The incidence and role of p53 abnormalities have not been reported in splenic lymphoma with villous lymphocytes (SLVL), the leukemic counterpart of splenic marginal zone lymphoma. Because p53 abnormalities correlate with progressive and refractory disease in cancer and isochromosome 17q has been described in SLVL, a low-grade lymphoma that behaves aggressively in a minority of patients, this study investigated p53 changes by molecular and immunophenotypic methods in samples from 59 patients. The p53 deletion was analyzed by fluorescence in situ hybridization, and p53 protein expression was assessed by immunocytochemistry in 35 of 59 cases and by flow cytometry in 20 of 35 patients. Ten patients (17%) had a monoallelic p53 loss, 3 (9%) of 35 nuclear protein expression by immunocytochemistry, and 2 (10%) of 20 by flow cytometry. Two patients had both deletion and protein expression. Direct sequencing of all p53 exons was used to delineate mutations in 9 of 11 patients with an identified abnormality. Mutations, both compromising p53 DNA binding, were identified in the 2 patients with deletion and protein accumulation. Kaplan-Meier analysis revealed a significantly worse survival for patients with p53 abnormalities. Although p53 abnormalities are infrequent in SLVL, they underlie a more aggressive disease course and poor prognosis.
Introduction
Splenic lymphoma with villous lymphocytes (SLVL) is an indolent, low-grade B-cell lymphoma characterized by splenomegaly and circulating villous lymphocytes. It is considered to be the leukemic counterpart of the splenic marginal zone lymphoma (SMZL). The immunophenotype,1 spleen histology,2 and clinical behavior3,4 have been established, but the pathogenesis of the disease remains unclear. SLVL is associated with a protracted clinical course, but behaves more aggressively in a minority of patients and the reasons for such differences in clinical evolution are unknown. The presence of isochromosome17q in a proportion of patients with SLVL5 suggests that monoallelic loss of p53 may be an important pathogenetic event in SLVL because thep53 gene is located on the short arm of chromosome 17 (17p13).1 However, the incidence and role of p53 abnormalities have not been reported in SLVL to date.
The p53 gene encodes for a protein with 393 amino acids, which acts as a multifunctional transcription factor involved in cell cycle arrest, apoptosis, cell differentiation, replication, DNA repair, senescence, and maintenance of genomic stability (reviewed in references 6 and 7). Cell cycle arrest and apoptosis are the most understood functions of p53 and both are fundamental for p53 functioning as a tumor suppressor gene. The p53 gene is the most frequently mutated gene in human and animal neoplasia8with over 10 000 reported mutations.6 Statistically significant correlations between p53 mutations and poor prognosis have been reported in colon cancer.9 Most mutations in thep53 gene have been found in the DNA binding domain (exons 5-8). However, the frequency of mutations in other regions of the gene may have been previously underestimated.
There are conflicting data in literature concerning the influence of p53 mutations on disease progression in non-Hodgkin lymphomas. It has been suggested that the presence of p53 mutations influences the pathologic grade10 but not the clinical behavior. Several studies showed a strong correlation between clinical progression and occurrence of p53 mutations,11-17 but others did not.18,19 Our aim was to establish the frequency and importance of abnormalities in p53 in SLVL. For this we screened a series of patients using fluorescence in situ hybridization (FISH) to detect loss of the p53 allele as well as immunocytochemistry and flow cytometry to examine protein expression. Direct sequencing of all 11 exons of the p53 gene was used to search for mutations in patients with p53 deletion or protein expression or both. The rationale for this strategy was based on the observation that in majority of cancers the presence of a p53 mutation is accompanied by loss of the other allele and, therefore, patients with a hemizygous p53 deletion may have a mutation in the remaining allele.20Furthermore, mutant p53 protein commonly accumulates in cancer cells and thus, combining FISH and immunocytochemistry allows the identification of patients with mutation but no deletion.
Patients and methods
Patients
Thirty women and 29 men were studied; their median age was 69 years (range, 39-95). Median white blood cell (WBC) count at diagnosis was 19.5 × 109/L (range, 2.3-138). All but 5 patients presented with splenomegaly; this information was not available in 3 patients, and 3 of 5 patients with no splenomegaly at diagnosis later developed this feature. The diagnosis of SLVL was established on the basis of immunophenotype, peripheral blood morphology, bone marrow histology in patients with follow-up data, and spleen histology2 in 27 patients. The immunophenotype was that of a mature B cell with moderate-to-strong surface immunoglobulin expression, strong expression of CD22 and FMC7.1Immunologically, differential diagnosis was conducted on the basis of the chronic lymphocytic leukemia (CLL)21 and hairy cell leukemia (HCL)22 scores. The CLL and HCL scores were low: 70% of patients had a CLL score of 0 or 1 and 95% an HCL score of 0 or 1. Analysis of peripheral blood films revealed the presence of medium-sized lymphocytes with scanty cytoplasm, short and fine cytoplasmic villi, sometimes polarized, and mature clumped chromatin. Patients were studied within 0 to 105 months from diagnosis (median, 1.5 mo). Diagnostic samples were studied in 18 patients and samples from another 21 patients were obtained within the first 6 months of the disease course. Forty-two patients (71%) received no treatment before testing for p53 abnormalities and another 4 had only had splenectomy before inclusion in the study. Therefore, a total 46 (78%) of 59 patients not previously treated with chemotherapy were tested for presence of p53 abnormalities. No patients were studied immediately after treatment. Follow-up information was available in 49 patients.
FISH analysis
The FISH analyses were performed using standard methods. All patients' samples contained at least 40% tumor cells (median, 70% tumor cells in the samples included in the study). Low-density (< 1.077 g/mL) cells were obtained by centrifugation of heparinized peripheral blood (51 cases), bone marrow (2 cases), or separated spleen cells (6 cases) over Histopaque (Sigma, Poole, United Kingdom), washed 2 times in Hanks balanced salt solution, fixed in methanol/glacial acetic acid (3:1), and stored at −20°C until needed. When fresh cells were not available (7 cases), frozen low-density cells were retrieved from liquid nitrogen and prepared in the same way. Slides were made by placing 15 μL cell suspension onto a microscope slide and drying overnight at room temperature. Hybridization was performed as follows: slides were incubated with RNAse A (0.1 mg/mL, Sigma) at 37°C for 1 hour, washed in 2 × standard sodium citrate (SSC) at room temperature for 5 minutes. This was followed by digestion in pepsin (0.1 mg/mL, Sigma) at 37°C for 10 minutes and washed with phosphate-buffered saline (PBS) at room temperature for 5 minutes and in 2 × SSC at 37°C for 30 minutes. Slides were then dehydrated in 70%, 90%, and 100% ethanol series (3 minutes in each) and dried. Denaturation took place on a dry-heat block at 72°C using 100 μL denaturing solution (70% formamide, 2 × SSC and 0.05 M sodium phosphate buffer, pH 7.0) under a 22 × 50-mm cover slip. Slides were then quickly rinsed in 2 × SSC, dehydrated, and dried. We used a p53 locus-specific probe (LSI p53 Spectrum Orange, Vysis, Richmond, United Kingdom) in combination with a probe specific for the chromosome 17 centromere (CEP17 Spectrum Green, Vysis) to exclude the possibility of monosomy 17 causing loss of hybridization signal for the p53-specific probe. Probes were prepared and denatured according to the manufacturer's instructions. Hybridization was carried out overnight at 37°C. Washes after hybridization consisted of 3 washes in 1 × SSC at 45°C followed by 3 washes in 0.1 × SSC at 60°C and 1 wash in 4 × SSC/Triton X-100 at room temperature, dehydrated in ethanol series, dried, and mounted with Vectashield mounting medium with DAPI (Vector Labs, Peterborough, United Kingdom).
Slides were assessed on a fluorescent microscope (Zeiss, Oberkochen, Germany) equipped with dual- and triple-band pass filters. Two hundred nuclei were scored per patient sample. Cells from 10 healthy donors prepared as above were used as normal controls. Five hundred nuclei were scored per control slide. All cases in whom hemizygous deletion of p53 was detected using the Vysis p53 probe were retested using another probe specific for the p53 locus (Oncor, Uxbridge, United Kingdom).
Immunocytochemistry
The immunoperoxidase method was used to test for p53 protein expression. Cytospin preparations from 35 patients were fixed in acetone for 10 minutes and air-dried. Two murine monoclonal antibodies were used for the staining: DO7 (Novocastra, Newcastle-upon-Tyne, United Kingdom) recognizing the first 45 amino acids of both wild-type and mutant p5323 and P1801 (Santa Cruz Biotechnologies, Santa Cruz, CA) against the amino terminal epitope consisting of amino acids 32 to 79, both at 1:20 dilution. Cytospins were incubated with anti-p53 antibody for 30 minutes, then washed and incubated with peroxidase-conjugated rabbit antimouse antibody (Dako, Ely, United Kingdom) diluted 1:20 for 30 minutes. Following a further wash peroxidase antiperoxidase mouse monoclonal antibody diluted 1:40 was applied for 40 minutes. The reaction was developed with 3,3′-diaminobenzidine tetrachloride chromogen and slides were counterstained with hematoxylin. A T-lymphoblastic leukemia cell line, CEM, with 2 missense mutations located on 2 separate alleles24 and strongly overexpressing p53 was included in each experiment as a positive control and lymphocytes from normal donors were used as a negative control. Cells were considered positive when the staining was present in the nucleus.
Flow cytometry
Low-density cells were fixed in cold 2% paraformaldehyde for 30 minutes at 4°C and permeabilized with ice-cold 80% ethanol added drop-wise while vortexing. At this stage cells were stored at −20°C for up to 2 weeks. Staining consisted of a 15-minute incubation with 5 μL monoclonal antibody recognizing amino acids 11 to 25 of both wild-type and mutant p53 (clone DO1, Novocastra) in 50 μL 2% AB serum. Following 3 washes in PBS-azide, cells were incubated with fluorescein isothiocyanate (FITC)-conjugated goat antimouse immunoglobulin G (IgG) F(ab)2 (1:25) (ICN Biomedicals, Basingstoke, United Kingdom). Isotypic control was included for each sample. Cells were analyzed on FACScan flow cytometer (Becton Dickinson, Oxford, United Kingdom).
Direct sequencing
Direct sequencing was conducted in 9 samples with deletion or protein expression or both, obtained at the same time as those used for FISH experiments. There was no material available in 2 patients with deletion. Available samples contained at least 60% tumor cells. Sequencing of all 11 exons of p53 was performed by polymerase chain reaction (PCR) amplification of exonic sequences from genomic DNA followed by fluorescent automated cycle sequencing of both DNA strands.
PCR amplification.
High-molecular-weight DNA was extracted from cryopreserved cells by standard methods. The quality of DNA was checked by electrophoresis of DNAs digested with EcoRI. PCR was performed in a total volume of 25 μL using 0.5 μL of genomic DNA (1 mg/mL), 2.5 μL 10 × Thermoprime Plus Taq buffer, 1.5 μL MgCl2 (25 mM), 1 μL dNTP (10 mM), 0.5 μL Thermoprime Plus Taq polymerase (1 U/μL) (all reagents from ABgene, Epsom, United Kingdom), 1 μL of each primer (5 μM), and water. The cycling program comprised 35 cycles of denaturation (94°C; 1 minute), annealing (exons 1, 2/3, 4, and 10: 55°C; exons 5, 6, 9, and 11: 61°C; exon 7: 62°C; exon 8: 59°C; 1 minute), and extension (72°C, 2 minutes). Exons 2 and 3 were amplified together because they and the intron dividing them are very short. Because of the CG-richness of exon 11 and to avoid multiple products, we used a touch-down protocol with 10 extra cycles starting with annealing temperature of 71°C and lowering by 1°C per cycle until the desired 61°C was reached. The sequences of the primers were as follows: exon 1: forward 5′GAGAATCCTGACTCTGCACC3′, reverse 5′AGCCGAGAGCCCGTGACTCA3′; exon 2/3: forward 5′ATGCTGGATCCCCACTTTTC3′, reverse 5′AAGAGCAGTCAGAGGACCAG3′; exon 4: forward 5′GACCTGGTCCTCTGACTGCT3′, reverse 5′GCATTGAAGTCTCATGGAAG3′; exon 5: forward 5′ACTTGTGCCCTGACTTTCAACT3′, reverse 5′CAATCAGTGAGGAATCAGAGGC3′; exon 6: forward 5′TCAGATAGCCGATGGTGAGCAG3′, reverse 5′GCCACTGACAACCACCCTTA3′; exon 7: forward 5′AGGCGCACTGGCCTCATCTT3′, reverse 5′GAAATCGGTAAGAGGTGGGC3′; exon 8: forward 5′GGAGTAGATGGAGCCTGGTTT3′, reverse GGTGATAAAAGTGAATCTGAGGC3′; exon 9: forward 5′GGAGACCAAGGGTGCAGTTAT3′, reverse 5′GTTAGTTAGCTACAACCAGGAGCC3′; exon 10: forward 5′CAATTGTAACTTGAACCATC3′, reverse 5′ATGAGAATGGAATCCTATGG3′; and exon 11: forward 5′CTCACTCATGTGATGTCATC3′, reverse 5′CAAAATGGCAGGGGAGG3′.
Sequencing reaction.
The same primers as the ones used for PCR were used for the sequencing. Sequencing reaction was performed using 4 μL reaction premix (dRhodamine Terminator Ready Reaction Kit; ABI, Perkin Elmer, Warrington, United Kingdom), 4 μL primer, and 4 μL PCR product. The following cycling conditions were used: denaturation (96°C, 30 seconds), annealing (50°C, 15 seconds), and extension (60°C, 60 seconds) for 25 cycles. Excess of dye terminator was removed by precipitation of DNA in 100 μL 80% ethanol and 6 μL sodium acetate (3 M, pH 5.2) for 10 minutes on ice. All PCR products were sequenced in both directions. All mutations were resequenced from a different PCR product. Obtained DNA sequences were analyzed using Sequence Analysis software (version 3.0) (ABI) and aligned and compared to published p53 sequence using Sequence Navigator software (ABI).
Results
By combining FISH and immunocytochemistry, 11 of 59 patients were identified with a monoallelic deletion in the p53 locus (with or without accumulation of p53 protein in the nucleus) or with protein expression alone. Of the 42 previously untreated patients, 9 had p53 abnormalities (Table 1).
Summary of p53 abnormalities in splenic lymphoma with villous lymphocytes obtained by different techniques
| Abnormality: Technique . | Patients studied . | Patients with p53 abnormalities . | ||
|---|---|---|---|---|
| All . | Untreated prior to testing . | All . | Untreated prior to testing . | |
| p53 deletion: | ||||
| FISH | 59 | 42 | 10 (17%) | 8 (19%) |
| p53 protein expression: | ||||
| Immunocytochemistry | 35/59* | 22/42* | 3 (9%)1-153 | 3 (13.6%)1-153 |
| Flow cytometry | 20/35† | 15/22† | 2 (10%) | 2 (13.3%) |
| p53 mutations: | ||||
| Direct sequencing | 9/11‡ | 8/9‡ | 2 (22%) | 2 (25%) |
| Abnormality: Technique . | Patients studied . | Patients with p53 abnormalities . | ||
|---|---|---|---|---|
| All . | Untreated prior to testing . | All . | Untreated prior to testing . | |
| p53 deletion: | ||||
| FISH | 59 | 42 | 10 (17%) | 8 (19%) |
| p53 protein expression: | ||||
| Immunocytochemistry | 35/59* | 22/42* | 3 (9%)1-153 | 3 (13.6%)1-153 |
| Flow cytometry | 20/35† | 15/22† | 2 (10%) | 2 (13.3%) |
| p53 mutations: | ||||
| Direct sequencing | 9/11‡ | 8/9‡ | 2 (22%) | 2 (25%) |
FISH indicates fluorescence in situ hybridization.
Cytospins available in 35 of 59 (22 of 42 for untreated) patients in whom FISH was performed.
Cells for flow cytometry available in 20 of 35 (15 of 22 for untreated) analyzed by immunocytochemistry.
DNA available in 9 of 11 (8 of 9 for untreated) patients with p53 deletion and/or p53 protein accumulation.
Protein expression was found in 2 of 10 (2 of 8 for untreated) patients with a p53 deletion, one patient had protein expression without accompanying deletion and was only tested by immunocytochemistry.
FISH analysis
The cut-off level for the confident detection of a deletion in the p53 locus for both p53 probes was 5%, calculated as mean plus 3 SDs of cells with one hybridization signal seen in 10 normal controls. FISH analysis revealed the presence of hemizygous deletion in 10 of 59 cases (8 of 42 previously untreated or 10 of 46 when including patients treated only with splenectomy prior to testing). The deletion was present in 15% to 91% of nuclei (Figure1). All cases with p53 deletion according to the Vysis probe were confirmed using the Oncor probe. Two patients were studied at different times during the course of the disease. One of them was studied at presentation, at disease progression, and in the final terminal phase. There was no p53 deletion at diagnosis, the second sample contained 15% cells with hemizygous deletion, and the last sample showed 60% cells with one hybridization signal. The other patient had hemizygous deletion in 80% nuclei at presentation and 88% 2 years later at the time of transformation into a high-grade lymphoma.
FISH analysis of
p53 gene copy number in SLVL. Double-color FISH analysis using a probe for chromosome 17 centromere (green signal) and locus-specific probe for p53 (red signal) allowing for identification of cases with one signal for the p53 probe due to monosomy 17. (A) Normal control showing 2 signals for centromere 17 and 2 signals for p53. (B) A patient with hemizygous deletion of p53 (2 green and 1 red signals).
FISH analysis of
p53 gene copy number in SLVL. Double-color FISH analysis using a probe for chromosome 17 centromere (green signal) and locus-specific probe for p53 (red signal) allowing for identification of cases with one signal for the p53 probe due to monosomy 17. (A) Normal control showing 2 signals for centromere 17 and 2 signals for p53. (B) A patient with hemizygous deletion of p53 (2 green and 1 red signals).
p53 expression
Cytologic preparations for immunocytochemical analysis were available in 35 of 59 patients, which included all patients with deletion. Three (9%) of 35 had nuclear accumulation of p53 (Figure2). Two patients had p53 deletion and p53 protein expression. One patient had protein expression and no p53 deletion. Both monoclonal antibodies used gave the same results. Flow cytometric analysis of p53 protein expression was performed in 20 patients and the protein was detected in 2 (Figure 2) who were also positive by immunocytochemistry; the third patient expressing p53 protein by immunocytochemistry was not studied by flow cytometry.
Analysis of p53 protein expression in SLVL.
(A) Positive staining with anti-p53 antibody (clone DO7) using peroxidase-antiperoxidase method on a cytospin preparation of separated spleen cells from a patient with SLVL. p53 is visualized as brown precipitate in the nucleus of the cells. (B) Flow cytometric analysis of p53 protein expression in a patient with SLVL; cells were fixed with paraformaldehyde and permeabilized with 80% ethanol. In an indirect staining protocol, a monoclonal antibody (clone DO1) recognizing an epitope comprising amino acids 11 to 25 of the p53 molecule was used for detection of p53 expression or an isotypic control was used as first layer followed by a second layer—FITC-conjugated goat antimouse IgG F(ab)2.
Analysis of p53 protein expression in SLVL.
(A) Positive staining with anti-p53 antibody (clone DO7) using peroxidase-antiperoxidase method on a cytospin preparation of separated spleen cells from a patient with SLVL. p53 is visualized as brown precipitate in the nucleus of the cells. (B) Flow cytometric analysis of p53 protein expression in a patient with SLVL; cells were fixed with paraformaldehyde and permeabilized with 80% ethanol. In an indirect staining protocol, a monoclonal antibody (clone DO1) recognizing an epitope comprising amino acids 11 to 25 of the p53 molecule was used for detection of p53 expression or an isotypic control was used as first layer followed by a second layer—FITC-conjugated goat antimouse IgG F(ab)2.
Direct sequencing
Material for direct sequencing was available in 9 of 11 with p53 deletion or protein expression or both. The results of the sequencing are shown in Table 2. Two (22%) of 9 patients studied had a p53 mutation. In both patients, the mutation was located in exon 8. In one patient, we found a heterozygous point missense mutation, involving a transition in codon 273, where CGT (Arg) was replaced by CAT (His). In the second patient, a heterozygous 6-bp deletion was detected at the very beginning of exon 8 (GGTAAT, codons 262-263).
Results of direct sequencing performed in patients with p53 deletion or protein expression or both
| Patient code . | p53 deletion (% nuclei) . | Protein expression . | Direct sequencing . | |||
|---|---|---|---|---|---|---|
| Exon . | Codon . | Mutation/polymorphism . | Amino acid substitution . | |||
| A | 50 | No | 4 | 72 | CGC to CCC | Arg to Pro |
| B | No deletion | Yes | No mutation found | |||
| C(1994) | No deletion | No | No mutation found | |||
| (1997) | 15 | No | No mutation found | |||
| (1998) | 60 | No | No mutation found | |||
| D | 83 | No | No DNA available | |||
| E | 75 | No | No DNA available | |||
| F | 65 | No | 4 | 32 | CCG to CCA | Not altered |
| 4 | 72 | CGC to CCC | Arg to Pro | |||
| G | 70 | No | No mutation found | |||
| I | 44 | No | 4 | 72 | CGC to CCC | Arg to Pro |
| J(1997) | 80 | Yes | 8 | 273 | CGT to CAT* | Arg to His |
| (1999) | 88 | Yes | 8 | 273 | CGT to CAT† | Arg to His |
| K | 91 | No | No mutation found | |||
| L | 56 | Yes | 8 | 262-263 | 6-bp deletion (GGTAAT)* | Gly-Asn deleted |
| Patient code . | p53 deletion (% nuclei) . | Protein expression . | Direct sequencing . | |||
|---|---|---|---|---|---|---|
| Exon . | Codon . | Mutation/polymorphism . | Amino acid substitution . | |||
| A | 50 | No | 4 | 72 | CGC to CCC | Arg to Pro |
| B | No deletion | Yes | No mutation found | |||
| C(1994) | No deletion | No | No mutation found | |||
| (1997) | 15 | No | No mutation found | |||
| (1998) | 60 | No | No mutation found | |||
| D | 83 | No | No DNA available | |||
| E | 75 | No | No DNA available | |||
| F | 65 | No | 4 | 32 | CCG to CCA | Not altered |
| 4 | 72 | CGC to CCC | Arg to Pro | |||
| G | 70 | No | No mutation found | |||
| I | 44 | No | 4 | 72 | CGC to CCC | Arg to Pro |
| J(1997) | 80 | Yes | 8 | 273 | CGT to CAT* | Arg to His |
| (1999) | 88 | Yes | 8 | 273 | CGT to CAT† | Arg to His |
| K | 91 | No | No mutation found | |||
| L | 56 | Yes | 8 | 262-263 | 6-bp deletion (GGTAAT)* | Gly-Asn deleted |
Heterozygous mutation.
Homozygous mutation.
In 2 cases samples from several time points were included. In one, no mutations were found at any stage of the disease despite a rising percentage of cells with p53 deletion. In the other patient, a heterozygous mutation was detected in the presentation sample. A homozygous mutation in the same codon was found in a sample from 2 years later (Figure 3A).
Mutational analysis of p53 by direct sequencing in SLVL.
Sequencing was performed in 9 patients with p53 deletion or protein expression or both. p53 mutations were identified in 2 patients. (A) A point missense transition mutation in codon 273 in patient J changing CGT (Arg) to CAT (His), heterozygous (*) at diagnosis, which became homozygous (**) 2 years later. (B) A heterozygous 6-bp deletion (2 entire codons: 262-263), leading to a protein lacking 2 amino acids (Gly-Asn), at the beginning of exon 8 in patient L.
Mutational analysis of p53 by direct sequencing in SLVL.
Sequencing was performed in 9 patients with p53 deletion or protein expression or both. p53 mutations were identified in 2 patients. (A) A point missense transition mutation in codon 273 in patient J changing CGT (Arg) to CAT (His), heterozygous (*) at diagnosis, which became homozygous (**) 2 years later. (B) A heterozygous 6-bp deletion (2 entire codons: 262-263), leading to a protein lacking 2 amino acids (Gly-Asn), at the beginning of exon 8 in patient L.
There were no mutations found outside of the highly conserved region of p53 (exons 5-9). However, we found p53 polymorphisms in exon 4 in 3 patients. These included a change of CCG (Pro) to CCA (Pro) in codon 36 in one patient and CGC (Arg) to CCC (Pro) in codon 72 in 3 patients.
Clinical correlations
Table 3 shows clinical features in patients with and without p53 abnormality. There were no differences between the groups regarding age, degree of lymphocytosis, duration of disease, and number of treatments received. Peripheral blood morphology, immunophenotype, and bone marrow infiltration patterns at diagnosis were not different either. There was, however, a higher number of men in the group with p53 abnormalities and more cases with disease progression and lymphoma-related death (Table 3).
Clinical features of splenic lymphoma with villous lymphocytes patients with and without p53 abnormality
| . | Patients . | Significance . | |
|---|---|---|---|
| Normal p53 . | Abnormal p53 . | ||
| No. of patients | 48 | 113-150 | |
| Median age at diagnosis | 69 | 69 | |
| Patient sex | 30F, 19M | 2F, 9M | χ2P < .025 |
| WBC count at diagnosis: mean, median, range (×109/L) | 24.8, 18.0, 5.0-138.0 | 26.8, 21.0, 2.5-53.0 | Wilcoxon rank sum test: not significant |
| Lymphocyte count at diagnosis: mean, median, range (×109/L) | 18.5, 12.9, 1.8-107.6 | 21.5, 20.0, 2.1-43.5 | Wilcoxon rank sum test: not significant |
| Duration of disease at the time of p53 study (mo): mean, median, range | 17.9, 1.7, 0-103.0 | 14.0, 1.5, 0-105.0 | |
| No. of patients with follow-up data | 38 | 11 | |
| No. of patients with progressive disease | 8 | 8 | χ2P < .01 |
| No. of patients dead of lymphoma | 7 | 8 | χ2P < .001 |
| No. of patients with no treatment | 7 | 1 | χ2 not significant |
| No. of patients with ≤ 1 treatment | 14 | 3 | χ2 not significant |
| . | Patients . | Significance . | |
|---|---|---|---|
| Normal p53 . | Abnormal p53 . | ||
| No. of patients | 48 | 113-150 | |
| Median age at diagnosis | 69 | 69 | |
| Patient sex | 30F, 19M | 2F, 9M | χ2P < .025 |
| WBC count at diagnosis: mean, median, range (×109/L) | 24.8, 18.0, 5.0-138.0 | 26.8, 21.0, 2.5-53.0 | Wilcoxon rank sum test: not significant |
| Lymphocyte count at diagnosis: mean, median, range (×109/L) | 18.5, 12.9, 1.8-107.6 | 21.5, 20.0, 2.1-43.5 | Wilcoxon rank sum test: not significant |
| Duration of disease at the time of p53 study (mo): mean, median, range | 17.9, 1.7, 0-103.0 | 14.0, 1.5, 0-105.0 | |
| No. of patients with follow-up data | 38 | 11 | |
| No. of patients with progressive disease | 8 | 8 | χ2P < .01 |
| No. of patients dead of lymphoma | 7 | 8 | χ2P < .001 |
| No. of patients with no treatment | 7 | 1 | χ2 not significant |
| No. of patients with ≤ 1 treatment | 14 | 3 | χ2 not significant |
WBC indicates white blood cell.
Ten patients with p53 deletion (including 2 with coinciding deletion and protein expression) and 1 patient with p53 protein expression without a deletion.
Management of all patients included splenectomy (27 of 49 patients with follow-up information), often as first-line treatment and chemotherapy, both either alone or in combination. One patient received splenic irradiation. Both groups of patients received the same therapeutic agents when treated: alkylating agents (chlorambucil or cyclophosphamide), fludarabine, and combination therapies such as cyclophosphamide, adriamycin, vincristine, prednisolone (CHOP); cyclophosphamide, vincristine, prednisolone (COP); or prednisolone, mitoxantrone, cyclophosphamide, etoposide, bleomycin, methotrexate (PMitCEBOM). However, more patients with abnormal p53 received combination chemotherapy when their condition deteriorated.
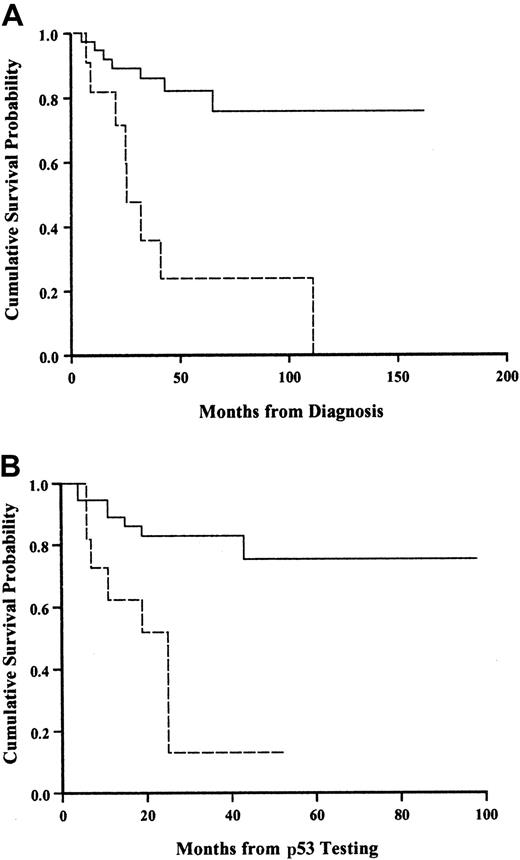
Figure 4 shows the survival curve for patients with and without a p53 abnormality. The presence of a p53 abnormality strongly predicted for shorter survival. To ensure that these findings were not an artifact reflecting a different time from diagnosis to testing for p53 abnormalities, we calculated the median time from diagnosis in both groups and showed that there was no difference (Table 3).
Kaplan-Meier survival analysis in SLVL patients with and without p53 abnormality.
(A) Calculated from diagnosis. (B) Calculated from the time of testing. Solid line, normal p53; broken line, abnormal p53. Log-rank,P < .001.
Kaplan-Meier survival analysis in SLVL patients with and without p53 abnormality.
(A) Calculated from diagnosis. (B) Calculated from the time of testing. Solid line, normal p53; broken line, abnormal p53. Log-rank,P < .001.
Eight of 11 patients with p53 alterations in this series died after a median of 28 months from diagnosis (range, 10-108 months). One of the 2 patients with mutations died within 25 months from diagnosis with a WBC count of 416 × 109/L. The other patient with a 6-bp deletion mutation is alive 25 months from diagnosis although disease has progressed.
Discussion
Various techniques have been used to study p53 in cancers. Direct sequencing is a sensitive procedure, which can detect mutations in samples with as few as 20% abnormal cells. Single-stranded conformational polymorphism (SSCP) analysis is a relatively simple technique with 10% sensitivity for DNA fragments up to 400 bp.25 However, it requires confirmation by direct sequencing to rule out polymorphisms and silent mutations. Immunocytochemistry allows identification of p53 protein accumulation at a single-cell level. Nevertheless, detection of immunoreactive p53 does not always correlate with presence of a mutation (patient B from this series18,26,27), and mutations leading to no expression or synthesis of a truncated p53 molecule will not be detected by immunocytochemistry.28 Loss of heterozygosity (LOH) analysis has been used as an indirect marker of p53 mutation for this is often accompanied by deletion of the other allele. However, it is possible to have LOH in the polymorphic loci on 17p without p53 deletion26 and, if allelic loss and the occurrence of a mutation are separated in time, LOH may not invariably indicate a mutation. Moreover, loss of the intact allele is not required for p53 function disruption because some mutants inactivate wild-type protein in a dominant-negative fashion by inducing conformational change.29 Locus-specific FISH is a direct way of revealing p53 allele deletion overcoming some of the limitations of LOH. No studies have compared directly the incidence of p53 deletion by FISH and p53 mutations by direct sequencing to date.
In this study of SLVL (n = 59), we identified 11 subjects with a p53 abnormality: 8 with hemizygous p53 deletion alone, 2 with both hemizygous p53 deletion and protein expression, and 1 with protein expression alone. Direct sequencing performed on 9 patients showed mutations in 2, both of whom had hemizygous p53 deletion and p53 protein expression.
In 4 of 6 cases with deletion but no p53 mutations (Table 3), the studies were conducted on diagnostic samples. A possibility therefore exists that a single allele deletion may represent the first mutational event in the process of losing p53 function and that this is followed by a second step—a mutation in the other allele leading to a total inactivation of wild-type p53 in tumor cells. Such a sequence of events has been demonstrated in chronic myeloid leukemia in which loss of p53 allele had preceded the occurrence of a detectable mutation.30 Substantiating our hypothesis are findings in patient J (Table 2) in whom FISH showed a deletion in 80% of nuclei and we found a heterozygous mutation in codon 273. To explain this result, we speculated that the mutation was present in a subclone emerging from within the clone with the deletion. We therefore repeated direct sequencing on a later sample, which exhibited 88% hemizygous deletion, and found only mutant p53 by direct sequencing. Thus, the change from a heterozygous to a homozygous mutant p53 status occurred in the presence of a persistently high frequency of cells bearing hemizygous p53 deletion, consistent with the notion that the mutated allele was present in a subclone that arose in the deleted population. Because this study was conducted retrospectively on cases referred to us for diagnosis, we were unable to pursue this point further.
One of the 2 patients (patient J) with mutations had a point missense mutation in codon 273. This codon is located within a CpG island and is the most frequently mutated site (> 7% of all mutation) within p53 with mutations resulting in defective DNA binding and therefore loss of ability of p53 to act as a transcriptional factor. Arg273 is directly involved in phosphate-backbone contact in the major groove of DNA helix.31 Single base substitutions at CpG islands are the commonest mutations in cancers and constitute 23% of all p53 changes. The second patient (patient L) with a mutation (Table 2) had a heterozygous 6-bp deletion of 2 full codons not altering the reading frame in exon 8. This involved codons 262 to 263 and also changed the DNA binding of p53 as it caused loss of 2 amino acids, Gly and Asn, leading to changes in the tertiary structure of the protein. This to our knowledge is the first description of this mutation (not in the IARC p53 mutations database32). Deletions involving complete codons and therefore not causing a frameshift are the rarest of the mutation types seen in p53.
It is not known whether loss of one p53 allele without a detectable mutation in the other (haploid insufficiency) is sufficient for the cell to gain a growth advantage. In a study of p53 gene deletions in myeloma,33 hemizygous p53 deletions were associated with shorter patient survival. Hemizygous p53 deletions were also shown to correlate with disease progression and resistance to treatment in CLL34 and in other B-cell disorders with p53 deletions due to gross chromosomal abnormalities.35However, one study on a large series of patients with non-Hodgkin lymphoma found that p53 deletion did not influence treatment outcome or prognosis.19 Our data suggest that the presence of a deletion in the p53 locus is significant for the clinical course of SLVL. Six of 7 cases with p53 deletion and no detectable mutation had progressive disease, which in 4 cases was resistant to treatment with alkylating agents.
Although, there are no published data concerning p53 alterations in SLVL, there are some reports of p53 mutations in SMZL,36-39 a disease sharing spleen histology with SLVL.2 The reported frequency of p53 abnormalities in SMZL ranges from 10% to 40%. Using SSCP and direct sequencing of exons 5 to 8, Sol Mateo and colleagues39 found p53 gene mutations in 10% (2 of 20) of SMZL cases. It is worth noting that the 2 mutations identified by these authors were found in codons 245 and 272. Codon 245, along with codon 273, is another of the 6 mutational “hotspots” at CpG islands, and Gly at this position plays a role in stabilizing the structure of the DNA binding surface of p53.31 Codon 272 belongs to the highly conserved region V of the p53 (residues 270-286) and is also part of the DNA binding domain.31 Lloret and coworkers38 studied 6 patients with SMZL and aggressive clinical course including 4 with villous cells in peripheral blood; 2 of these 6 patients (both with villous cells) had inactive p53. Baldini and coworkers36found a p53 mutation in 6 of 15 cases and more recently, they suggested that p53 mutations correlate with an aggressive disease course.37
The frequency of p53 abnormalities in the present series was 17%. The difference in incidence compared to previous reports in SMZL may be explained by differences in patient selection, group size, and methodology. We have studied patients with the diagnosis of SLVL, whereas Lloret and coworkers38 restricted their study to patients with aggressive clinical course and others36 39applied the technique of SSCP, which does not detect allelic loss of p53.
Our study suggests that p53 deletions are relatively infrequent in SLVL and that mutations are present in about 22% of patients with deletions. The mutations in SLVL/SMZL appear to be mostly located within DNA binding domain of the p53 protein, especially at CpG islands and are associated with altered DNA binding of p53 (2 cases from this series and 2 cases from the literature39). However, more cases with mutations need to be studied to confirm this finding. The presence of an abnormality in p53 predicts poor prognosis and is associated with progressive disease.
Supported by the Arbib Foundation and the Kay Kendall Leukaemia Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Daniel Catovsky, Academic Department of Haematology and Cytogenetics, Institute of Cancer Research/Royal Marsden NHS Trust, Fulham Rd, London, SW3 6JJ United Kingdom; e-mail:d.catovsky@icr.ac.uk.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal