HLA class I expression is controlled by several regulatory pathways. The X2 box is a crucial element of the SXY regulatory module, which controls the transactivation of HLA class I and β2-microglobulin and of HLA class II and their accessory genes.1,2 Critical in this class II transactivator (CIITA)–induced transactivation is the cooperative binding of a multiprotein complex consisting of RFX, ATF/CREB, and NFY on the SXY regulatory module.2
Recently, Girdlestone presented evidence to suggest that nucleotide variation in the X2 box would be responsible for a differential regulation between HLA-A and HLA-B.3 It was concluded that the X2 box of HLA-A did not bind ATF/CREB factors, leading to a lack of or weaker induction of this locus by CIITA and that an interaction and synergy between CIITA and RelA would compensate for the lack of ATF/CREB binding. Here we present data that, similar to HLA-B, the X2 box of HLA-A binds ATF/CREB and mediates, as part of the SXY regulatory module, CIITA-induced transactivation.4 5 This demonstrates that the X2 box is not the basis for locus-specific regulation of HLA-A and HLA-B genes. Our methods were as follows:
Plasmids: Luciferase reporter plasmids used contained a 228 bp BglII-AhaII HLA-A*0201promoter fragment (pGL3-A230), a 140 bpPpuMI-AhaII HLA-A*0201 promoter fragment (pGL3-A140), or a 269 bp AspI-AhaIIHLA-B*0702 promoter fragment (pGL3-B250) cloned into pGL3-Basic (Promega, Madison, WI), as described.4 The X2 box mutant constructs (pGL3-AmX2 and pGL3-BmX2) contained a 4 bp mutation in the X2 box region (HLA-A2: TCACGACGCG>TCAAACAGCG; HLA-B7: TCGTGACGCG>TCGGACAGCG) were generated by overlap extension polymerase chain reaction (PCR), as described previously.5The expression vectors pRSV0-RelA (p65) and pREP4-CIITA were described previously.4 6
Transient transfection: Cell lines and the calcium phosphate precipitation method of transfection were described previously.5 In each of 4 wells of a 6-well plate, 0.2 × 106 Tera-2 cells were transfected with a DNA precipitate containing 1 μg firefly luciferase pGL3 reporter plasmid, 0.5 μg pREP4-CIITA, and/or 1 μg pRSV0-RelA (p65) and 0.2 μg Renilla luciferase control plasmid (pRL-SV40 or pRL-RSV). Cells were harvested 3 days after transfection. Luciferase activity was determined using the dual luciferase reporter assay system (Promega) and a luminometer (Tropix, Badford, MA).
Electrophoretic mobility gel shift assay (EMSA): Preparation of nuclear extracts and EMSA was performed as described previously.5 The nucleotide sequence of the X2 probe of HLA-A*0201 and HLA-B*0702 were: X2A, GGATACTCACGACGCGGACC; X2B, GGATACTCGTGACGCGTCCC. For the supershift assays, 1 μg of each antibody (Ab) was added 30 minutes after adding the probe and incubated for 1 hour at 4°C. The Abs used were directed against ATF/CREB (sc-270x), ATF1 (sc-243x; Santa Cruz Biotechnology, Santa Cruz, CA). The specific antisera against CREB (anti-CREB1) was previously described.7
Based on this, we achieved the following results:
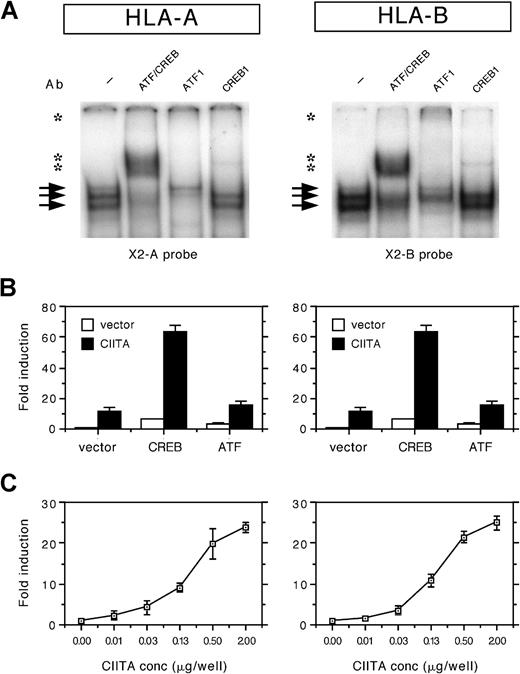
The X2 box of both HLA-A and HLA-B is an ATF/CREB binding site:The X2 box is highly conserved and displays relatively little nucleotide variation amongst the different HLA class I loci.1 However, the X2 box region of HLA-B (CTCGTGACGCG) is divergent in HLA-A (CTCACGACGCG).
The X2 probes of both loci were found to form a number of protein/DNA complexes using nuclear extracts of HeLa cells in EMSAs, albeit that the complex was slightly weaker to the X2 box of HLA-A compared to HLA-B (Figure 1A). The complexes binding to the X2 box of HLA-A and HLA-B were supershifted with the general ATF/CREB Ab and with the Abs specific for ATF1 and CREB1 (Figure 1A). Complex formation was similar in Tera-2 and in B cells (not shown). This suggests that there is no significant difference in binding of these ubiquitously expressed factors.
CREB1 and ATF1 bind the X2 box of HLA-A and HLA-B and are partners in the CIITA-induced transactivation.
(A) EMSA analysis of nuclear extracts from HeLa cells incubated with the X2 probes of HLA-A and HLA-B (X2A and X2B probes). Arrows indicate protein/DNA complexes. Supershifts were observed for ATF/CREB, ATF1, and CREB1 (indicated by asterisks). (B) Transient cotransfection assay of HLA-A and HLA-B promoter–driven luciferase reporter constructs (1 μg/well) with Rc/RSV-CREB (1 μg/well), Rc/RSV-ATF1 (1 μg/well), and/or pREP4-CIITA (0.5 μg/well) in Tera-2 cells. Normalized luciferase activity values are expressed as mean fold induction ± SD of n = 4. (C) Transient cotransfection assay of HLA-A and HLA-B promoter constructs with a range of CIITA concentrations (0, 0.01, 0.03, 0.13, 0.5, and 2 μg/well) in Tera-2 cells. Normalized luciferase activity values are expressed as mean fold induction ± SD of n = 4.
CREB1 and ATF1 bind the X2 box of HLA-A and HLA-B and are partners in the CIITA-induced transactivation.
(A) EMSA analysis of nuclear extracts from HeLa cells incubated with the X2 probes of HLA-A and HLA-B (X2A and X2B probes). Arrows indicate protein/DNA complexes. Supershifts were observed for ATF/CREB, ATF1, and CREB1 (indicated by asterisks). (B) Transient cotransfection assay of HLA-A and HLA-B promoter–driven luciferase reporter constructs (1 μg/well) with Rc/RSV-CREB (1 μg/well), Rc/RSV-ATF1 (1 μg/well), and/or pREP4-CIITA (0.5 μg/well) in Tera-2 cells. Normalized luciferase activity values are expressed as mean fold induction ± SD of n = 4. (C) Transient cotransfection assay of HLA-A and HLA-B promoter constructs with a range of CIITA concentrations (0, 0.01, 0.03, 0.13, 0.5, and 2 μg/well) in Tera-2 cells. Normalized luciferase activity values are expressed as mean fold induction ± SD of n = 4.
ATF/CREB contribute to the CIITA induced transactivation of HLA-A and HLA-B: The crucial dependency of CIITA transactivation on the X2 box of HLA-A and HLA-B is revealed from mutational analysis (not shown; see Gobin et al5). In order to evaluate the role of ATF/CREB factors in the transactivation of HLA-A and HLA-B genes, transient transfection assays were performed with CREB1, ATF1, and CIITA. Expression of exogenous CREB1 or ATF1 enhanced CIITA-induced transactivation of HLA-A and HLA-B (Figure 1B). Both CREB1 and ATF1 also enhanced the basal level of HLA-A and HLA-B transactivation (Figure 1B). This demonstrates that CREB1 and ATF1 are functional partners of the CIITA route of transactivation and that the contribution of ATF/CREB to CIITA-induced transactivation is similar in both loci.
Similar transactivation kinetics of HLA-A and HLA-B by CIITA:In order to test whether the CIITA-induced transactivation of HLA-A and HLA-B displayed locus-specific differences, promoter activity was tested with a range of CIITA concentrations. In Tera-2 cells, HLA-A and HLA-B were induced by CIITA to a similar extent at all concentrations tested (Figure 1C). It should be noted that, in transient transfection experiments in several other cell types, frequently a slightly lower fold induction by CIITA was found for the HLA-A promoter (maximal 50% of HLA-B). However, this difference was generally related to the higher basal level of promoter activity of HLA-A, whereas the absolute luciferase values after CIITA induction were similar for HLA-A and HLA-B. Thus there are no significant locus-specific differences in HLA-A and HLA-B transactivation by CIITA.
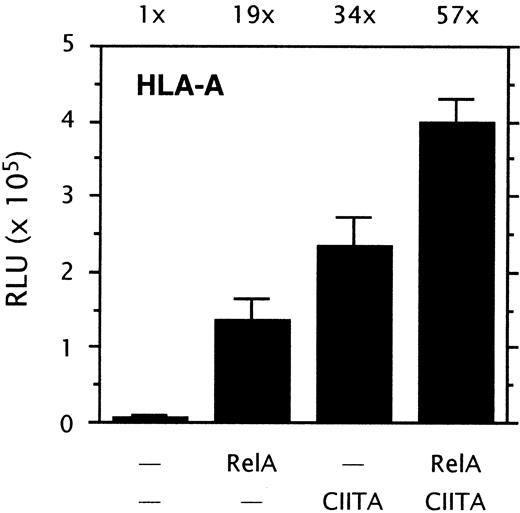
The NF-κB and CIITA induced routes of HLA-A transactivation are not interdependent: A possible interdependency between the NF-κB and CIITA-induced routes of HLA-A transactivation was tested in transient transfection experiments. RelA strongly transactivated HLA-A, and cotransfection of RelA with CIITA further increased the level of transactivation (Figure2). The transactivation of HLA-A by RelA and CIITA was generally additive rather than synergistic. Transactivation of HLA-A was not reduced by mutation of the X2 box, since the X2 box mutant construct pGL3-AmX2 was induced by NF-κB to a similar extent as the wild-type promoter construct pGL3-A230 (not shown). Cotransfection of CIITA with a HLA-A promoter construct lacking the upstream regulatory sequences (enhancer A, ISRE) showed that CIITA transactivation was not impaired by the lack of the upstream κB sites of enhancer A (not shown; see Gobin et al4). Together, this demonstrates that the NF-κB and CIITA-induced routes of HLA-A transactivation are not interdependent.
NF-κB and CIITA-induced HLA-A transactivation.
Transient cotransfection assay of HLA-A promoter driven luciferase reporter constructs pGL3-A230 (1 μg/well) with pRSV0-RelA (1 μg/well), and/or pREP4-CIITA (0.5 μg/well) in Tera-2 cells. Normalized luciferase activity values are expressed as mean ± SD of n = 4. Stimulation indexes are indicated above the graph.
NF-κB and CIITA-induced HLA-A transactivation.
Transient cotransfection assay of HLA-A promoter driven luciferase reporter constructs pGL3-A230 (1 μg/well) with pRSV0-RelA (1 μg/well), and/or pREP4-CIITA (0.5 μg/well) in Tera-2 cells. Normalized luciferase activity values are expressed as mean ± SD of n = 4. Stimulation indexes are indicated above the graph.
In conclusion, the regulation of HLA class I expression is mediated through several regulatory pathways. The SXY regulatory module is the mediator of transactivation by the MHC enhanceosome.1,2,4,5 Here we demonstrate that the X2 box of HLA-A binds several proteins of the ATF/CREB family of transcription factors and, as such, participates in the formation of the multiprotein complex on XY DNA. This enables CIITA-induced transactivation of HLA-A to levels that are similar to HLA-B. Transactivation by NF-κB through the upstream κB sites of enhancer A is strengthened by cotransfection of CIITA, which is suggestive of a cooperative transactivation. However, these 2 routes of HLA-A transactivation are not interdependent. Thus our data demonstrate that there are no significant locus-specific differences in HLA-A and HLA-B transactivation by CIITA. However, the upstream regulatory region containing the enhancer A and ISRE displays important structural and functional differences that contribute to the locus-specific expression of HLA class I molecules.6 8-10
Regulation of HLA class I loci by CIITA
The widespread expression of MHC class I genes might lead them to be considered as “housekeeping” in comparison with other immune response genes, but it is evident that they are subject to complex controls that provide for a wide spectrum of expression levels according to locus, cell type, developmental stage, and the cytokine milieu. Several groups reported the interesting finding that CIITA, an IFNγ-induced transcription factor identified through its regulation of MHC class II genes, is also a potent activator of class I genes.1-1,1-2 The action of CIITA was found to be mediated by a class I promoter region that had been shown previously to be homologous to the class II promoter1-3 and to have the properties of a locus control region1-4. But Gobin et al reported that, while the promoter was sufficient to mediate virtually full transactivation by CIITA, the upstream enhancer (ENH) and interferon response element (IRE) were required in addition to the promoter α site for induction by IFNγ.2(figs 2,5)Although one interpretation is that CIITA is not involved in the IFNγ response, a more plausible hypothesis was that under normal conditions CIITA acts in cooperation with factors binding to the ENH/IRE, since cytokine stimulation does not result in the high levels of CIITA achieved by overexpression in transient transfections. To test this, I performed titration series of the candidate factors and found greater than additive effects with IRF-1 and particularly with RelA.1-5 Gobin and van den Elsen's letter depicts findings with only one dose of each vector, which is maximal for CIITA and undefined for RelA; so it is not possible to make meaningful conclusions regarding interactions. Their use of internal controls driven by viral promoters that are likely to be subject to the effects of the introduced factors also complicates comparison of expression levels.
One aspect of the original paper dealt with the differential regulation of HLA-A and -B. Despite their strong homologies, these loci differ in key bases that have been shown by me and others to affect the binding and function of a number of transcription factors (see Girdlestone1-6). In light of the critical role of the promoter α site, it was striking to me that the sequence of the HLA-A site differs from that of other HLA and H2 class I genes, and does not match any empirically determined binding sites for CREB/ATF factors (TRANSFAC). Under my EMSA conditions I did not find appreciable binding of nuclear factors to the HLA-A sequence,1-5 but Gobin and van den Elsen have now reported its interaction with ATF1/CREB1. They do mention that the complex was “slightly weaker,” but standard EMSA conditions are not quantitative. It appears that they are using probe concentrations about 10-fold higher than used for my experiments, and this would serve to drive the formation of lower affinity complexes. Cold-competition titration series or binding studies with purified proteins will be required to assess the differences in affinity of the CREB/ATF factors for the HLA-A and -B α sites.
I certainly did not mean to suggest in the original paper that CREB/ATF factors were not involved in HLA-A expression; indeed, I mentioned as “data not shown” that a dominant-negative CREB1 blocked expression of both HLA-A and -B reporters.5(p3807) I also reported that HLA-A and -B loci are expressed similarly in HeLa cells,5(fig2E) a finding reproduced in Gobin and van den Elsen's letter. However, the inverted CAAT box was also found to be relatively unimportant in HeLA but absolutely required for promoter activity in MOLT4 cells,1-5 providing another indication that that there are differences in the nature of the class I “enhanceosome” that forms in different cell types.
What I hoped to convey in the original paper was that expression levels of class I genes represent the collective actions of numerous factors and that their relative binding and/or transcriptional activities differ according to the locus and cell type. What is important now is to address the functional relevance of these differences and to determine how they may contribute to the autoimmune destruction of tissues expressing inappropriate levels of class I proteins or to the preferential role of HLA-B in graft rejection.
References
Supported by the Netherlands Organisation for Research (NWO grant 900-09-200) and the Netherlands Foundation for the Support of Multiple Sclerosis Research (96-248 MS). S.J.P.G. is a fellow of the Royal Netherlands Academy of Arts and Sciences.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal