Tumor cells of patients with cutaneous T-cell lymphoma (CTCL) have the cell surface phenotype of mature T-helper lymphocytes, and it may be impossible to differentiate them from nonmalignant lymphocytes in skin and blood. Until now, no specific cell membrane marker of CTCL has been reported. In the current study, it is reported for the first time that CTCL cells express the major histocompatibility complex class I binding p140–killer cell immunoglobulin-like receptor, which has been described on a minor subset of natural killer lymphocytes and on a marginal circulating CD8+ T lymphocyte subset. Interestingly, the molecular characterization of this KIR expressed by CTCL allowed us to isolate a novel allelic form of p140–KIR3DL, resulting in 4 amino acid substitutions, 3 in the extracellular immunoglobulin-like domain of the protein and one in the cytoplasmic region. This finding is likely to be important both for the pathophysiology and for the clinical treatment of patients with CTCL.
Introduction
Cutaneous T-cell lymphomas (CTCL) are a heterogeneous group of lymphomas primarily involving the skin.1 Mycosis fungoides (MF) is characterized by skin invasion of clonally derived malignant CD4+ T lymphocytes that phenotypically resemble mature T-helper cells. A more aggressive form of CTCL develops when the malignant cells become nonepidermotropic and are associated with extracutaneous involvement. Sézary syndrome (SS) is a more aggressive form of CTCL characterized by a clonal expansion of CD4+/CD45RO+ T cells and the appearance of these malignant T cells in the blood.2,3 The biology of the disease remains poorly understood; it is difficult to identify the malignant cell because of the lack of specific cell surface markers. Thus, in cutaneous lesions, it is difficult to distinguish CD4+ CTCL cells from reactive infiltrating CD4+T lymphocytes.4-6 We previously reported a unique CD4+ T-cell line derived from CTCL lesions.6We demonstrated that the cell line and the in vivo tumor cells expressed an identically sized, complementarily determining region 3 of T-cell receptor (TCR)-Vβ transcripts. More recently, we functionally characterized an IL-7–dependent CD4+CD8αα+tumor T-cell line isolated from the blood of another patient with cutaneous erythrodermic CTCL.7 This tumor T-cell line was identical to the major circulating T-cell populations, as demonstrated by the expression of TCR-Vβ22 and the identity of the TCRβ-VDJ sequences.
Here, we show that these 2 different CTCL lines express the p140–KIR3DL2 inhibitory receptor for HLA-A alleles. Importantly, this receptor was detected on freshly isolated tumor cells derived from the same patients. Moreover, the p140 was also co-expressed by a major subset of CD4+ lymphocytes in 7 other patients with SS, and by tumor skin CD4+ cells in 2 additional patients with advanced MF. The p140–KIR3DL2 is detected in healthy persons on a minor natural killer (NK) cell subset8-10 and on rare peripheral blood CD3+ CD8+cells.11,12 P140 is a member of the killer immunoglobulin-like receptor (KIR) family known to negatively modulate NK-mediated cytotoxicity on recognition of different groups of HLA class I alleles. In particular, p140 in normal NK cells has been shown to inhibit NK-mediated lysis after interaction with some HLA-A alleles, such as HLA-A3 and HLA-A11. T cells obtained from skin in other dermatologic diseases, such as inflammatory skin diseases and toxic epidermal necrolysis, did not express this receptor.13Thus, our current findings suggest that p140 represents a suitable marker on CD4+ cells for the identification of CTCL.
Patients, materials, and methods
Patients
After informed consent from patients and approval by an ethics committee were obtained, we obtained skin and blood samples from 11 patients with CTCL. Eight patients had Sézary syndrome, with 10% to 45% circulating Sézary cells in the blood. In 7 patients, the phenotype of tumor cells was CD3+, CD4+, CD8−. In one patient (patient Pno), the phenotype was CD3+, CD4+, CD8αα+. Three patients had transformed mycosis fungoides (Lez, Cou, Bic) with disseminated skin tumors with a CD3+, CD4+, CD8− phenotype. No patients were previously treated with chemotherapy.
Isolation of tumoral lymphocytes
Fresh CTCL tumor cells were obtained from tumor fragments mechanically dispersed into single-cell suspensions.6Mononuclear cells were then washed and frozen in human serum plus 10% dimethyl sulfoxide for later use. For patients with SS, the mononuclear blood cells were isolated by Ficoll-Isopaque (Pharmacia Fine Chemicals, Piscataway, NJ).
Long-term culture of tumor cell lines
We established the long-term culture of Pno cell line (TCRVβ22+, CD3+, CD4+, CD8αα+, major histocompatibility complex (MHC) class I+, MHC class II−) in vitro from the peripheral blood of the patient as previously described.6,7 The human leukocyte antigen (HLA) haplotype of patient Pno is HLA-A1, A2, and HLA-B18, B57. We demonstrated that both the malignant clone circulating in the patient blood and the derived cultured T-cell line were identical for their cell surface phenotype and for their size and sequence of the TCRβ VDJ region.7 The Cou-L cell line (TCRVβ13+) was cultured in vitro with rIL-2 for more than 3 years. It corresponds to a subclone of the CD4+ Cou-LS CTCL line previously described.6 The Cou-L cell line, the original TCRVβ13+, CD4+ Cou-LS CTCL, and the tumor cells freshly isolated from the skin shared the same size and sequence of the TCRβ VDJ region.6 The HLA haplotype of patient Cou is HLA-A1, A2 and HLA-B5 (51) B35.
Monoclonal antibodies and flow cytometry studies
One- and 2-color immunofluorescence analysis was performed as previously described.6 The monoclonal antibodies (mAbs) anti-CD3, anti-CD4, anti-MHC class I, and anti-MHC class II were produced locally. The anti-TCRVβ13+ mAb was purchased from BIOadvance (Emerainville, France), and the anti-TCRVβ22+ mAb was obtained from Beckman-Coulter (Marseille, France). The anti-CD8 αβ 2ST8.5H7 mAb was kindly provided by Dr E. L. Reinherz (DFCI, Boston, MA). Q66 (IgM, anti-p140),8 AZ158 (IgG2a, recognizing both p70–NKB1 and p140), Z27 (IgG1, anti-p70/NKB1), EB6 (IgG1, anti-p58.1/p50.1), GL183 (IgG1, anti-p58.2/p50.2), XA185 (IgG1, anti-CD94), and Z199 and Z270 (IgG2b and IgG2a respectively, anti-NKG2A) were produced in one of our laboratories.11 Dr B. Malissen (INSERM-CNRS, Marseille Luminy, France) provided the B9.4 (IgG2b, anti-CD8) mAb.
Biochemical characterization
Then 20 × 106 cells were incubated (15′ at 20°C) in 1 mL PBS pH 8 containing 250 μg EZ-Link Sulfo-NHS-LC-LC-Biotin (Pierce, Rockford, IL) and washed 3 times in washing buffer (10 mM Tris pH 8, 0.14 M NaCl). Cells were lysed in 1% NP-40 and immunoprecipitated with Sepharose–PA (Pharmacia Biotech, Piscataway, NJ)-coupled AZ158 (IgG2a, anti-p70/p140). Samples were analyzed by discontinuous SDS-PAGE either undigested or digested withN-glycosidase F (Boehringer Mannheim GmbH, Mannheim, Germany) and transferred to Immobilon P (Millipore, Bedford, MA). After staining the cells with Neutravidin (Pierce), we used the Renaissance Chemiluminescence Kit (NEN, Boston, MA) for detection. NK cell clones were obtained by limiting dilution as previously described.8
RT-PCR analysis
Total RNA was extracted from CTCL cell lines Pno and Cou-LCD8 αα using RNA-Clean System (AGS GmbH, Heidelberg, Germany). cDNA synthesis was performed using oligo-dT priming. Primers used for cDNA amplification of the complete open-reading frame of KIR displaying 3 immunoglobulin-like domains (1395 bp) were the following: 5′CATGT(CT)GCTCA(CT)GGTCGTC (Ig3 UP) and 5′ GGTTTTGAGACAGGGCTG (Ig3 DOWN). Amplification was performed for 30 cycles (30 seconds at 94°C, 30 seconds at 55°C, 30 seconds at 72°C), followed by a 7-minute incubation at 72°C, using AmpliTAQ (PerkinElmer Applied Biosystems, Foster City, CA). PCR products were subcloned into pcDNA3.1/V5-His-TOPO vector (Invitrogen, Carlsbad, CA). DNA sequencing was performed using d-Rhodamine Terminator Cycle Sequencing Kit and a 377 Applied Biosystems Automatic Sequencer (PerkinElmer Applied Biosystems).
Transient transfections
COS-7 cells were transfected with pcDNA3.1 TOPO-KIR3D cl.24 or with pCR3-cl.1.18 using Fugene 6 (Roche, Monza, Italy). Briefly, cells were seeded at 5 × 105/plate; 24 hours later they were incubated with 6 μg plasmid and 10 μL Fugene-6 reagent in Dulbecco minimum essential medium/10% fetal calf serum. After 48 or 72 hours, transfected cells were used for cytofluorometric analysis. Cell transfectants were stained with Q66 and AZ158 mAbs, followed by a phycoerythrin-conjugated goat antibody to mouse IgG2a or IgM and analyzed by flow cytometry using FACSort (Becton Dickinson, San Jose, CA).
Results
CTCL cell lines are stained by mAbs to the p140
Two long-term CTCL tumor lines Pno (labeled with anti-TCR-Vβ22 mAb)7 and Cou-L (labeled with anti-CD3 mAb)6were analyzed for reactivity with different anti-KIR mAbs. We found that both cell lines were reactive with mAbs Q66 (Figure1) and AZ158 (not shown), both recognizing p140. In contrast, these cell lines did not express other inhibitory receptors specific for HLA class I molecules, including p58.1, p58.2, p70 KIRs, and the CD94/NKG2A lectin-like receptor (data not shown and 7).
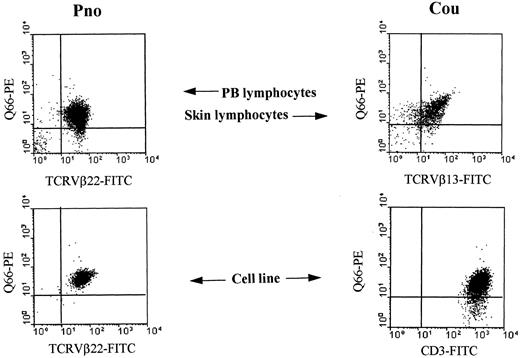
Detection of p140/KIR3DL2 molecules on CTCL cell lines and on fresh tumor lymphocytes.
Two long-term CTCL tumor lines (Pno, Cou-L) and fresh tumor cells isolated from the blood of patient Pno (PB lymphocytes–Pno) and from tumoral skin fragments of patient Cou (skin lymphocytes) were analyzed by 2-color immunofluorescence and FACS analysis with the indicated mAbs followed by isotype-specific fluorescein isothiocyanate or phycoerythrin-conjugated goat antimouse second reagents.6 7 Contour plots were divided into quadrants representing unstained cells (lower left), cells with only red fluorescence (upper left), cells with red and green fluorescence (upper right), and cells with only green fluorescence (lower right).
Detection of p140/KIR3DL2 molecules on CTCL cell lines and on fresh tumor lymphocytes.
Two long-term CTCL tumor lines (Pno, Cou-L) and fresh tumor cells isolated from the blood of patient Pno (PB lymphocytes–Pno) and from tumoral skin fragments of patient Cou (skin lymphocytes) were analyzed by 2-color immunofluorescence and FACS analysis with the indicated mAbs followed by isotype-specific fluorescein isothiocyanate or phycoerythrin-conjugated goat antimouse second reagents.6 7 Contour plots were divided into quadrants representing unstained cells (lower left), cells with only red fluorescence (upper left), cells with red and green fluorescence (upper right), and cells with only green fluorescence (lower right).
Tumor T lymphocytes freshly isolated from patients with CTCL are stained by anti-p140 mAbs
To determine whether p140 was expressed by freshly isolated tumor cells, we tested the reactivity of Q66 mAb with uncultured tumor cells isolated from the blood of SS patient Pno and from tumoral skin fragments of MF patient Cou. We found that most tumor cells were stained by this mAb (Figure 1). In particular, we observed that most TCRVβ22+ tumor lymphocytes isolated from the blood of patient Pno were reactive with Q66 antibody and that most of the TCRVβ13+ tumor lymphocytes isolated from the skin of patient Cou were stained by the same antibody (Figure 1).
Next, we studied the phenotype of tumor T lymphocytes from the blood of 7 additional patients with SS, with malignant cells representing 10% to 45% of circulating CD4+ lymphocytes, and tumor T lymphocytes isolated from skin tumors of 2 other patients with a MF. Remarkably, all patients tested exhibited a significant population co-expressing CD4 and p140 (Table 1). It should be noted that all Q66+ cells were included in the CD4+ cell population (data not shown). Thus, the expression of p140, which in healthy persons is restricted to subsets of lymphocytes from the NK and CD8+ populations,6appears to be a characteristic of CTCL tumor CD4+ T lymphocytes, both in the skin and in the blood. As control, skin T lymphocytes derived from another dermatologic disease, toxic epidermal necrolysis, which were shown to contain small percentages of various KIR-expressing T lymphocytes,13 failed to express p140 (data not shown).
Anti-Q66 mAb stained CD4+ lymphocytes isolated from the skin of patients with transformed mycosis fungoides and from blood of patients with Sézary syndrome
| Patient samples . | Percentage of positive cells with . | |
|---|---|---|
| anti-CD4 mAb . | anti-CD4 mAb + anti-Q66 mAb . | |
| Mycosis fungoides | ||
| Lez | 65 | 29 |
| Bic | 53 | 44 |
| Sézary syndrome | ||
| Bri | 85 | 35 |
| Bar | 90 | 18 |
| Att | 95 | 35 |
| Ros | 98 | 15 |
| Can | 78 | 36 |
| Pet | 45 | 9 |
| Riv | 98 | 19 |
| Patient samples . | Percentage of positive cells with . | |
|---|---|---|
| anti-CD4 mAb . | anti-CD4 mAb + anti-Q66 mAb . | |
| Mycosis fungoides | ||
| Lez | 65 | 29 |
| Bic | 53 | 44 |
| Sézary syndrome | ||
| Bri | 85 | 35 |
| Bar | 90 | 18 |
| Att | 95 | 35 |
| Ros | 98 | 15 |
| Can | 78 | 36 |
| Pet | 45 | 9 |
| Riv | 98 | 19 |
Molecular characterization of the p140 receptor expressed by CTCL
The Pno and Cou-L cell lines were surface labeled with biotin, and cell lysates were immunoprecipitated with an anti-p140 (AZ158 mAb). As shown in Figure 2, this mAb immunoprecipitated, from an NK clone and from the Pno and Cou-L cell lines, a molecule with a molecular mass of approximately 70 kd under reducing conditions. Treatment with N-glycosidase revealed a protein backbone of approximately 50 kd with a slightly higher mobility for Pno and Cou-L than the NK clone. These data suggested that the p140 inhibitory receptor expressed by these CTCL tumor cell lines could be almost similar to that previously detected on normal NK cells.8
Biochemical analysis of p140 molecules.
The NK clone AM61 (p140+/p70−), derived from a healthy donor, and the cell lines Pno and Cou-L were surface labeled with biotin and immunoprecipitated with anti-p140 mAb. Samples were treated (+) or not (−) with N-glycosidase F and analyzed in 8% SDS-PAGE under reducing conditions. Molecular weight markers (kd) are indicated on the right.
Biochemical analysis of p140 molecules.
The NK clone AM61 (p140+/p70−), derived from a healthy donor, and the cell lines Pno and Cou-L were surface labeled with biotin and immunoprecipitated with anti-p140 mAb. Samples were treated (+) or not (−) with N-glycosidase F and analyzed in 8% SDS-PAGE under reducing conditions. Molecular weight markers (kd) are indicated on the right.
Next, we determined whether the cDNA encoding the molecule recognized by Q66 and AZ158 mAbs on Pno and Cou-L CTCL cell lines corresponded to one of the already described cDNAs encoding p140. To this end, RT-PCR was performed on RNA derived from these cell lines using a set of primers able to amplify all cDNA encoding KIR with 3 immunoglobulin-like domains. From the Pno cell line, we isolated a full-length cDNA, termed KIR3D cl.24. Comparison of its nucleotide sequence with DNA sequences coding for all KIR characterized by 3 extracellular immunoglobulin-like domains revealed that KIR3D cl.24 represents a novel transcript for the p140 gene. In particular, its nucleotide sequence displays 5 differences compared to the previously described cl. 1.1 cDNA,8 resulting in 4 amino acid substitutions in the mature protein (Figure3). Three of the 4 substitutions are found in the extracellular immunoglobulin-like domain (positions 20, 92, and 111 of the mature protein), whereas the other is located in the cytoplasmic region (position 401). RT-PCR performed on Cou-L CTCL cell line revealed 2 different allelic forms of p140, one corresponding to cl. 1.1 cDNA and the other identical to KIR3D cl.24 (isolated from Pno cell line). The cDNA derived from the CTCL cell lines were then transiently transfected in COS-7 cells. As expected, all cell transfectants were brightly stained by Q66 and AZ158 mAbs, whereas they were unreactive with the p70/KIR3DL1-specific Z27 mAb used as a negative control (data not shown). Finally, RT-PCR was performed on RNA extracted from PBL derived from 3 patients with SS (including patient Pno) and from skin-derived T cells of one additional patient with MF. In addition, in these samples one or another allelic isoform of p140 described above could be identified.
Amino acid sequence alignment of KIR3D cl. 24- and KIR3DL2 cl. 1.1-encoded proteins.
Amino acids corresponding to the signal peptide are in lower case letters, and the transmembrane region is underlined in the consensus sequence. Amino acids identical to the consensus sequence are indicated by dots. The GenBank/EMBL accession number for KIR3D cl. 24 isAJ276125.
Amino acid sequence alignment of KIR3D cl. 24- and KIR3DL2 cl. 1.1-encoded proteins.
Amino acids corresponding to the signal peptide are in lower case letters, and the transmembrane region is underlined in the consensus sequence. Amino acids identical to the consensus sequence are indicated by dots. The GenBank/EMBL accession number for KIR3D cl. 24 isAJ276125.
KIR3D cl.24 may represent an additional polymorphism in the p140 gene and could be considered a novel allelic form of p140 receptor. However, further investigation is required to assess the allelic nature of KIR3D cl.24 and to exclude the possibility that somatic mutations may play a role in the generation of KIR3D cl.24 sequence. On the other hand, this seems unlikely because, as mentioned above, KIR3D cl.24 sequence could be amplified from different unrelated patients.
Discussion
Skin lesions in CTCL contain a heterogeneous lymphocytic infiltrate composed of malignant T cells, which are most often CD4+, and nonneoplastic tumor-infiltrating T lymphocytes. We previously reported CD4+ cytotoxic tumor-infiltrating lymphocytes specifically directed against the autologous malignant CTCL CD4+ cell line.6 However, because no tumor-restricted cell surface structure, aside from the clonotypic TCR expressed by tumor cells, has been identified on CTCL, it is difficult to use standard methods to distinguish malignant from nonmalignant, reactive CD4+ lymphocytes.
In the current study, we report for the first time that tumor cells from patients with MF and SS express p140/KIR3DL2. This receptor has been identified in skin and blood tumor cells from patients with CTCL and in 2 long-term culture CTCL lines. Two-color fluorescence analysis indicated that p140 expression is restricted to T cells characterized by a given TCRβ-VDJ previously identified on tumor cells. Thus, p140 allows rapid identification of tumor cells from tumor-reactive cells among the T lymphocyte CD4+ population. This could be particularly useful in patients with SS, in whom the tumor T-cell clone is not always easily identified within the peripheral blood CD4+ lymphocyte population. Moreover, during the course of the disease or after treatment, it is crucial to assess whether an increase of the CD4+ population results from an expansion of the tumor cell population or of reactive T lymphocytes. Because the anti-p140 mAbs appear to recognize tumor cells in all patients with CTCL analyzed, they may be considered a suitable tool for the direct identification of CTCLs. In addition, the use of p140 co-expression on CD4+ CTCL cells provides a unique tool to distinguish malignant cells from normal cells in every patient. This is in contrast to TCR determination that is unique to each patient. Moreover, p140 may represent a possible target for the development of novel specific immunotherapy for CTCL.
Previous studies indicated that this receptor was able to generate inhibitory signals on recognition of HLA-A3 and HLA-A11 alleles.8 It is of note, however, that p140 expression in the various patients analyzed is apparently independent of their own HLA class I haplotype. It is possible that the product of the novel transcript for p140 may recognize HLA-A alleles different from those reacting with the described p140 receptor. Additional studies will be required to verify this possibility. Nevertheless, the actual role of p140 in the pathophysiology of CTCL is an important feature to be studied by taking into account the potential role of this receptor in the tolerance to self.14 In conclusion, the current study demonstrates for the first time the expression of p140 in CD4+ CTCL cells and the isolation of a novel transcript encoding this receptor in tumor cells. This finding is likely to be an important new issue, both for the pathophysiology and the clinical management of patients with CTCL.
Supported by grants from Inserm, Paris XII University, ARC, Société Française de Dermatologie, Laboratoires La Roche Posay, Dermatologiques Evaux, and Académie de Médecine, Associazione Italiana per la Ricerca sul Cancro, Istituto Superiore di Sanità, Ministero della Sanità, and Ministero dell' Università e della Ricerca Scientifica e Tecnologica, Consiglio Nazionale delle Ricerche, Progetto Finalizzato Biotecnologie, and by grant E.0892 from Telethon-Italy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Martine Bagot, Faculté de Médecine, Inserm U448, 8 Avenue du Général Sarrail, 94010 Créteil, France; e-mail:martine.bagot@hmn.ap-hop-paris.fr.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal