Epstein-Barr virus (EBV)–encoded latent membrane protein-1 (LMP1) is highly expressed in Hodgkin and Reed-Sternberg (H-RS) cells from patients with EBV-associated Hodgkin disease. It was previously demonstrated that CD99 can be negatively regulated by LMP1 at the transcriptional level, and the decreased expression of CD99 in a B lymphocyte cell line generates H-RS–like cells. In this study, detailed dissection of the CD99 promoter region was performed to search regulatory factor(s) involved in the expression of the gene. Using various mutant constructs containing deletions in the promoter region, it was revealed that the maximal promoter activity was retained on 5′-deletion to the position −137 from the transcriptional initiation site. Despite the presence of multiple putative Sp1-binding sites in the promoter region, the site located at −95 contributes heavily as a positive cis-acting element to its basal promoter activity. However, on examination of the involvement of the positive-acting Sp1-binding site of the promoter for the repressive activity of LMP1, it appeared to be dispensable. Instead, the repressive effect was mapped to the nuclear factor (NF)-κB activation domains in the cytoplasmic carboxyl terminus of LMP1 despite the absence of the NF-κB consensus sequences in the CD99 promoter region. Furthermore, the decreased CD99 promoter activity by LMP1 was markedly restored when NF-κB activity was inhibited. Taken together, these data suggest that Sp1 activates, whereas LMP1 represses, transcription from the CD99 promoter through the NF-κB signaling pathway, and they might aid in the understanding of the molecular mechanisms of viral pathogenesis in EBV-positive Hodgkin disease.
Introduction
Human CD99 is a glycosylated transmembrane protein with a molecular mass 32 kd, and it is expressed on most cell surfaces.1-3 Although its function is not fully understood, it has been suggested that CD99 is involved in multifactorial cellular events such as cell-cell adhesion during hematopoietic cell differentiation, apoptosis of immature thymocytes and neuronal cells, and T-cell activation.4-8 The functional role of CD99 in B lymphocytes is also reported; the in vitro down-regulation of CD99 generates cells with Hodgkin and Reed-Sternberg (H-RS) phenotypes as seen in the lymph nodes of patients with Hodgkin disease (HD), implicating the association of loss of CD99 with HD pathogenesis.9
The mic2 gene encoding CD99 is the first human gene isolated from the pseudo-autosomal region located at the X- and Y-chromosome short arms and is required for the correct pairing of these morphologically different chromosomes during male meiosis.10,11 Subsequent analysis of this gene defined 10 exons that are considerably smaller than average for mammalian genes.12 At the 5′ end of the mic2 gene, there is a G+C-rich promoter region containing a great number of Sp1-binding core sequences with no identifiable TATA box or CAAT element, suggesting its role as a housekeeping gene.13-15
Despite extensive molecular and biochemical analysis of CD99, study of the mechanism by which gene expression is regulated has been limited. Thus, no cellular or viral factor affecting CD99 expression has been identified until recently. Because Epstein-Barr virus latent membrane protein 1 (EBV LMP1) is highly expressed in H-RS cells from patients with EBV-associated HD and because the down-regulation of CD99 produces H-RS–like cells, we investigated the possibility of LMP1 association in the regulation of CD99 expression and reported that LMP1 acts as a transcriptional repressor on CD99, subsequently generating cells with H-RS phenotypes.16
LMP1 has transforming properties in rodent fibroblasts, highlighting the importance of this viral oncoprotein in cellular transformation associated with EBV infection.17 Expression of LMP1 in B lymphocytes induces the transcription of many genes, including those encoding activation antigens such as CD23 and CD40, adhesion molecules such as LFA-1, LFA-3, and ICAM-1, and inhibitor molecules of programmed cell death such as Bcl-2 and A20.18-23 Induction of these genes is likely to play an important role in the transformation by LMP1.
LMP1 is an integral membrane protein consisting of 386 amino acids. Six transmembrane-spanning domains (162 amino acids) connect a short N-terminal stretch (24 amino acids) with a long C-terminal domain (200 amino acids), both of which are located in the cytoplasm.24 Mutational analysis of LMP1 with respect to activation of cellular signaling pathways divides the carboxy-terminal domain into 3 regions. Carboxyl-terminal activation region 1 (CTAR1), located between amino acids 187 and 231, is a relatively weak activator of nuclear factor (NF)-κB and interacts with members of the tumor necrosis factor (TNF) receptor-associated factors (TRAF) family.25-27 CTAR2, located between amino acids 352 and 386, is not only the major NF-κB activation domain but also is responsible for the activation of c-Jun N terminal kinase (JNK) and interacts with the TNF receptor-associated death domain protein (TRADD).26-28 The amino acids separating these 2 regions was recently reported to contain a third functional domain, CTAR3, at amino acids 275 to 330, required for the activation of the Jak/Stat pathway.29
The relative contribution of each of the LMP1 signaling pathways to affecting the various phenotypes associated with LMP1 expression is unclear. Furthermore, though many researchers have documented the association of LMP1 with the activation of cellular genes, its role as a negative regulator was also recently reported for CD99. In the present study, we analyzed the 5′ flanking region of the CD99 to obtain insights on the possible mechanisms by which the promoter is transcriptionally regulated.
Materials and methods
Cell culture and transient transfection
Cell lines used in this study—such as BJAB (human Burkitt lymphoma cell line), 293, and 293T (human embryonic kidney cell lines)—were obtained from American Type Culture Collection (ATCC; Rockville, MD) and were maintained in Dulbecco modified Eagle medium (DMEM) (GIBCO BRL, Rockville, MD), supplemented with 10% heat-inactivated fetal bovine serum, antibiotics, and 2 mM glutamine in the humidified atmosphere of a 5% CO2 incubator. Nonlymphoid cells were plated at a density of 1 × 106cells/60-mm dish the day before transfection and were transfected by calcium phosphate precipitation using various amounts of reporter and effector plasmids. To normalize transfection efficiencies, we included in the cotransfection either a thymidine kinase promoter (TK)-drivenRenilla luciferase plasmid (pRLTK, 0.1 μg; Promega, Madison, WI) as an internal control plasmid or a chloramphenicol acetyl transferase (CAT) expression plasmid containing the BiP promoter (pBiP670CAT,30 1 μg), in the event of cotransfection with LMP1 instead of pRLTK, to avoid using promoters that could be modulated by coexpressed LMP1. Transiently transfected LMP1 displayed little effect on the expression of BiP, which was confirmed elsewhere by comparison of the levels of Bip expression between control and LMP1-transfected B lymphoid cell lines.16 Simultaneously, pNF-κB CAT was cotransfected instead of pBiP CAT in another set of experiments to assess NF-κB activity induced by LMP1. BJAB cells were transfected by the lipofectamine (GIBCO BRL) transfection method. In addition, a promoterless and enhancerless luciferase reporter vector, p(0)luc, modified from pGL-2/Basic (Promega), was also transfected in each experiment as a negative control. For cotransfection studies, 2 μg effector plasmid was transfected together with 0.5 μg promoter-fused reporter plasmid, unless otherwise indicated. After overnight incubation, cells were washed and cultured in complete culture media for 36 hours.
Drosophila melanogaster SL2 cells were grown on Schneider medium M3 (Sigma, St Louis, MO) supplemented with 10% IMS (Sigma) and transfected by the dimethyldioctadecylammonium bromide method, as described elsewhere.31
Western blot analysis
Cells were washed twice in cold phosphate-buffered saline and solubilized in lysis buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 10 μg/mL aprotinin, and 10 μg/mL leupeptin) for 30 minutes on ice. After centrifugation, the protein concentration was determined using the Bradford method (Bio-Rad Laboratories, Hercules, CA). Lysates were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto nitrocellulose filters for probing with monoclonal antibodies. Antibodies were obtained from hybridoma cultures or were purchased as follows: LMP1 (CS1-4; DAKO, Caterpillar, CA), FLAG (M5; Sigma), Sp1 (D-20; Santa Cruz Biotechnology, Santa Cruz, CA), Sp3 (1C6; Santa Cruz Biotechnology), CD99 (DN16; DiNonA Inc, Korea), and Calnexin (Transduction Laboratories, San Diego, CA). Specifically bound antibodies were detected using peroxidase-conjugated goat antimouse IgG except for Sp3, goat antirabbit IgG (Zymed, San Francisco, CA), and visualized with the enhanced chemiluminescence detection system (Amersham, Arlington Heights, IL).
Flow cytometric analysis
Samples of 5 × 105 cells were incubated with 10 μL R-phycoerythrin (PE)–conjugated antihuman CD99 (Pharmingen, San Diego, CA) for 30 minutes at 4°C. These cells were then washed with phosphate-buffered saline twice. Flow cytometric analysis was performed on a FACScan (Becton Dickinson, Franklin Lakes, NJ).
Gene constructs
Cloning of the 5′ flanking region of human CD99 has been reported previously.16 The full sequence of the region is available in the GenBank (accession number, AF310969). The region from −1641 to +123 relative to the transcriptional initiation site was amplified by PCR using primers 5′-CCCATGGTCACTCATATGTGGCTCAG-3′ (sense strand) and 5′-GGGGTACCGAAGGCGGCAGGACAGATAC-3′ (antisense strand) and subsequently ligated into p(0)luc. A series of 5′-, 3′-, and internal GC box-deleted CD99 promoter segments was generated by polymerase chain reaction (PCR). PCR was performed with a reaction mixture containing 250 μM deoxynucleotides, 0.1 μg primers, 5% formamide, and 0.5 U Taq polymerase (Life Technologies). Cycling parameters were 1 minute at 94°C, 1 minute at 50°C, and 1 minute at 72°C for 30 cycles. To generate 5′- and 3′-deleted CD99 promoter constructs, 10 promoter segments—amplified by using 25-mer primers that placed SmaI and KpnI sites at the 5′ and 3′ ends, respectively—were cloned between the SmaI and the KpnI sites of the p(0)luc reporter. Internal deletion mutants were generated using pairs of 40-bp overlapping oligonucleotides with the desired deletion, as follows: pΔ(−137/−78)luc for a deletion between −137 and −78, pΔ(−77/−38)luc for a deletion between −77 and −38, and pΔ(−37/+2)luc for a deletion between −37 and +2. The point mutation derivatives of the CD99 promoter region, such as p1mGC, p2mGC, p3mGC, and p123mGC, were generated by using oligonuclotides containing the mutated Sp1-recognition sites (TTCGGTT,AACCGAA,AACCGAA at −93, −50, −7, and the 3 mutated sequences at −93, −50, and −7, respectively). The DNA sequence of each promoter segment was confirmed by sequencing before transfection.
An LMP1 cDNA-containing pcDNA3-LMP1 and its negative control construct, pcDNA3-1-PML, were gifts from Dr C. V. Paya (Mayo Clinic, Rochester, MN). Dr G. Suske (Philipps-Universitat Marburg, Germany) kindly provided human Sp1 and Sp3 expression vectors (pEVR2/CMV/Sp1 and pRC/CMV/Sp3). Dr A. Courey (University of California at Los Angeles) provided various mutant Sp1 constructs driven by Drosophilaβ-actin promoter (pPac327C and pPac168C), and the full-length Sp1 construct (pPacSp1) was from Dr K. Han (Hallin University, Korea). All the LMP1 deletion mutants were constructed as described elsewhere.32 The pLMP1Δ187-351 mutant was constructed by removal of the 495-bp NcoI restriction fragment in the LMP1 cDNA. The pLMP1Δ187-386 and the pLMP1Δ232-386 mutants were constructed by blunt-end ligation of a stop codon containing double-stranded oligonucleotide (5′-CTAGTCTAGACTAG-3′) into the Klenow end-filled NcoI site and into the NaeI site in the LMP1 cDNA, respectively. An LMP1 mutant construct containing point mutations at amino acid 208, 210, and 212 in CTAR1 and at amino acid 384 in CTAR2 was generated by site-directed mutagenesis. The reporter constructs, pNF-κB CAT and pAP-1 CAT, were kindly provided by Dr S. Kim (Seoul National University, Korea). pcDNA3-FLAG-JNK1 and pcDNA3-FLAG-JNK1 (apf) and pcDNA3-FLAG-IκBαS32/36A were gifts from Dr R. Davis (University of Massachusetts, Worcester) and Dr E. Kieff (Harvard University, Cambridge, MA), respectively.
Luciferase and chloramphenicol acetyl transferase assays
After 48 hours of transfection, cells were harvested and extracts were prepared for luciferase and chloramphenicol acetyl transferase (CAT) assays using passive cell lysis buffer provided by the manufacturer (Promega). Luciferase activity was measured for a 15-second time course using a luminometer (Turner Designs, Sunnyvale, CA) and dual luciferase assay system (Promega) with one tenth of the total volume of cell extract. CAT assays were made as previously described. Protein concentrations of cell extracts were measured using the bicinchoninic acid protein assay reagent kit (Pierce, Rockford, IL). The luciferase activity of each sample was subsequently adjusted according to protein concentration and either TK-drivenRenilla luciferase or CAT activity per sample.
Results
The region up to position −95 from the transcriptional initiation site is required for the core promoter activity of CD99
We previously reported the cloning of a genomic fragment containing the region from −1641 to +123 relative to the transcriptional initiation site of CD99.16 According to the sequencing data from others and our laboratory, the 5′-flanking region of CD99 contains no recognizable TATA element but a large number of GC boxes that are putative binding sites for transcription factor Sp1.13 Although Sp1 has been found to play a major role in the positive regulation of many TATA-less promoters,33 34its regulation of CD99 expression has not been demonstrated. Thus, to search for cellular factor(s) regulating CD99 expression, we performed detailed dissection of the promoter region.
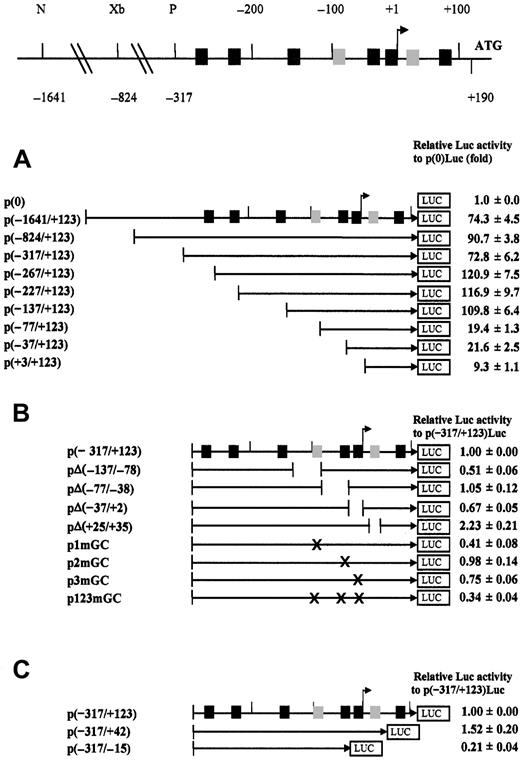
We initially constructed 8 plasmids of 5′ deletions with 3′ fixed-end (+123) and 2 plasmids of 3′ deletions with 5′ fixed-end (−317) that were transiently transfected into 293 cells. Transient transfection with the 3′-deletion construct, p(−317/+42)luc, revealed that the sequence between positions +42 and +123, possibly the GC boxes at position +74, acted negatively so that the promoter activity was increased up to 50% when deleted (Figure1C). In addition, the adjacent GC box, located near the transcription start site between +25 and +35, also played a role as a negative cis-acting element so that its deletion increased the promoter activity more than 2-fold (pΔ(+25/+35)luc in Figure 1B). On the other hand, the promoter activity was decreased 5-fold on further deletion, to −15, at which the transcription start site is located, possibly because of the loss of the natural transcription initiation site. Transient transfection with 5′ deletion constructs linked to luciferase reporter revealed that the region between positions −317 and −268 contains one more negative cis-element. The high transcriptional activity was maintained until deleted to the position −137, and subsequent deletions beyond the site decreased the promoter activity. The contribution of 2 regions at positions −137/−78 and −37/+2 on the promoter activity were more dramatic so that deletions in the 2 regions led to a 50% and a 30% decrease, respectively (Figure 1B). To limit the responsible sequences for the activity in those regions, the point mutations were introduced at the GC boxes, located at the positions −95 and −5 (p1mGC and p3mGC in Figure 1B). In p3mGC, point mutations were carefully designed so they would not disturb the transcription start site. Although positive roles of the GC boxes at −95 and −5 on the CD99 promoter activity were confirmed, the mutation at −95 displayed a more profound effect; p1mGC showed almost 60% decreased activity of the wild-type construct. Thus, this analysis suggested that multiple GC boxes between −1641 and +123 unevenly contribute to the CD99 promoter activity and that the GC box located at −95 from the transcription initiation site is most crucial for a maximal core promoter activity whereas GC boxes at +24, and possibly at +74 and −274, function as the negative elements.
Deletion and point mutation analysis of the CD99 gene promoter region.
(Top) Linear diagram of the promoter region is shown. Putative SP1-binding sites, core sequences (GGGCGG, complement CCGCCC, or both) and the decanucleotide (GGGGCGGGGC) consensus sequences, are located at −274, −242, −151, −95, −48, −5, +24, and +74 and are indicated as black and gray rectangles, respectively. Nucleotide numbers are relative to the transcription start site (+1) published previously14 and are indicated by the arrowhead. (Bottom) Various promoter-luciferase fusion constructs and the promoterless vector as a negative control were transiently transfected into 293 cells. Eighteen CD99 promoter variants 8 5′ deletion derivatives, 4 internal deletion and 4 point mutation derivatives, and 2 3′ deletion derivatives. (A) Relative activities of the 5′ deletion derivatives of the CD99 promoter region. Plasmid names of eight 5′ deletion derivatives and the full-length CD99 promoter construct are listed to the left of the diagram in the middle. The included CD99 promoter region is denoted in the name of each construct. Relative luciferase activity, shown to the right of each construct, is expressed as the -fold of the luciferase activities obtained from the promoter-luciferase fusion construct over the promoterless vector, p(0)luc. To normalize plate-to-plate variations, the construct containing the TK promoter-driven Renilla luciferase (pRLTK) was cotransfected, and the activities of firefly and Renillaluciferases were measured sequentially from a single sample using a dual-luciferase reporter assay system. Data represent averages from 3 separate transfections performed with each construct. (B) Relative activities of the internal deletion and the point mutation derivatives of the CD99 promoter region. The name of internal deletion mutant indicates the position of the deleted region. p1mGC, p2mGC, p3mGC, and p123mGC contain the mutated GC boxes at −95, −48, −5, and −95/−48/−5, respectively. (C) Relative activities of the 3′ deletion derivatives of the CD99 promoter region. The included CD99 promoter region is denoted in the name of each construct. Transfection and analysis were performed as previously described in panel A.
Deletion and point mutation analysis of the CD99 gene promoter region.
(Top) Linear diagram of the promoter region is shown. Putative SP1-binding sites, core sequences (GGGCGG, complement CCGCCC, or both) and the decanucleotide (GGGGCGGGGC) consensus sequences, are located at −274, −242, −151, −95, −48, −5, +24, and +74 and are indicated as black and gray rectangles, respectively. Nucleotide numbers are relative to the transcription start site (+1) published previously14 and are indicated by the arrowhead. (Bottom) Various promoter-luciferase fusion constructs and the promoterless vector as a negative control were transiently transfected into 293 cells. Eighteen CD99 promoter variants 8 5′ deletion derivatives, 4 internal deletion and 4 point mutation derivatives, and 2 3′ deletion derivatives. (A) Relative activities of the 5′ deletion derivatives of the CD99 promoter region. Plasmid names of eight 5′ deletion derivatives and the full-length CD99 promoter construct are listed to the left of the diagram in the middle. The included CD99 promoter region is denoted in the name of each construct. Relative luciferase activity, shown to the right of each construct, is expressed as the -fold of the luciferase activities obtained from the promoter-luciferase fusion construct over the promoterless vector, p(0)luc. To normalize plate-to-plate variations, the construct containing the TK promoter-driven Renilla luciferase (pRLTK) was cotransfected, and the activities of firefly and Renillaluciferases were measured sequentially from a single sample using a dual-luciferase reporter assay system. Data represent averages from 3 separate transfections performed with each construct. (B) Relative activities of the internal deletion and the point mutation derivatives of the CD99 promoter region. The name of internal deletion mutant indicates the position of the deleted region. p1mGC, p2mGC, p3mGC, and p123mGC contain the mutated GC boxes at −95, −48, −5, and −95/−48/−5, respectively. (C) Relative activities of the 3′ deletion derivatives of the CD99 promoter region. The included CD99 promoter region is denoted in the name of each construct. Transfection and analysis were performed as previously described in panel A.
The minimal sequence within a promoter to drive the normal activity is called the core promoter. Figure 1A shows that a 280-bp fragment (−137 to +123) has complete promoter activity. In fact, this core promoter is almost 50% higher in activity than the full-length CD99 promoter. The core promoter identified was active in all cell lines tested (data not shown).
Transcription of the CD99 promoter is positively regulated by Sp1 through its recognition sites
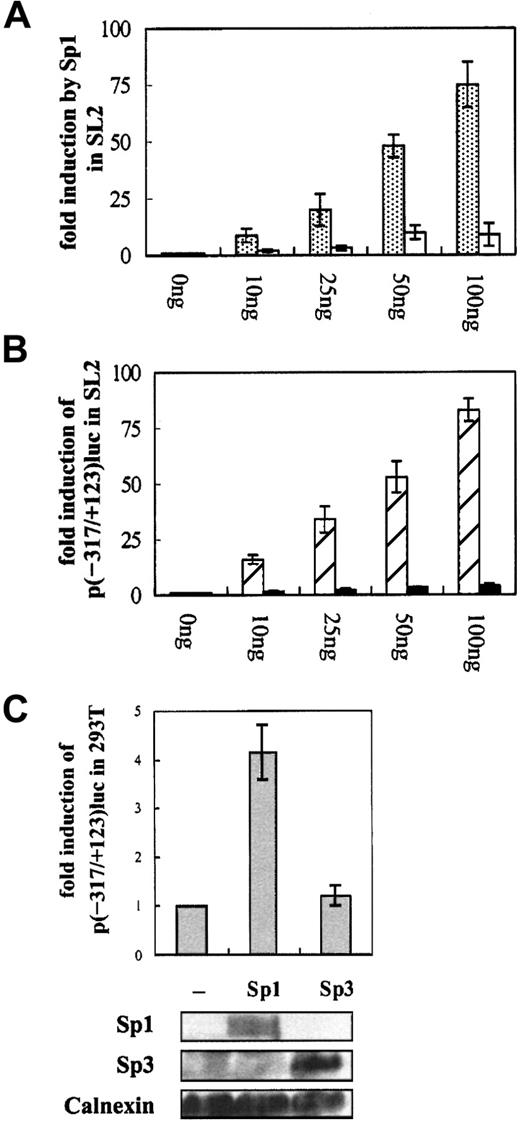
Our current study demonstrated that at least 2 GC boxes located upstream of the transcription initiation site play roles in the positive transcriptional activity of CD99, whereas 2 other GC boxes located downstream act as negative cis-acting elements. Thus to examine whether Sp1 interacts with hexanucleotide Sp1 recognition sites in the CD99 promoter and actually regulates the transcription of CD99, we adopted Drosophila tissue culture cells, which lack endogenous Sp1, as a host system. To achieve optimal protein expression in D melanogaster SL2 cells, we used Sp1 constructs driven by the Drosophila β-actin promoter. When transiently transfected with various amounts of the construct containing the full sequence of Sp1, pPacSp1, the CD99 promoter region from −317 to +123 was activated in a dose-dependent manner. On the other hand, dose-dependent transactivation disappeared when the construct containing mutations at the GC boxes in the promoter, p123mGC, was cotransfected (Figure 2A). The role of Sp1 on the CD99 promoter was confirmed by analysis with 2 amino-terminal deletion mutants of Sp1 that contain 327 and 168 carboxyl terminal amino acid residues of Sp1, pPac327C, and pPac168C, respectively. Previous studies reported that carboxyl terminal 168 amino acid residues of Sp1 only encode 3 zinc fingers for sequence-specific binding to DNA and thus are insufficient for transcriptional activity because of the lack of activation domains, whereas carboxyl terminal 327 amino acid residues of Sp1 maintain a high level of transcriptional activity.35 36 In the cotransfection experiments, the CD99 promoter activity was markedly increased by pPac327C but not by pPac168C (Figure 2B), indicating that the promoter requires both the activation and the DNA recognition domains of Sp1 for its activation. Therefore, results from the transfection experiments of the CD99 promoter constructs intoDrosophila SL2 cells indicate that Sp1 functions as a positive transactivator of the CD99 promoter through specific recognition.
Sp1 positively regulates the CD99 promoter in
D melanogaster SL2 cells and in 293T. (A) Activation of the CD99 promoter by Sp1 in D melanogaster SL2 cells. To achieve sufficient protein expression in transient transfection assays, various amounts of Drosophila β-actin promoter-driven Sp1 (pAcSp1) were used. One microgram wild-type promoter construct p(−317/+123)luc (░) or the construct containing mutated Sp1 recognition sites in the promoter, p123mGC (■), was transfected into SL2 cells. (B) Effect of the activation-domain deletion mutant of Sp1 on the CD99 promoter activation. A construct containing either 327 or 168 carboxyl-terminal amino acid residues of Sp1, pPac327C (▨) and pPac168C (▪), respectively, was cotransfected with the wild-type CD99 promoter construct, p(−317/+123)luc. (C) Effects of Sp1 and Sp3 on the CD99 promoter in 293T. The wild-type promoter-luciferase fusion construct, p(−317/+123)luc was cotransfected with expression plasmid for Sp1 or Sp3 into 293T cells. The internal control, 1 μg pBip670CAT, was cotransfected. Transfected cells were prepared in 2 separate ways for luciferase and CAT assay and for Western blot analysis. For luciferase and CAT assay, cells were prepared with passive lysis buffer solution. For Western blot analysis, cells were washed and solubilized in lysis buffer as described in “Materials and methods.” After protein quantitation, the lysates were separated by 10% SDS PAGE and electroblotted onto the nitrocellulose filters. The same filter was hybridized with monoclonal antibodies such as D-20, 1C6, and anti-Calnexin for Sp1, Sp3, and Calnexin, respectively. Relative luciferase activity is expressed as relative activity over the negative control transfection with pcDNA3.
Sp1 positively regulates the CD99 promoter in
D melanogaster SL2 cells and in 293T. (A) Activation of the CD99 promoter by Sp1 in D melanogaster SL2 cells. To achieve sufficient protein expression in transient transfection assays, various amounts of Drosophila β-actin promoter-driven Sp1 (pAcSp1) were used. One microgram wild-type promoter construct p(−317/+123)luc (░) or the construct containing mutated Sp1 recognition sites in the promoter, p123mGC (■), was transfected into SL2 cells. (B) Effect of the activation-domain deletion mutant of Sp1 on the CD99 promoter activation. A construct containing either 327 or 168 carboxyl-terminal amino acid residues of Sp1, pPac327C (▨) and pPac168C (▪), respectively, was cotransfected with the wild-type CD99 promoter construct, p(−317/+123)luc. (C) Effects of Sp1 and Sp3 on the CD99 promoter in 293T. The wild-type promoter-luciferase fusion construct, p(−317/+123)luc was cotransfected with expression plasmid for Sp1 or Sp3 into 293T cells. The internal control, 1 μg pBip670CAT, was cotransfected. Transfected cells were prepared in 2 separate ways for luciferase and CAT assay and for Western blot analysis. For luciferase and CAT assay, cells were prepared with passive lysis buffer solution. For Western blot analysis, cells were washed and solubilized in lysis buffer as described in “Materials and methods.” After protein quantitation, the lysates were separated by 10% SDS PAGE and electroblotted onto the nitrocellulose filters. The same filter was hybridized with monoclonal antibodies such as D-20, 1C6, and anti-Calnexin for Sp1, Sp3, and Calnexin, respectively. Relative luciferase activity is expressed as relative activity over the negative control transfection with pcDNA3.
The finding was further confirmed by the effect of Sp1 on the CD99 promoter in mammalian cells. In the experiment, a wild-type CD99 promoter construct, p(−317/+123)luc, was cotransfected with a mammalian Sp1 expression plasmid into 293T cells. Similar to the results obtained with SL2 cells, the overexpression of Sp1 greatly enhanced the CD99 promoter activity up to 4-fold (Figure 2C). Because it is known that the hexanucleotide Sp1 recognition site can interact with several human transcription factors other than Sp1, we examined whether Sp1 recognition sites present in the CD99 promoter region could interact with Sp3 expressed ubiquitously as Sp1 among Sp1-related proteins. Although it is well known as a bifunctional transcriptional regulator that can repress and activate transcription depending on the cell and promoter type,37 Sp3 had no effect on CD99 transcription when cotransfected. Thus, these results indicate that the GC boxes in the CD99 promoter are recognized as acis-acting element specifically by Sp1, but not by Sp3, at least among those human transcription factors that are known to interact with the Sp1 recognition sequence.
Down-regulation of the core promoter activity of CD99 correlates with LMP1 expression
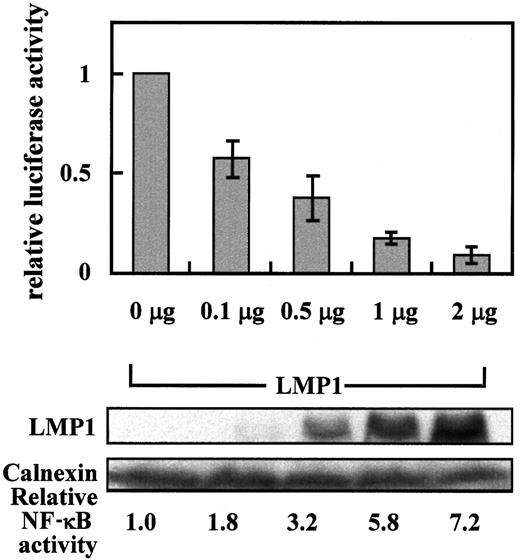
Given that LMP1 has been reported as another transcriptional modulator that can suppress transcription from the 1.65-kb promoter region of the CD99 gene,16 we tested whether this repression event also appears with the core promoter region of the gene. Thus, to see whether the event was specific to the expression level of LMP1, we transfected the construct containing the sequence from −137 to +123 of the CD99 promoter, p(−137/+123)luc, into 293 cells with cytomegalovirus immediate-early (CMV IE) promoter-driven LMP1 in increasing amounts. Although B-cell lines are relevant models for HD, we used 293 or 293T cells because the system is not particularly suitable for elucidating LMP1-mediated CD99 down-regulation with the markedly low transfection efficiency of lymphoid cells and the consistent suppression by LMP1 in nonlymphocytic cells and lymphocytic cells. As a control, NF-κB was measured in parallel transfections using a pNF-κB CAT reporter. As shown in Figure 3, the activity of the CD99 core promoter was still inhibited by LMP1 in a dose-dependent manner, whereas NF-κB became activated, indicating that a factor recognizing the core region is associated with the LMP1-mediated repression of CD99.
LMP1 represses the core promoter activity of CD99 in a dose-dependent manner.
Relative luciferase activity was measured in 293 cells cotransfected with 0.5 μg CD99 promoter-driven luciferase construct p(–137/+123)luc, together with various amounts of the LMP1 expression plasmid (pcDNA3-LMP1). Transfected cells were prepared as previously described. After protein quantitation, lysates were separated by 10% SDS-PAGE and electroblotted onto the nitrocellulose filters. The same filter was probed with monoclonal antibodies such as CS1-4 and anti-Calnexin for LMP1 and Calnexin, respectively. One microgram pNF-κB CAT was cotransfected to confirm the activity of the cotransfected LMP1. Relative luciferase activity values represent mean ± SD of 3 independent transfections, whereas CAT values are the averages of 2 transfections among them.
LMP1 represses the core promoter activity of CD99 in a dose-dependent manner.
Relative luciferase activity was measured in 293 cells cotransfected with 0.5 μg CD99 promoter-driven luciferase construct p(–137/+123)luc, together with various amounts of the LMP1 expression plasmid (pcDNA3-LMP1). Transfected cells were prepared as previously described. After protein quantitation, lysates were separated by 10% SDS-PAGE and electroblotted onto the nitrocellulose filters. The same filter was probed with monoclonal antibodies such as CS1-4 and anti-Calnexin for LMP1 and Calnexin, respectively. One microgram pNF-κB CAT was cotransfected to confirm the activity of the cotransfected LMP1. Relative luciferase activity values represent mean ± SD of 3 independent transfections, whereas CAT values are the averages of 2 transfections among them.
Sp1 plays an important role in the activation of the CD99 core promoter (Figure 1). Therefore, we examined the effect of LMP1 on the CD99 promoter containing mutated Sp1 recognition site p123mGC to see whether the Sp1 interaction is directly involved in LMP1-mediated down-regulation. Despite its crucial role in transcriptional activity, the Sp1-binding sequences of the CD99 promoter appeared not to mediate the repression of CD99 gene transcription by LMP1 so that the mutant construct displayed nearly the same level of inhibition by LMP, as shown in the wild-type promoter (data not shown). Similar results were obtained with the constructs containing Sp1 mutations downstream of the transcription start site (data not shown), indicating that the inhibition might not have been mediated through the Sp1-binding sites examined.
Involvement of the C-terminal domains of LMP1 in its repressive activity
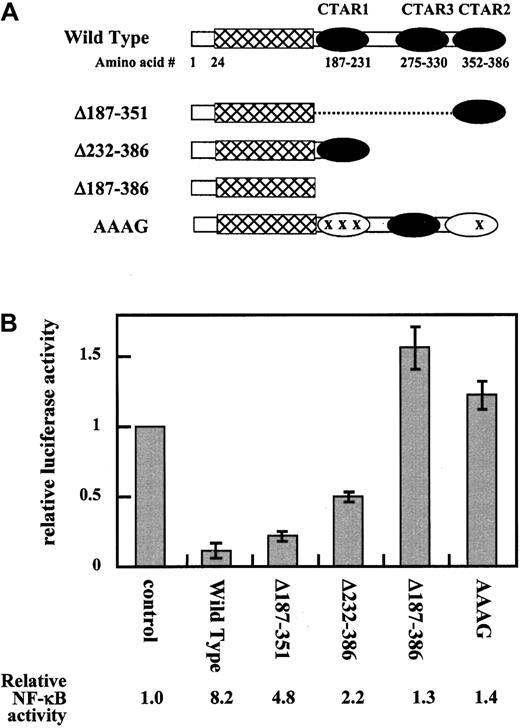
Because cis-acting elements by which LMP1 down-regulates transcription from the CD99 promoter were unidentifiable, we examined whether cellular factors previously reported to be induced by LMP1 would be involved in this event. To identify the region of LMP1 essential for its inhibitory function on the full-length CD99 promoter, a panel of LMP1 mutants was assayed in 293 cells, such as pAAAG (point mutations at amino acid 208, 210, and 212 in CTAR1 and at amino acid 384 in CTAR2), pLMP1Δ187-351 (deletion of CTAR1 and CTAR3), pLMP1Δ232-386 (deletion of CTAR2 and CTAR3), and pLMP1Δ187-386 (deletion of CTAR1, CTAR2, and CTAR3) (Figure4A).
C-terminal domain of LMP1 is required for its repressive activity.
(A) Diagram of wild-type and mutant LMP1 proteins. The 386-amino acid wild-type LMP1 consists of 3 major regions: a 24-amino acid cytoplasmic N-terminus (■), a transmembrane region (▩), and a 200-amino acid cytoplasmic carboxyl-terminal region. The carboxyl-terminal region can be further divided into 3 CTAR regions ( ). All the LMP1 point and deletion mutants are described in “Results” in detail. (B) Inhibition of CD99 promoter activity by C-terminal domain of LMP1. Various LMP1 mutants were cotransfected with p(−317/+123)luc to identify the region essential for its inhibitory function. A negative control vector pcDNA3-1-PML, a wild-type pcDNA3-LMP1, pLMP1Δ187-351, pLMP1Δ231-386, and pLMP1Δ187-386, and pAAAG are shown as control, wild-type, Δ187-351, Δ232-386, and Δ187-386 on the x-axis of the graph, respectively. The bar graph is expressed as the relative luciferase activity that is total luciferase activity divided by control transfection after the normalization with CAT activities from cotransfected pBip670CAT. Relative luciferase activity values represent mean ± SD from 3 separate transfections performed with each construct, whereas CAT values are the averages of 2 transfections among them.
). All the LMP1 point and deletion mutants are described in “Results” in detail. (B) Inhibition of CD99 promoter activity by C-terminal domain of LMP1. Various LMP1 mutants were cotransfected with p(−317/+123)luc to identify the region essential for its inhibitory function. A negative control vector pcDNA3-1-PML, a wild-type pcDNA3-LMP1, pLMP1Δ187-351, pLMP1Δ231-386, and pLMP1Δ187-386, and pAAAG are shown as control, wild-type, Δ187-351, Δ232-386, and Δ187-386 on the x-axis of the graph, respectively. The bar graph is expressed as the relative luciferase activity that is total luciferase activity divided by control transfection after the normalization with CAT activities from cotransfected pBip670CAT. Relative luciferase activity values represent mean ± SD from 3 separate transfections performed with each construct, whereas CAT values are the averages of 2 transfections among them.
C-terminal domain of LMP1 is required for its repressive activity.
(A) Diagram of wild-type and mutant LMP1 proteins. The 386-amino acid wild-type LMP1 consists of 3 major regions: a 24-amino acid cytoplasmic N-terminus (■), a transmembrane region (▩), and a 200-amino acid cytoplasmic carboxyl-terminal region. The carboxyl-terminal region can be further divided into 3 CTAR regions ( ). All the LMP1 point and deletion mutants are described in “Results” in detail. (B) Inhibition of CD99 promoter activity by C-terminal domain of LMP1. Various LMP1 mutants were cotransfected with p(−317/+123)luc to identify the region essential for its inhibitory function. A negative control vector pcDNA3-1-PML, a wild-type pcDNA3-LMP1, pLMP1Δ187-351, pLMP1Δ231-386, and pLMP1Δ187-386, and pAAAG are shown as control, wild-type, Δ187-351, Δ232-386, and Δ187-386 on the x-axis of the graph, respectively. The bar graph is expressed as the relative luciferase activity that is total luciferase activity divided by control transfection after the normalization with CAT activities from cotransfected pBip670CAT. Relative luciferase activity values represent mean ± SD from 3 separate transfections performed with each construct, whereas CAT values are the averages of 2 transfections among them.
). All the LMP1 point and deletion mutants are described in “Results” in detail. (B) Inhibition of CD99 promoter activity by C-terminal domain of LMP1. Various LMP1 mutants were cotransfected with p(−317/+123)luc to identify the region essential for its inhibitory function. A negative control vector pcDNA3-1-PML, a wild-type pcDNA3-LMP1, pLMP1Δ187-351, pLMP1Δ231-386, and pLMP1Δ187-386, and pAAAG are shown as control, wild-type, Δ187-351, Δ232-386, and Δ187-386 on the x-axis of the graph, respectively. The bar graph is expressed as the relative luciferase activity that is total luciferase activity divided by control transfection after the normalization with CAT activities from cotransfected pBip670CAT. Relative luciferase activity values represent mean ± SD from 3 separate transfections performed with each construct, whereas CAT values are the averages of 2 transfections among them.
Wild-type LMP1 strongly repressed the promoter activity by more than 7-fold over that of the vector control, as previously shown16 (Figure 4B). The mutant form pLMP1Δ187-386, however, completely lacked the repressive ability, in which the entire carboxyl terminus was deleted, including CTAR1, CTAR2, and CTAR3. The mutant pLMP1Δ187-351, with deletion in CTAR1 and CTAR3, had nearly wild-type inhibition (20% of the vector control). In addition, the mutant pLMP1Δ232-386, deleted in CTAR2 and CTAR3, had reduced but clearly evident activity (50% of the vector control). These results indicate that either CTAR1 or CTAR2 alone is sufficient for the down-regulation of CD99 but that the Jak-binding CTAR3 domain is dispensable. This was confirmed in the experiment using a construct with point mutations in CTAR1 and CTAR2, pAAAG. The construct was unable to activate NF-κB because of the point mutations and, simultaneously, because repressive activity on CD99 was lacking. The activation level of NF-κB with the various mutated LMP1 genes was measured (at the bottom of Figure 4B), and the LMP1-associated NF-κB activation was inversely related to the ability to down-regulate CD99 so that the deletion of either CTAR1 or CTAR2 had little effect, whereas inactivation of the 2 regions completely abolished NF-κB activation. Taken together, these experiments indicated that the domains responsible for the repressive activity of LMP1 colocalize those sequences important for NF-κB activation in 293. Similar repressive patterns of the various LMP1 deletions on the transcriptional regulation of CD99 were also demonstrated in a human B cell line, BJAB (data not shown).
Decreased CD99 promoter activity by LMP1 was markedly restored when cotransfecting a constitutively active form of the κB inhibitory protein, IκB αS32/36A
The carboxyl terminal domains of LMP1, which are important for NF-κB activation, were found to be responsible for its repressive activity on CD99. Hence, we tested whether the event is mediated by the NF-κB signaling pathway. Under normal conditions, NF-κB exists in a cytoplasmic complex with an inhibitor protein IκB.38-40The activation of NF-κB requires phosphorylation of IκB-α at serines 32 and 36.41 This phosphorylation targets IκB-α for ubiquitination and proteosome-mediated degradation, thereby releasing NF-κB to enter the nucleus and activate a series of genes.42 Therefore, we chose to inhibit NF-κB complexes specifically by transfection of constitutively active IκB-α mutated on serines 32 and 36 (IκBαS32/36A) to examine the involvement of an NF-κB signaling pathway in CD99 down-regulation.
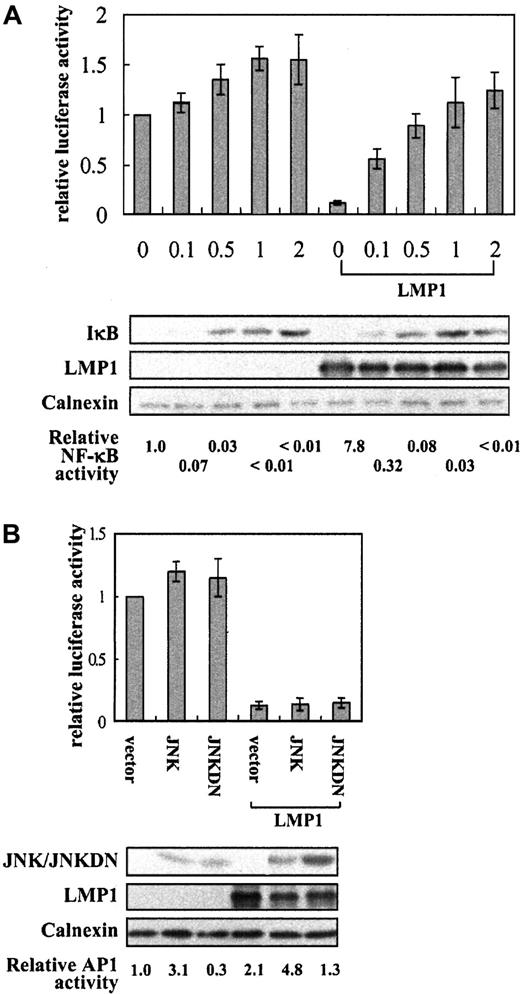
We transiently cotransfected the CD99 promoter construct with increasing amounts of IκBαS32/36A tagged with the FLAG sequence (pcDNA3-FLAG-IκBαS32/36A) in the presence or absence of LMP1 (Figure 5A). Use of IκB to investigate the role of NF-κB was technically difficult because a dose that efficiently inhibits inducible NF-κB may substantially down-regulate the expression of the cotransfected LMP1. Because we used the LMP1 expression construct driven by the CMV promoter known to be sensitive to IκB activity, we monitored LMP1 expression and assayed the effect on NF-κB CAT reporter. Transfected IκBαS32/36A was expressed in a dose-dependent manner. In the absence of LMP1, it was observed that the CD99 promoter activity was enhanced (up to 1.5-fold) by the forced expression of IκBαS32/36A, which was in contrast to the decrease of NF-κB activity. Although the effect was not marked, the result indicated that a molecule in NF-κB signaling pathway might play a role in regulating endogenous CD99 expression. In the presence of LMP1, the repressed CD99 promoter activity was remarkably enhanced (up to 9-fold) and completely restored by the cotransfected IκBαS32/36A. Albeit a slight reduction, LMP1 expression was steadily maintained at a high level when transfected with up to 2 μg IκBαS32/36A, indicating that the restoration of CD99 promoter function even in the presence of LMP1 was not caused by the depletion of LMP1.
Inhibition of CD99 by LMP1 is markedly restored by a constitutively active form of the κB inhibitory protein, IκBαS32/36A.
(A) Effect of IκBαS32/36A on the CD99 promoter activity in the presence or absence of LMP1. A half microgram of the reporter construct, p(−317/+123)luc, was cotransfected with increasing amounts (μg) of the construct expressing FLAG-IκBαS32/36A. The same filter was probed with monoclonal antibodies such as CS1-4, M5, anti-Calnexin for LMP1, FLAG- IκBαS32/36A, and Calnexin, respectively. One microgram pNF-κB CAT was cotransfected to confirm the functions of cotransfected LMP1 and FLAG-IκBαS32/36A. Values represent the mean of 3 independent transfections. (B) Relative CD99 promoter activities with various JNK constructs in the presence or absence of LMP1. M5 monoclonal antibody was used for the detection of the wild-type JNK (pcDNA3-FLAG-JNK) and its dominant-negative form, shown as JNKDN (pcDNA3-FLAG-JNK(apf)). The reporter construct, AP-1 CAT, was cotransfected to confirm the activity of cotransfected JNKs. Relative luciferase activity values represent mean of 3 independent transfections, whereas CAT values are the averages of 2 transfections among them.
Inhibition of CD99 by LMP1 is markedly restored by a constitutively active form of the κB inhibitory protein, IκBαS32/36A.
(A) Effect of IκBαS32/36A on the CD99 promoter activity in the presence or absence of LMP1. A half microgram of the reporter construct, p(−317/+123)luc, was cotransfected with increasing amounts (μg) of the construct expressing FLAG-IκBαS32/36A. The same filter was probed with monoclonal antibodies such as CS1-4, M5, anti-Calnexin for LMP1, FLAG- IκBαS32/36A, and Calnexin, respectively. One microgram pNF-κB CAT was cotransfected to confirm the functions of cotransfected LMP1 and FLAG-IκBαS32/36A. Values represent the mean of 3 independent transfections. (B) Relative CD99 promoter activities with various JNK constructs in the presence or absence of LMP1. M5 monoclonal antibody was used for the detection of the wild-type JNK (pcDNA3-FLAG-JNK) and its dominant-negative form, shown as JNKDN (pcDNA3-FLAG-JNK(apf)). The reporter construct, AP-1 CAT, was cotransfected to confirm the activity of cotransfected JNKs. Relative luciferase activity values represent mean of 3 independent transfections, whereas CAT values are the averages of 2 transfections among them.
In addition to its ability for NF-κB induction, CTAR2 of LMP1 has been also reported to trigger AP-1 activity through the SEK/JNK signaling pathway.28 Therefore, to exclude the possibility of involvement of JNK on the CD99 promoter activity, we transiently transfected either the wild-type or the dominant-negative JNK construct (shown as JNK and JNKDN, respectively, in Figure 5B) in the presence or absence of LMP1. Given that CTAR1 does not trigger AP-1 activity but still induces CD99 inhibition, we found that inhibiting JNK by transfecting a dominant-negative form does not impair the ability of LMP1 to inhibit CD99. In contrast, AP-1 activity was clearly affected by the cotransfected JNK constructs.
Discussion
In more than 40% of patients with HD, the disease is known to be associated with the expression of EBV antigens and H-RS cells are frequently LMP1-positive.43,55 Moreover, it has been recently reported in this laboratory that development of HD could be caused by the down-regulation of CD99.9 Therefore, based on these observations, we previously examined whether there was any correlation between the expression of CD99 and LMP1, particularly in terms of the pathogenesis of HD, and we found that LMP1 induces the repression of CD99 mainly at the transcriptional level.16Although these results implied that LMP1 signaling events might suppress CD99 promoter activity through the regulation of some molecules responsible for the transcription of CD99, the mechanism of the down-regulation of CD99 molecules by LMP1 was not certain. In the present study, the 5′ proximal region of the CD99 promoter was characterized to search for factors that regulate the expression from the CD99 promoter. Results from this study demonstrate that Sp1 positively mediated the core promoter activity. However, CD99 inhibition by LMP1 was not an Sp1-dependent activity. Rather, transient transfection demonstrated that the NF-κB activation domains in the cytoplasmic terminus of LMP1 were associated with its inhibition ability. Because there was no evidence of the presence of NF-κB responsive consensus sequences in the CD99 promoter region, it is likely that the inhibition ability is indirect.
The 5′-flanking sequence of the gene is GC-rich and lacks consensus TATA and CCAAT elements.13 Using the TFSEARCH program (version 1.3, threshold: 85.0 point), we found that it contains potential binding sites for a number of transcription factors such as GATA, CREB, STAT, and an NF-κB element, as well as Sp1, which has been implicated in the transcription of some TATA-less genes.44-46 However, those factors (except Sp1) are distally localized and not in the region between −137 and +23, which was mapped to be important in this study for the positive transcriptional activity of the CD99 gene. The program has identified one more putative Sp1-binding site at −119, in addition to the 3 previously reported sites in the region.13 Although the importance of the additional site should be further determined, little effect was shown when internally deleted (data not shown).
Many eukaryotic promoters contain multiple binding sites for sequence-specific DNA-binding proteins interacting synergistically to activate transcription. However, studies have shown that in some promoters Sp1 can discriminate between different consensus Sp1-binding sites, indicating that certain Sp1-binding sites contribute more to overall activity than do other sites. Typical examples of promoters that rely predominantly on only one of multiple potential Sp1 sites are the transforming growth factor β type I receptor promoter and the c-kit promoter.47,48 Deletion and site-specific mutation analyses of multiple Sp1 sites throughout the CD99 promoter also establish that one upstream site at position −95 contributes heavily to basal promoter activity compared to other sites. This result is consistent with the previous footprinting data showing that the Sp1 site at position −95 is 1 of 3 DNase I protected footprints between nucleotides −122 and +34 by a recombinant Sp1.14 One of the other 2 protected sequences localized downstream in the decanucleotide Sp1-binding site at +24 acts as a negativecis-element for the CD99 promoter activity. Although the third one is a putative Sp1-binding site at −119 identified by the computer program, no effect was detected when internally deleted, as previously mentioned (data not shown).
Transcriptional activation and repression are important mechanisms of transcriptional regulation. Usually, a GC box acts as a positive element in both TATA-containing and TATA-less promoters. In TATA-less promoters, the GC box near the initiation sites (36-70 bp upstream) often plays an important activation role in both transcription initiation and efficiency. Among many GC box-recognizing factors, Sp1 is a trans-acting transcription factor that has been critically linked to the process of normal development.49Although ubiquitously expressed, there was an unexpected difference of at least 100-fold in the expression of Sp1 among different cell types in mice. Substantial variations in Sp1 expression were also found during different stages of development in some cell types. Sp1 levels appeared to be highest in developing hematopoietic cells, fetal cells, and spermatids, suggesting that an elevated level of the Sp1 molecule is tightly associated with the differentiation process. These results indicate that Sp1 has a regulatory function during cellular development in addition to its general role in the transcription of housekeeping genes. In this context, our finding that the expression of CD99 is critically dependent on Sp1 seems to be reasonable considering the fact that CD99 is highly expressed on cortical thymocytes, pancreatic islet cells, ovarian granulosa cells, and Sertoli cells in testis,50 which also show high levels of Sp1 expression. It is, therefore, likely that Sp1 is involved in directing the expression of CD99, resulting in its variation among cell types. In addition, cotransfection studies using an Sp1 or an Sp3 expression plasmid revealed that though the expression of the transfected Sp1, in addition to the endogenous Sp1, still caused stimulation, Sp3 had little effect on Sp1-mediated transactivation of CD99. These results support the idea that tissue- and cell-specific expression of the CD99 gene may be controlled by the relative amounts of Sp1. However, the levels of CD99 expression were not exactly correlated with those of Sp1 in the cell lines with Western blot analysis (data not shown), indicating the presence of a cellular factor(s) other than Sp1 that may play an additional role in directing the regulation of CD99.
LMP1 immortalizes B lymphocytes on infection in vitro.17,51 Expression of LMP1 in B lymphocytes induces the transcription of many genes, including those encoding activation antigens, adhesion molecules, and molecules that inhibit apoptosis. In addition, the expression of LMP1 in epithelial cells has been reported to induce expression of the EGFR and the A20 molecule.23,32 Induction of these genes by LMP1 is likely to play an important role in cellular transformation. In contrast to the well-documented functions of LMP1, which are involved in the activation of many cellular signal transduction pathways, it has been recently reported that LMP1 acts as a negative transcriptional regulator on the CD99 promoter, suggesting a novel mechanism for the development of HD by LMP1.16
LMP1 is a powerful inducer of NF-κB–mediated transcription.31 Transcription factor NF-κB is a mediator of inducible gene expression in response to inflammatory cytokines and pathogens, and it controls the expression of a number of growth-promoting cytokines.38,40 Activated NF-κB is one of the common characteristic properties of H-RS cells.52Despite apparent heterogeneity in the cellular origins of H-RS cells, the clinical and histopathologic features of HD, such as the deregulated expression of various cytokines, growth factors, and cell-surface receptors are similar,53 suggesting a common pathogenic mechanism. In an experiment with a dominant-negative inhibitor of NF-κB stably introduced into H-RS cell lines, it was found that constitutive NF-κB activation is essential for both proliferation and survival of HD tumor cells.54 These data demonstrate a direct requirement of NF-κB in a human neoplastic disease, suggesting that Hodgkin lymphoma is a malignancy of deregulated NF-κB.
Because all naturally occurring LMP1 deletion variants isolated from patients with HD fully stimulate NF-κB–mediated transcription,56 the integrity of LMP1-dependent NF-κB–mediated transcriptional activation seems important for EBV-associated HD. Moreover, forced expression of the LMP1 in an EBV-negative HD cell line induced an increased number of RS cells,57 supporting the hypothesis that identical signal transduction pathways are associated with the generation of RS cells of EBV-negative and EBV-positive HD. The result suggests that the deregulation of cellular factors associated with the NF-κB signaling pathway may be also acting in LMP1-negative HD.
Based on the previous findings that CD99 down-regulation by LMP1 in B cells generates cells with an H-RS phenotype16 and that LMP1 induces NF-κB activation,31 the result of this study showing that CD99 down-regulation by LMP1 is an NF-κB-dependent event seems relevant. Although the molecular mechanism by which CD99 is down-regulated in patients with EBV-negative HD remains to be solved, some evidence supports the linkage between the loss of CD99 and the development of HD. Gokhale et al58 reported families coinheriting both HD and Leri-Weill dyschondrosteosis, whose gene is localized to the short-arm pseudo-autosomal region (PAR) of the X and Y chromosomes. In addition, it was recently revealed by linkage analysis that a putative gene for HD is localized in the PAR.59Because the mic2 gene encoding CD99 is located in Xp22.32-pter and Yp11-pter of PAR, their data provide indirect evidence of linkage between CD99 and HD.
So far, 3 independent carboxyl-terminal activation regions of LMP1—CTAR1, CTAR2, and CTAR3—are identified as related to LMP1 signaling. According to the deletion analysis of LMP1 from our results, CTAR1 and CTAR2 are responsible for, but can act independently of, each other on the transcriptional down-regulation of CD99. This result exactly mirrors the situation with regard to LMP1-mediated activation of NF-κB; the repressive effect was mapped to the NF-κB activation domains in the cytoplasmic carboxyl terminus of LMP1. Taken together, if not all then at least some of the loss of CD99 through the NF-κB pathway is likely to play a critical role in the pathogenic sequence to the formation of H-RS cells. The detailed dissection of the signaling pathway in mediating negative signals to the CD99 promoter is under investigation and will provide molecular insight into the role of CD99 in the generation of H-RS cells.
Supported in part by research funding from DiNonA Inc and from the 2000 BK21 project for Medicine, Dentistry and Pharmacy. M.R. and A.M. were funded by the Leukemia Research Fund, London.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Seong Hoe Park, Department of Pathology, Seoul National University College of Medicine, 28 Yongon-dong Chongno-gu, Seoul 110-799, Korea; e-mail: pshoe@plaza.snu.ac.kr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal