as shown previously.54 Human:mouse DNA mixture controls shown were used to measure the level of engraftment. (B) Multilineage differentiation of human repopulating cells engrafting the BM of NOD/SCID mice following hDelta-1 treatment and control-treated repopulating cells. Cells from the BM of positively engrafted mice were stained with various human-specific monoclonal antibodies and analyzed by flow cytometry. (i) Forward- and side-scatter properties were used to gate live cells in R1 for analysis. (ii) Isotype control for nonspecific IgG staining for PE and FITC fluorescence. Cells expressing the human pan-leukocyte marker CD45 were gated (not shown) and analyzed for expression of multiple-lineage human hematopoietic markers. (iii) Pan–B-cell markers CD19 and CD20 for cells of the lymphoid lineage. (iv) Myeloid markers CD15 and CD33 for immature subsets. (v) Cell surface markers CD34 and CD38 for immature subsets.
Discussion
A major impediment to successful expansion and gene transfer of human repopulating cells has been the lack of defined conditions capable of inducing the proliferation of human hematopoietic stem cells.41 The ability of Notch signal activation to maintain cells in a primitive state suggests that Notch ligands may represent ideal candidates for the self-renewal and expansion of hematopoietic stem cells.17 In the present study, the use of soluble Notch ligands to expand human hematopoietic progenitors and SRCs has important implications in the optimization of ex vivo culture conditions used for stem cell expansion and gene transfer procedures. Our study indicates that hDelta ligands are able to expand primitive CD34+CD38− subsets and multilineage hematopoietic progenitors, illustrating that Delta ligands are capable of acting like hematopoietic growth factors. Furthermore, hDelta-1 was capable of increasing the number of human pluripotent repopulating cells (SRCs) while maintaining the proliferation of progenitor populations. Our group has recently demonstrated that an additional Notch ligand, Jagged-1, is capable of expanding the number of human SRCs.42 However, in contrast to the effects of human Delta-1, the expansion of human SRCs induced by soluble forms of Jagged-1 are only detectable after 12 to 15 days of ex vivo culture.42 In the murine system, expression of a constitutively active form of the Notch receptor causes cytokine-dependent immortalization of murine hematopoietic stem cells.43 The combined role for Notch signal activation, together with hematopoietic cytokines, is supported by previous studies demonstrating that activated forms of the Notch receptors expressed in hematopoietic progenitor cell lines are capable of inhibiting differentiation and allowing the proliferation of cells in an immature state.18,21,44 Similar to what was found in our current work, the role of activated Notch signals functions in concert with regulatory signals of hematopoietic cytokines used in serum-free ex vivo cultures.21,43 On the basis of the effects of Delta ligands shown in our current study and in studies from other groups, we suggest that ex vivo expansion or gene transfer of human repopulating cells may benefit from the addition of soluble forms of human Delta proteins together with modulating hematopoietic cytokines. In contrast to hDelta-1 function, hDelta-4 treatment appeared to reduce the number of human SRCs after 6 days of ex vivo culture. While the reason for the differences between hDelta-1 and hDelta-4 is presently unclear, it is possible that the 2 forms of Delta have distinct roles in Notch signal activation of primitive human hematopoietic cells. This may be due to a variety of reasons, including differences in binding affinities of Notch receptors expressed by human stem cells, as well as differential regulation by a host of other Notch modifiers including Fringe and Numb.45-49 Alternatively, the function of hDelta-4 may influence endothelial and vascular tissue development as reported previously,30 50 thereby eliminating SRCs prior to intravenous transplantation by instructing endothelial development.
Previous reports have shown that membrane-bound Delta ligands are expressed by adult stromal or endothelial cells.30,51Regulated expression of these ligands among stromal cells and endothelial cells is thought to create niches within the BM and to activate Notch signals within adjacent hematopoietic cells that express Notch receptors.17 21 In addition to the existence of instructive signals provided by stromal or endothelial cells alone, the expression of Delta-1 and Delta-4 in hematopoietic cells reported here suggests that Delta ligands may mediate activation of Notch in a multidirectional manner among hematopoietic progenitors. This is supported by the ability of chimeric forms of hDelta-1 and hDelta-4 ligands to competitively bind hematopoietic cells in the absence of stromal support. In addition, differences in the binding capacity to Delta-1 or Delta-4 ligands were demonstrated with the use of primitive human CB Lin− cells. These differences in binding capacity may represent heterogeneity of immature hematopoietic cells at the level of Notch receptor expression. It may be possible to use chimeric forms of human Delta ligands together with FACS analysis to provide an approach for the functional characterization of primitive human subsets isolated from the Lin− population that is independent of cell differentiation (CD) markers.
On the basis of the proliferative effects of soluble Deltas on human hematopoietic repopulating cells illustrated in this study, we suggest that other tissue-specific stem cells may share common embryonic programs in response to environmental stimuli and therefore may respond similarly to human Delta proteins. Therefore, our results provide a basis for investigating whether soluble Delta ligands represent regulators of recently identified neural or myogenic stem cells.52 53 Experiments attempting to elucidate the fundamental mechanisms involved in Notch receptor and Delta ligand interactions that permit the expansion and maintenance of tissue-specific stem cells derived from human neural, hematapoietic, or muscle tissue is currently ongoing in our laboratory.
Amgen, Thousand Oaks, CA, for cytokines; the staff of the labor and delivery departments of St. Joseph's Hospital and London Health Sciences, London, ON, Canada, and especially Marlene Watson and Jane Popma for providing cord blood specimens; and Drs Michael Underhill, Joseph Verdi, and David Kelvin for critically reviewing this manuscript.
Supported by a grant (MT-15063) and a scholarship award (MSH-35681) to M.B. from the Canadian Institutes of Health Research, Ottawa, ON, Canada.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Mickie Bhatia, The John P. Robarts Research Institute, Developmental Stem Cell Biology, 100 Perth Dr, London, ON, N6A 5K8; e-mail: mbhatia@rri.on.ca.


![Fig. 4. Functional and phenotypic analysis of human CD34+CD38−Lin− cells cultured in serum-free conditions containing soluble forms of hDelta-1 and hDelta-4. / Highly purified CD34+CD38−Lin−cells (purity greater than 99%; data not shown) were seeded in 96-well culture plates containing serum-free BIT medium and growth factors as described in “Materials and methods.” Cells were harvested at the indicated times for proliferative and differentiative analysis by fluorescence-activated cell sorting (FACS) or assayed for functional progenitors in methylcellulose containing human cytokines. (A) The fold increase in total cell number relative to cells seeded on Day 0. Cells from individual wells were counted, and the mean fold increase in total cell number was calculated. (B) Changes in the total number of primitive CD34+CD38− cells in culture. Total number of primitive CD34+CD38− cells was determined for each well from the frequencies of CD34+CD38− cells examined by FACS analysis. Mean frequencies of CD34+CD38− cells expressed as a percentage of the total cells in the culture is indicated for each treatment. (C) Effect of hDelta-1 and hDelta-4 treatment on the total number of clonogenic progenitors. Aliquots of cells were plated in methylcellulose cultures as described in “Materials and methods.” Hematopoietic progenitors (colony-forming units [CFUs]) were scored, and the frequency of progenitors was determined from the input cell number. The frequency of CFUs and the total number of cells harvested from each well were used to calculate the total number of clonogenic progenitors. Values shown are the mean ± SEM (n = 6). *Significant difference; P < .01.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/7/10.1182_blood.v97.7.1960/5/m_h80710855004.jpeg?Expires=1765953692&Signature=YrufCOw3O2GJuvn02wvNibMyg~0674FL3JAra-PwT3hhrDqvJ-dZCy63SiH4UCmZCaUG3dzh7mt~AL6mqi~PqCZJVzyiAnmD-oNRYWIJpCX4DJ6Frvg9dNk8vAjjbQ0jepMFCbAdJmz2TG69X4yo-S-rShZMm5mm1aAYe7d9geIxc9xypjXtZDu-loX4LzuGhJSqHI0RHcMhEipG95RdawL2IM1hNXIAtKBXqOSmYZxBt-GbydOBmheITW7nte5DOrHMqvdVSxnyJgIWwpn6PdnxNloPpXqffO~Q8a~RFMvIhtBZMHWu3tfJKgR1df27yTZcPfsIARi9JP6waN6Z~Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal