Abstract
Chromosome 11q deletions are frequently observed in chronic lymphocytic leukemia (CLL) in association with progressive disease and a poor prognosis. A minimal region of deletion has been assigned to 11q22-q23. Trinucleotide repeats have been associated with anticipation in disease, and evidence of anticipation has been observed in various malignancies including CLL. Loss of heterozygosity at 11q22-23 is common in a wide range of cancers, suggesting this is an unstable area prone to chromosome breakage. The location of 8 CCG-trinucleotide repeats on 11q was determined by Southern blot analysis of a 40-Mb YAC and PAC contig spanning 11q22-qter. Deletion breakpoints in CLL are found to co-localize at specific sites on 11q where CCG repeats are located. In addition, a CCG repeat has been identified within the minimal region of deletion. Specific alleles of this repeat are associated with worse prognosis. Folate-sensitive fragile sites are regions of late replication and are characterized by CCG repeats. The mechanism for chromosome deletion at 11q could be explained by a delay in replication. Described here is an association between CCG repeats and chromosome loss suggesting that in vivo “fragile sites” exist on 11q and that the instability of CCG repeats may play an important role in the pathogenesis of CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by a clonal proliferation of mature B-lymphocytes increasing in incidence logarithmically with age in adults.1 Sporadic CLL may have a hereditary component because several studies report an elevated risk for hematologic malignancy2,3 and other cancers in first-degree relatives. Familial clustering of CLL does occur; when it does, it is multigenerational and displays vertical transmission. The phenomenon of anticipation, originally reported for inherited neurodegenerative disease, results from the expansion of trinucleotide repeats and describes an increase in severity or earlier age of onset of a disease occurring with each subsequent generation in an autosomal dominant manner.4 However, it is now known to occur in other disorders, including familial leukemias, cancers, and CLL.2,3,5 6
In patients with sporadic CLL, 11q deletions are a common chromosomal abnormality. This subset of patients displays an aggressive form of the disease7-9 with distinct characteristics, including early age of onset and bulky lymphadenopathy9 that constitute rapidly progressive disease with a high mortality rate.7-9Previous reports describe a minimal region of deletion of 2 to 3 Mb within 11q22-23.10,11 This region includes theATM gene, which has been proposed as a tumor suppressor gene in CLL, based on mutations in a proportion of patients.12-14 In mantle cell lymphoma15-17and other lymphoid and myeloid malignancies,18 11q21-23 is also a frequent region of loss. Kobayashi et al19 have mapped the inv(11) breakpoint associated with certain myeloid malignancies to the same region. This involves the DDX10gene.20,21 Loss of heterozygosity (LOH) at 11q21-23 is commonly reported in many solid tumors.22-27 These observations beg the question of why the distal portion of chromosome 11 is an unstable region, prone to breakage and rearrangement, in so many cancers.
Genetic instability is a common feature of many human cancers. Microsatellite instability was first described in hereditary nonpolyposis colon cancer and is attributable to defective mismatch repair causing mutations at microsatellites harboring dinucleotide, trinucleotide, or tetranucleotide repeat sequences.28 This form of genomic instability has been commonly reported in sporadic solid tumors29 but is rare in adult leukemia.30 Mutational expansion of unstable trinucleotide repeat sequences, as another form of instability, has also been described and is the cause of anticipation in several diseases.31 Briefly, trinucleotide repeats are inherently unstable, particularly when they exceed a certain length, and are prone to expansion during vertical transmission.4
Folate-sensitive fragile sites are inherited cytogenetic phenomena that appear as poorly condensed regions of metaphase chromosomes32 and are susceptible to breakage under specific experimental conditions.32,33 Five fragile sites have been characterized at the molecular level, and their expression is due to the extensive expansion of CCG-trinucleotide repeats and an associated hypermethylation of adjacent CpG islands.31Expression of a fragile site in chromosome band 11q23.3 (FRA11B) has been shown to cause partial chromosome loss in a chromosome deletion syndrome (Jacobsen syndrome).34 In some of these patients, chromosome 11 is truncated near the site ofFRA11B, and the deleted chromosome derives from a parent carrying an expanded CCG repeat. Chromosome truncations at an unstable CCG repeat demonstrate an inherited component in the mechanism by which chromosome breakage occurs in vivo.
Trinucleotide repeats are associated with anticipation, which has been observed in malignancy. Chromosome deletion has also been described in association with trinucleotide repeats. These 2 findings raise the possibility that CCG repeats have a role in the development of neoplasia through instability leading to chromosome deletion. Some candidate leukemia oncogenes contain CCG repeats, including theBCR gene35 and theCBFB36 gene involved in inversion 16 of acute myeloid leukemia. In addition, an increased incidence of spontaneous breakage of chromosomes and fragile site expression has been demonstrated in sporadic CLL.37 This study investigates a potential role for CCG repeats in the pathogenesis of CLL. A previously constructed YAC38 and PAC contig39 spanning 11q22 to the telomere was used to analyze the chromosome 11 deletions of patients with sporadic CLL and low-grade non-Hodgkin lymphoma to localize potential tumor suppressor genes and to identify mechanisms for chromosome breakage.
Materials and methods
Patient samples
Metaphase chromosomes were prepared from B-lymphocytes after the stimulation of peripheral blood with phorbol-12-myristate-13-acetate (final concentration, 0.05 μg/mL) (Sigma, St Louis, MO) and by blocking dividing cells in metaphase by the addition of Colcemid (Karyomax; Gibco, Paisley, United Kingdom). Cells were harvested according to standard cytogenetic techniques, fixed in methanol–acetic acid (3:1), and dropped onto glass slides.
DNA was purified from whole blood, the malignant B-cell population, and buccal mouthwashes of patients with chronic lymphoproliferative disorders and from the whole blood of ethnically matched control subjects of similar age, using Qiagen columns, according to the manufacturer's instructions. Malignant B cells were isolated from the blood of 137 patients with CLL and 3 patients with mantle cell lymphoma using CD19+ Dynabeads (Dynal, Oslo, Norway).
Probes
Whole chromosome 11 paints (Vector Laboratories, Peterborough, United Kingdom) were hybridized to the patient material according to manufacturer's instructions. A digoxigenin-conjugated chromosome 11-specific α-satellite probe (Oncor, Gaithersburg, MD) was used to confirm the identity of normal and derivative chromosome 11. Cosmid, PAC, and YAC probes were used in dual-color fluorescence in situ hybridization (FISH) analysis; DNA was labeled with either biotin-16-dUTP or digoxigenin-11-dUTP by nick-end translation.40 Probe detection was performed using fluorescein isothiocyanate-conjugated avidin D and biotinylated goat anti-avidin antibody (both Vector Laboratories) and anti-digoxigenin rhodamine antibody (Boehringer Mannheim, Mannheim, Germany).
Fluorescence in situ hybridization
Metaphase chromosomes were denatured in 70% formamide–2×SSC, and probes were hybridized in a 50% formamide–10% dextran sulfate solution overnight at 37°C. Then 200 ng PAC DNA, 300 ng YAC DNA, or 100 ng cosmid DNA was hybridized per slide in the presence of COT-1 DNA (Gibco). Post-hybridization washes were performed at 45°C in 50% formamide–2×SSC. The chromosomes were counterstained with DAPI (Sigma) in phosphate-buffered saline–anti-fade (Citifluor, London, United Kingdom), and images were captured with a cooled charged-coupled device camera linked to an Apple Macintosh computer with IP Lab Spectrum software (Digital Scientific, Cambridge, United Kingdom).
Cloning and characterization of a CCG-trinucleotide repeat
Southern blot analysis of the YAC contig with a CCG probe was performed using standard protocols.40 Briefly, a 15-base pair oligonucleotide probe (CCG)5 was end-labeled with γ32P-ATP using polynucleotide kinase and hybridized to nylon membranes in a 10% dextran sulfate–5× Denhardt hybridization buffer40 overnight at 50°C. Membranes were washed in 2×SSC–0.1% sodium dodecyl sulfate at room temperature and in 0.2×SSC–0.1% sodium dodecyl sulfate at 55°C for 15 minutes each. A PAC contig was constructed between D11S1897 and D11S2105 by polymerase chain reaction (PCR) of the RPCI1 PAC library pools (provided by the Human Genome Mapping Project Resource Centre, Hinxton, Cambridge, United Kingdom41) with microsatellite markers. Further defining of the position of some PACs within the contig was performed by hybridization of PAC end clones, derived using a previously described PCR-based technique called bubble PCR.42
A 2.8-kb PstI restriction fragment containing a CCG repeat from YAC y975H06 was subcloned into PUC18 and transfected into XL1-blue competent cells (Stratagene, La Jolla, CA). The fragment was sequenced, and the following PCR primers flanking the CCG repeat were constructed (Cruachem, Glasgow, United Kingdom): forward, 5′-GCAGAAAGCGTGGAGTGAAT; reverse, 5′-CTCTGGCAATAGGCTGCGAG. PCR reactions were carried out in a 10μL volume containing 50 ng genomic DNA; 1 μM each primer; 1.5 mM MgCl2; 250 μM each of dATP, dCTP, deaza-dGTP (Boehringer), and dTTP; 10% dimethyl sulfoxide; 2.5 U Taq DNA polymerase (Qiagen, Hilden, Germany); and 2 μCi α32P-dCTP (Amersham, Amersham, United Kingdom). PCR cycling was performed in a Biometra Uniblock thermocycler as follows: 94°C for 5 minutes, followed by 35 cycles of 94°C for 30 seconds; 60°C for 30 seconds; 72°C for 30 seconds, and a final elongation at 72°C for 10 minutes. Samples were loaded onto 6% polyacrylamide denaturing gels and run alongside size markers that consisted of YAC DNA containing (CCG)12 and PAC DNA containing (CCG)6. After electrophoresis, the gel was exposed to X-OMAT AR x-ray film (Kodak, Rochester, NY). Haplotype analysis using the microsatellite markers D11S1302, D11S1294, D11S2106, D11S2003, D11S2221, D11S1300, D11S1347, and D11S1885 was carried out in a similar way.
Results
Definition of a minimal region of deletion on 11q
With a selection of YAC probes covering the 11q21-qter region of chromosome 11,38 FISH was used to define a minimal region of deletion by hybridization to the metaphase chromosome spreads of CLL cells. These were taken from patients with known 11q deletions defined by cytogenetic analysis. Nine patients with B-CLL (identified by morphology and immunophenotyping) were studied. For many patients with CLL, no cytogenetic data were available. To try and further refine the minimal deleted region by analyzing a larger set of patient material, haplotype (microsatellite PCR) analysis was performed. Tumor DNA and DNA from the whole blood or buccal mouthwashes from 60 patients with CLL were examined.
Analysis of the metaphase material using overlapping YAC clones and LOH data of 60 patients with the microsatellite markers D11S1302, D11S1294, D11S2106, D11S2003, D11S2221, D11S1300, D11S1347, and D11S1885 (Figure1) defined a minimum region of 11q deletion in patients with CLL. The size of this deletion is approximately 4 Mb, it involves bands 11q22.3 to 11q23.1, and it lies between D11S2106 and YAC y797E07 (Figure 1). LOH analysis of patient 5 revealed heterozygosity in both the tumor and buccal samples for the marker D11S2106. However, analysis with an adjacent marker D11S2221 (more telomeric) showed heterozygosity in the buccal sample but homozygosity in the tumor sample (Figure 1). The detection of LOH with D11S2221 in this tumor further refines the minimal region of deletion. Our data are in concordance with previously published results10,11 (Figure 1) for the 11q-deleted region in CLL (all of which include the trinucleotide repeat TNR/11q#1; see below). Taken together with the published data, we further reduced the size of the minimal deleted region for CLL. The published data for mantle cell lymphoma16 17 show less concordance (Figure 1).
Localization of the minimal region of deletion and patients' breakpoints on the 40-Mb 11q contig.
The locations of the trinucleotide repeats and the FRA11Bfragile site are indicated by bold type. The various genes and D11S markers in the region are also indicated at the top of the figure, from centromeric to telomeric and from left to right. Below this the location of various YACs and PACs and their relative sizes are indicated by gray boxes together with the relevant identification number, corresponding to the identification number in the text. Patient identification numbers are indicated on the left of black horizontal boxes that correspond to the portion of chromosome 11 present in the patient CLL material, as determined by FISH and haplotype analysis. (+) The nearest retained marker to the deleted region. The unfilled area between the boxes represents the area of deletion. Grey boxes represent the maximum extent of the region in which the breakpoint must be located with the most proximal deleted marker, denoted by (−). Proximally our minimal region of deletion is defined by patient 5, and distally it is defined by patients 3 and 4. The previously published data on the minimal region of 11q deletion for CLL and mantle cell lymphoma are indicated in the same manner, and the disease type (CLL, chronic lymphocytic leukemia; MCL, mantle cell lymphoma) is indicated in the gap representing the deleted region. The hatched box represents an area that was not analyzed.
Localization of the minimal region of deletion and patients' breakpoints on the 40-Mb 11q contig.
The locations of the trinucleotide repeats and the FRA11Bfragile site are indicated by bold type. The various genes and D11S markers in the region are also indicated at the top of the figure, from centromeric to telomeric and from left to right. Below this the location of various YACs and PACs and their relative sizes are indicated by gray boxes together with the relevant identification number, corresponding to the identification number in the text. Patient identification numbers are indicated on the left of black horizontal boxes that correspond to the portion of chromosome 11 present in the patient CLL material, as determined by FISH and haplotype analysis. (+) The nearest retained marker to the deleted region. The unfilled area between the boxes represents the area of deletion. Grey boxes represent the maximum extent of the region in which the breakpoint must be located with the most proximal deleted marker, denoted by (−). Proximally our minimal region of deletion is defined by patient 5, and distally it is defined by patients 3 and 4. The previously published data on the minimal region of 11q deletion for CLL and mantle cell lymphoma are indicated in the same manner, and the disease type (CLL, chronic lymphocytic leukemia; MCL, mantle cell lymphoma) is indicated in the gap representing the deleted region. The hatched box represents an area that was not analyzed.
Whole chromosome 11 paints were used to determine whether the deleted portion of chromosome 11q had been simply deleted from the malignant cell or had been part of a more complex rearrangement, with the 11q region translocated to another chromosome. The results showed a simple deletion of chromosome 11, with no transfer of chromosome 11 material to other regions of the genome.
Identification of a CCG repeat within the minimal deleted region
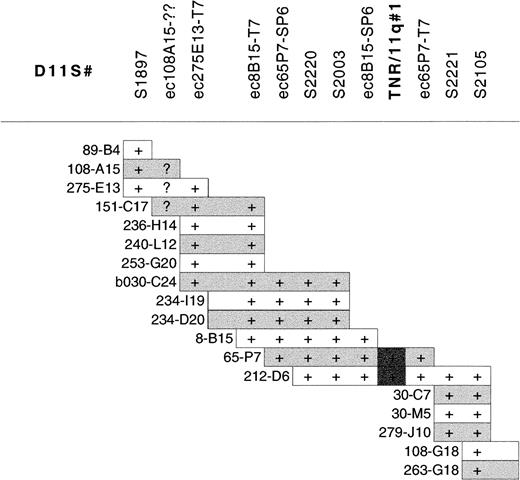
Southern blot analysis of the 40-Mb 11q YAC contig38was performed with a CCG-trinucleotide probe to identify sites of CCG-repeat sequences.39 Nine overlapping YACs (y986C07, y984E06, y891F09, y952H06, y921B11, y795F05, y975H06, y793D09, y920C04), located within the CLL minimal deleted region, were found to contain the same trinucleotide repeat. The high density of microsatellite markers in this region of the YAC contig allowed the accurate localization of this CCG-repeat to between D11S1897 and D11S2105. A PAC contig was constructed between these markers (Figure2) to further refine the location of the repeat. Southern blot analysis of the PAC contig revealed that this CCG repeat was contained within PACs dJ65-P7 and dJ212-D6, localizing it between markers D11S2003 and D11S2221 (Figure 2). We have designated this CCG repeat TNR/11q#1 (trinucleotide repeat on chromosome 11q). TNR/11q#1 is contained within our defined minimal region of deletion in CLL and within previously published critical regions of 11q deletion in CLL10 11 (Figure 1).
PAC contig constructed between D11S1897 and D11S2105 within the minimal region of 11q deletion in CLL showing the localization of TNR/11q#1.
STS markers within the contig are listed without the D11 prefix for clarity. STSs were demonstrated within PACs by PCR. PAC end clones are given the prefix ec, together with the clone name and the end from which they are derived; they were screened against PACs by hybridization. (+) Positive results for PCR of STSs or hybridization with PAC end clones. Overlap of PAC clones in the contig was also determined by cross-hybridization of PACs against each other. (?) Unclear hybridization.
PAC contig constructed between D11S1897 and D11S2105 within the minimal region of 11q deletion in CLL showing the localization of TNR/11q#1.
STS markers within the contig are listed without the D11 prefix for clarity. STSs were demonstrated within PACs by PCR. PAC end clones are given the prefix ec, together with the clone name and the end from which they are derived; they were screened against PACs by hybridization. (+) Positive results for PCR of STSs or hybridization with PAC end clones. Overlap of PAC clones in the contig was also determined by cross-hybridization of PACs against each other. (?) Unclear hybridization.
A 2.8-kb PstI fragment from YAC y975H06 containing the repeat was cloned and sequenced, revealing the presence of (GCC)9TTC(GCC)2 trinucleotide repeat (referred to as (CCG)12 in the text). Oligonucleotide primers flanking TNR/11q#1 were constructed for use in PCR analysis of CCG-repeat length. The repeat was found to be polymorphic in the control population (n = 97; heterozygosity, 49.5%; Table1) with 6 or 12 copies of the CCG trinucleotide representing the most common alleles. Rarer allele sizes include 14 and 15 copies of the CCG trinucleotide and were present in only 5.67% of normal chromosomes.
Genotype data for (CCG)n within the control (normal) and patient populations
| Genotypes . | Controls . | Patients . |
|---|---|---|
| 6,6 | 10 | 11 |
| 6,12 | 37 | 42 |
| 6,13 | 1 | 0 |
| 6,14 | 5 | 3 |
| 12,12 | 38 | 67 |
| 12,13 | 1 | 0 |
| 12,14 | 4 | 11 |
| 12,15 | 0 | 1 |
| 14,14 | 0 | 5 |
| 15,15 | 1 | 0 |
| Total | 97 | 140 |
| Genotypes . | Controls . | Patients . |
|---|---|---|
| 6,6 | 10 | 11 |
| 6,12 | 37 | 42 |
| 6,13 | 1 | 0 |
| 6,14 | 5 | 3 |
| 12,12 | 38 | 67 |
| 12,13 | 1 | 0 |
| 12,14 | 4 | 11 |
| 12,15 | 0 | 1 |
| 14,14 | 0 | 5 |
| 15,15 | 1 | 0 |
| Total | 97 | 140 |
In the genotype column, the two numbers refer to the size of each of the 2 alleles for TNR/11q#1 (eg, 12,6 = [CCG]12, [CCG]6).
Analysis of TNR/11q#1 among patients with CLL
PCR analysis of TNR/11q#1 (Table 1) on DNA from the blood of 140 patients with chronic low-grade lymphoproliferative disease (137 with CLL and 3 with mantle cell lymphoma), with primers flanking the CCG repeat, was performed to examine repeat size within the patient population. A significant increase in the presence of the larger repeat (12 or more copies of the CCG trinucleotide) was found compared to the control group (χ2 with Yates correction;P = .036) (Table 2).
Comparison of TNR/11q#1 CCG-repeat length between patients with CLL and the control population
| Genotype . | Controls . | Patients . |
|---|---|---|
| At least one allele has a repeat of exactly 6 | 53 | 56 |
| Both alleles have repeats greater than 6 | 44 | 84 |
| Genotype . | Controls . | Patients . |
|---|---|---|
| At least one allele has a repeat of exactly 6 | 53 | 56 |
| Both alleles have repeats greater than 6 | 44 | 84 |
P = .036 (Yates correction).
Distribution of repeat size for different stages of disease was performed. Patients with Binet stage B or C disease (ie, patients with advanced-stage disease) and those with progressive disease (classified by a change in Binet stage) all had significant increases in the presence of alleles containing the larger repeats ([Chi]2with Yates correction; P = .021; Table3). Patients with stable stage A disease from presentation did not differ significantly from the control population in distribution of allele size (Table 4). These data suggest that the presence of the larger allele alone defines a poor prognosis for patients with CLL.
Comparison of TNR/11q#1 CCG-repeat length between patients with advanced or progressive disease and the control population
| Genotype . | Controls . | Patients . |
|---|---|---|
| At least one allele has a repeat of exactly 6 | 53 | 40 |
| Both alleles have repeats greater than 6 | 44 | 21 |
| Genotype . | Controls . | Patients . |
|---|---|---|
| At least one allele has a repeat of exactly 6 | 53 | 40 |
| Both alleles have repeats greater than 6 | 44 | 21 |
P = .021 (Yates correction).
Comparison of TNR/11q#1 CCG-repeat length between patients with stable stage A disease from presentation and the control population
| Genotype . | Controls . | Patients . |
|---|---|---|
| At least one allele has a repeat of exactly 6 | 53 | 26 |
| Both alleles have repeats greater than 6 | 44 | 25 |
| Genotype . | Controls . | Patients . |
|---|---|---|
| At least one allele has a repeat of exactly 6 | 53 | 26 |
| Both alleles have repeats greater than 6 | 44 | 25 |
P = .80 (Yates correction).
PCR across large CCG-repeat expansions is difficult and, as a result, cannot be relied on for identifying long repeats. Southern blot analysis was performed on a proportion of patients with a 0.7-kb fragment containing TNR/11q#1 to detect long repeats that might have been missed by PCR analysis. However, no additional bands of hybridization other than those containing repeat lengths previously detected by PCR were present (data not shown).
Localization of CLL deletion breakpoints to CCG repeats
A proportion of patients with CLL and cytogenetically defined 11q- were also analyzed by dual-color metaphase FISH to localize deletion breakpoints to identify potential mechanisms for chromosome breakage. FISH with YAC clones from the 11q contig mapped the distal breakpoints to regions containing CCG repeats. The breakpoint regions corresponded to those previously observed in Jacobsen syndrome.39 The mechanism of breakage in Jacobsen syndrome has been shown to directly relate to CCG repeats (eg, FRA11B).34 The effect of a trinucleotide repeat (chromosome breakage and deletion) may be manifested a number of kilobases away from the location of the repeat sequence (eg, fragile X syndrome43 and Jacobsen syndrome34). It is therefore more suitable to demonstrate CCG-related breakage by FISH with probes immediately flanking the repeat rather than by Southern blot analysis with a probe from the repeat region. Southern hybridization may not demonstrate a rearrangement because of the distance away from the breakpoint region. PACs from regions flanking CCG repeats39 were applied as probes in dual-color metaphase FISH to further refine the deletion mapping and to determine a possible involvement for CCG repeats in chromosome breakage (Figure 1).
FISH analysis of patient 1 (karyotype 46,XY, inv(1), del(11)[q22q23]) placed the distal deletion breakpoint between D11S2090 (PAC dJ36-N10) and D11S707 (PAC dJ177-H4) (Figure 1), which are less than 1 Mb apart. Within this region TNR/11q#7 has been located. We have also recently mapped the breakpoint of a patient with Jacobsen syndrome to the same repeat.39A further CCG repeat (TNR/11q#6) was located within PAC dJ36-N10 less than 1 Mb from the repeat TNR/11q#7.
The distal breakpoint of patient 2 (karyotype 46,XX, del(11)[q2?1q2?3]) mapped to a more centromeric region of 11q23. Dual-color FISH analysis with PAC probes dJ133-C4 and dJ7-L17 showed that the former was deleted and the latter retained (Figure 1). The distal breakpoint for this patient maps to between ATDC and D11S667, a region of no more than 250 kb that contains 2 CCG repeats (TNR/11q#3 and TNR/11q#4). Most Jacobsen syndrome breakpoints are within this region between D11S924 and D11S925.44
Patients 3 (karyotype 46,XY, del(11)[q21]) and 4 (karyotype 46,XY, del(11)[q2?2q2?3]) both map to the same distal breakpoint (Figure 1). YAC y966F02 was deleted in both patients, and the adjacent YAC y797E07 was retained. These YACs flank another CCG repeat (TNR/11q#2) and represent a total area of 2.8 Mb. This is an area of breakage described in both CLL and mantle cell lymphoma. In the latter, most patients' deletion breakpoints are clustered.16 Monni et al16 have recently mapped the distal breakpoint in 12 of 19 patients with mantle cell lymphoma to y785E12, which lies within 300 kb of the breakpoint we describe (Figure 1). Furthermore Zhu et al11 demonstrate that discontinuous deletions occur in patients with CLL specifically involving y785E12. This YAC contains TNR/11q#2 and the SMA and APOC3 genes.
Finally, in patients 1 and 2, a split signal was seen with PAC dJ13-H12, which lies approximately 100 kb from the ETS1 gene in 11q24 (Figure 1). This PAC also contains a CCG-repeat (TNR/11q#8). Overlapping probes on either side of the CCG repeat gave a normal retained signal. This raises the possibility that a more complex rearrangement, such as an inversion, has occurred in addition to the deletion.
Discussion
Deletions of chromosome 11q are common in low-grade lymphoproliferative disease.10,45 Patients with this chromosomal abnormality have aggressive disease and poor prognoses.7-9 A contig of this region has been used to identify a region of deletion that may contain disease-associated genes. In addition a possible insight into the mechanism of chromosome breakage may also be obtained. A region of deletion minimum of approximately 4 Mb, extending from 11q22.3 to 11q23.1, has been identified for CLL by metaphase FISH and LOH analysis with 8 microsatellite markers. This region lies within that previously described10,11 for CLL and includes the region for mantle cell lymphoma described by Monni et al,16 but it differs by a small distance from the region described by Stilgenbauer et al17 for mantle cell lymphoma (Figure 1). A polymorphic CCG repeat (TNR/11q#1) has been identified within our minimal region of deletion. PCR analysis has demonstrated that a repeat length of 12 to 15 copies of the CCG trinucleotide is more common among the patient group, particularly those with an 11q deletion, and is a marker of disease of worse prognosis. Analysis of the distal deletion breakpoint regions for patients with CLL reveals clustering at specific hot spots on distal 11q, where other CCG repeats are known to be located. CCG repeats, in a manner similar to that for microsatellite regions, may be polymorphic. These results suggest that the stability of CCG-repeat sequences may be an important genetic factor in the pathogenesis of CLL and some forms of low-grade lymphoma.
Two distinct and independent mechanisms for the potential role of CCG repeats in CLL can be proposed from this study. First, the association of a specific TNR/11q#1 allele with progression of the disease implies that this genetic element may be linked to a nearby tumor suppressor gene. Genes close to CCG repeats may have their patterns of expression altered by the presence of the repeat, particularly when the length is enlarged.31 It is possible that the length of the CCG-repeat TNR/11q#1 directly affects the function or expression of a tumor-suppressor gene, which in turn plays an important role in the pathogenesis of CLL. Specific alleles of a similarly sized CCG repeat within the PABP2 gene have been shown to cause oculopharyngeal muscular dystrophy.46 The normal population has 6 copies of the CCG trinucleotide; the addition of only 2 more copies is sufficient to cause the phenotype of the disorder. In this disease a relatively frequent allele among the normal population, (CCG)7, can act as a modifier of a dominant phenotype or as a recessive mutation. It is therefore clear that small increases in the length of a CCG repeat (as seen with the longer allele at TNR/11q#1) may be of pathologic significance.
Polymorphic trinucleotide repeats have already been identified in a number of known disease-associated genes.47 Some genes contain CCG repeats within the 5′ untranslated portion of the transcribed sequence and influence transcription. In the normal population, allele frequencies for these polymorphic trinucleotide repeats are usually in Hardy-Weinberg equilibrium. However, in some disease populations, the presence of Hardy-Weinberg disequilibrium for a particular repeat-length allele would represent linkage to a gene important in the pathogenesis of the disease.47 An example of this is glutathione-S-transferase and liver disease—repeat length may influence enzyme activity.47 Essentially persons with specific alterations of CCG-repeat length alleles may be more prone to specific diseases.
An alternative explanation for the correlation of CCG-repeat length with progression of the disease is the presence of a common founder chromosome harboring a (CCG)12 allele at the TNR/11q#1 locus and a mutation within a nearby tumor-suppressor gene. The CCG repeat may not be involved directly in the function or expression of this gene but may only represent a closely linked marker to the mutation that causes the disease. This hypothesis can be tested by a more thorough analysis of allelic haplotypes in the surrounding region. It is interesting to note that haplotype analysis of family members with CLL, though only with polymorphic markers close to theATM gene, did not show any evidence of genetic linkage at the ATM locus.48 However, this region is located approximately 2 Mb centromeric to TNR/11q#1; as such, the linkage data from the ATM region are not informative for the region we describe. The ATM gene is undoubtedly important in the pathogenesis of CLL, but mutations have only been found in a proportion of patients.12-14 Our minimal region of deletion, together with those of Monni et al16 and Zhu et al,11 excludes the ATM gene. These observations could suggest that mutations in ATM may predispose to chromosome deletions and that a second gene at 11q22.3 is important to the pathogenesis of the disease.
The co-localization of CCG repeats, with chromosome deletion breakpoints in patients with CLL, suggests a second role for these sequences in the pathogenesis of CLL. Of only 8 CCG repeats identified in the 40-Mb YAC contig, 4 are shown here to be close to chromosome deletion breakpoints in patients with CLL, and another is shown to be close to an additional chromosome rearrangement. Of importance, TNR/11q#2 is close to a cluster of deletion breakpoints found in patients with mantle cell lymphoma16; of 19 patients, 12 were found to have a deletion breakpoint within the same YAC (y785E12) that contains TNR/11q#2. This region is also involved in the discontinuous deletions described by Zhu et al11 in CLL. It could be speculated that TNR/11q#2 is a particularly potent hot spot for deletion breakpoints in CLL and mantle cell lymphoma. In addition, these distal CLL deletion breakpoints lie close to chromosome deletion breakpoints in the rare inherited 11q deletion syndrome (Jacobsen syndrome). Specifically, the repeats TNR/11q#3 and TNR/11q#4 are within the region containing most Jacobsen deletion breakpoints,44 whereas TNR/11q#7 and TNR/11q#8 are also breakpoint regions in the syndrome.39
The potential role of fragile sites in cancer and leukemia is a controversial issue, and statistical associations between the locations of fragile sites and both oncogenes and translocation breakpoints are contradictory.49-51 However, recent work has demonstrated that the aphidicolin-inducible fragile sites FRA3B andFRA7G are the sites of frequent chromosome rearrangements in many types of neoplasia,52-58 regardless of their significance to tumorigenesis. In particular an increased level of spontaneous breakage and fragile site expression has been observed for CLL.37
The clustering of breakpoints to CCG-repeat loci on 11q in patients with CLL and mantle cell lymphoma suggests a common mechanism of chromosome breakage in these diseases. The direct role of a CCG repeat in causing chromosome breakage has been described; expansion of theFRA11B CCG-repeat has been shown to cause chromosome deletions,34 though the breakage points do not occur directly at the repeat sequence. Detection of expansions of CCG repeats after deletion may be problematic because the repeat may be within the deleted region. However, FISH analysis provides good evidence for their involvement. It is possible that those CCG repeats, which co-localize with chromosome deletion breakpoints in patients with CLL, are the locations of previously unidentified folate-sensitive fragile sites on chromosome 11q. Fragile sites are expressed when cells are cultured under conditions that inhibit DNA replication.32 If fragile sites are to be considered regions of late replication, it is possible that CCG repeats mediate chromosome deletions by a mechanism involving the failure of DNA replication. The folate-sensitive fragile sites FRAXA and FRAXE are located in regions that have been identified as late-replication sites. For both these sites, replication is delayed even further on chromosomes carrying a CCG-repeat expansion compared to chromosomes with a normal CCG-repeat length.59-61
By contrast, though the mechanism of expression of the aphidicolin-inducible fragile sites, such as FRA3B andFRA7G, is not related to trinucleotide repeat sequences, the role of DNA replication timing in the expression of FRA3Bhas been established.62 In a manner similar to the folate-sensitive fragile sites, the aphidicolin-inducible fragile sites are also late-replicating regions of the genome. In vitro experimental evidence has demonstrated that, in the case of FRA3B, when cells are exposed to aphidicolin to inhibit DNA polymerase α and β, it induces expression of the fragile site by delaying DNA replication in regions that initiate this DNA replication late in the S phase. Furthermore these cells can enter G2 having failed to complete the replication of FRA3B and essentially having deleted this region of DNA from the cell. The result is a failure of replication as the cell continues cycling with a consequent loss of DNA in that region in the newly formed cell. If a tumor-suppressor gene is deleted, tumorigenesis may occur.
Previously, fragile sites have been characterized exclusively by in vitro cytogenetic criteria. However, the co-localization of CCG repeats with chromosome breakage in different diseases suggests that classification may be possible, based on these in vivo observations. Analysis of the timing of replication at the chromosome deletion breakpoints identified in this study will shed light on the potential role of CCG repeats in their etiology and would further justify their classification as fragile sites.
We have shown that the chromosome breakpoints of patients with CLL cluster at specific hot spots on distal 11q, where trinucleotide repeats are located. In addition, a polymorphic CCG repeat has been identified within the minimal region of deletion, and it is shown that this may have an effect on the pathogenesis and outcome of chronic LPD. Although we cannot rule out the possibility of an association by chance alone, the results offer evidence that trinucleotide repeats may be responsible for the chromosome breakage and rearrangement in malignancy and could underlie tumor predisposition and development.
Acknowledgments
We thank Sandra Merscher for her generous gift of 11q13 cosmid clones. We thank Michael James for the generous provision of YAC clones from his 40-Mb YAC contig. We thank the staff at the Regional Cytogenetics Department at the Birmingham Women's Hospital for their assistance in the preparation of patient metaphases.
Supported by the Leukaemia Research Fund, The Royal London Hospital special trustees, and the Sydney and Phyllis Goldberg Research Trust (R.L.A, C.J., F.E.C).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Finbarr E. Cotter, Department of Exp Haematology, St Bartholomew's and the Royal London School of Medicine and Dentistry, Turner St, London E12AD, United Kingdom; e-mail:f.e.cotter@mds.qmw.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal